Abstract
This article presents evidence that starvation for leucine in an Escherichia coli auxotroph triggers metabolic activities that specifically target the leu operon for derepression, increased rates of transcription, and mutation. Derepression of the leu operon was a prerequisite for its activation by the signal nucleotide, guanosine tetraphosphate, which accumulates in response to nutritional stress (the stringent response). A quantitative correlation was established between leuB mRNA abundance and leuB− reversion rates. To further demonstrate that derepression increased mutation rates, the chromosomal leu operon was placed under the control of the inducible tac promoter. When the leu operon was induced by isopropyl-d-thiogalactoside, both leuB mRNA abundance and leuB− reversion rates increased. These investigations suggest that guanosine tetraphosphate may contribute as much as attenuation in regulating leu operon expression and that higher rates of mutation are specifically associated with the derepressed leu operon.
Keywords: stringent response, transcription, mutations
The mechanisms of evolution have been the subject of many controversies and speculations for some 200 years (1, 2). Clearly, selection of the fittest occurs, but does the environment also play a role in generating the fittest? Do all of the variants selected result from mutations that are completely “random”? Background mutations are loosely referred to as random even though they do not occur with equal probability, but at different and characteristic rates because of DNA context and variables such as the intrinsic instability of cytosine, giving rise to the (most frequent) C-to-T transition mutations (3, 4) or the presence of tandem repeats, resulting in frameshift mutations (5). Moreover, environmental conditions such as thymidine starvation can selectively increase the rate of particular kinds of mutation (6). However, in an evolutionary context, “random” has a very specific meaning: Neo-Darwinism holds that the spectrum of background mutations and the frequency with which they occur are random (undirected) with respect to selective conditions of the environment. Another ambiguous word, “mechanism,” can mean one thing when applied to evolution and another when applied to mutations. There are mechanisms by which particular kinds of mutations occur (e.g., base substitution, deletion, frameshift), and there are mechanisms by which the rates of many kinds of background mutations are stimulated (e.g., replication, UV irradiation, defective repair, transcription). It is the latter sort of mechanism that applies to evolution because stimulating mutation rates increases the availability of variants on which evolution depends. Our data indicate that transcription (starvation-induced derepression) is unique in augmenting variant availability in a specific manner, i.e., by stimulating rates of transcription (and associated phenomena such as RNA polymerase pausing) in targeted operons, thereby increasing the concentration of single-stranded DNA (ssDNA), which is more vulnerable to mutations than double-stranded DNA. Although the mutations per se are random, as described above for background mutations, the mechanisms that target operons for increased rates of transcription are highly specific. This specificity is not compatible with current neo-Darwinian dogma. And yet, evidence in the literature supports the two major assumptions on which our hypothesis is based: (i) ssDNA is more vulnerable to mutagenesis than double-stranded DNA; increased rates of transcription will, therefore, increase rates of mutation; and (ii) derepression and activation of an amino acid biosynthetic operon occur specifically in response to starvation for that amino acid.
Evolution implies stress; a perfectly adapted organism in a stable environment would not need to evolve. In fact, evidence from marine fossil communities indicates that environmental stress accelerates the rate of evolution (7). In microorganisms, prolonged nutritional stress results in genome-wide hypermutation, i.e., high mutation rates in various genes located in chromosomes, episomes, transposons, or insertion elements (8–14). Such effects likely contribute to the evolutionary process by increasing the size of the mutant population. Indeed, hypermutable genes apparently have been selected during evolution as uniquely advantageous to the survival of the organism (15). The question then arises: By what mechanism(s) related to stress could specific genes become hypermutable? Transcription-enhanced hypermutation is a plausible mechanism by which a direct causal relationship can be established between higher mutation rates in specific genes and the selective conditions of stress that evoke them. There is an accepted biochemical basis for the stimulation of transcription by starvation (derepression); for example, the lack of an amino acid results in the derepression of the operon controlling its synthesis. Because increased rates of transcription can cause higher mutation rates in specific operons (see Discussion), a direct causal relationship appears to exist between starvation for a particular amino acid and higher mutation rates in genes of the operon encoding the enzymes that catalyze its synthesis.
The metabolism of starvation is called the stringent response (17). After deprivation of any essential nutrient, the cell restructures its metabolism from one geared to growth and cell division under favorable nutritional conditions to one allowing survival under conditions of starvation. This abrupt shift in metabolism results in the rapid accumulation (16, 17) of guanosine tetraphosphate (ppGpp), a signal nucleotide that appears to be the most dominant primary regulator of nutritional distress (17, 18). It is synthesized by ppGpp synthase I, the relA gene product, and is degraded by the spoT gene product. Recent investigations have demonstrated that this signal nucleotide binds to the β-subunit of RNA polymerase (19) and affects both transcription initiation (20) and polymerase pausing (21). By recognizing particular promoter sequences (17, 22), ppGpp serves three essential roles for the starving cells. The first two occur regardless of the starvation regimen and consist of the inhibition of DNA, rRNA, nucleotide, and phospholipid synthesis, thereby arresting cell division and the activation of metabolic pathways that protect the vulnerable cells from environmental extremes such as heat, desiccation, and oxidative damage (17, 18). The third essential role is starvation regimen-dependent in that ppGpp activates only those genes that are specifically derepressed by the type of starvation imposed. For example, the expression of amino acid biosynthetic operons requires both derepression (removal of end product inhibition by attenuation or repression) and activation by ppGpp. Starvation for tryptophan as well as other amino acids elicits the stringent response, but only tryptophan starvation increases trp expression (23); the absence of threonine, but not arginine or histidine, leads to derepression of the thr operon (24). However, regulation in some operons apparently may be triggered as a general consequence of the stringent response (17).
MATERIALS AND METHODS
Bacterial Strains and Mutation Rate Experiments.
The bacteria used in this study are two multiple auxotrophs of Escherichia coli K12, CP78 and CP79, isogenic except for relA, and a construct of CP78, called 78AL (see below). The conditions for growing CP78 and CP79 and for determining mutation rates in these strains has been described (25, 26). To determine mutation rates in 78AL, the minimal medium used was limiting for arginine or histidine, rather than leucine, to avoid selection of Leu+ revertants. When the cultures reached stationary phase (≈24 h), they were washed twice with leucine-free minimal medium to remove all residual leucine and isopropyl-d-thiogalactoside (IPTG) and were plated (at a concentration of ≈1 × 108) on the same medium. Thereafter, total cell numbers (determined at 24, 48, and 72 h) were constant and comparable, regardless of prior exposure to IPTG. Mutation rates were determined by the “zero” method of Luria and Delbruck (27). The values given in Tables 1 and 2 are averages of 5–8 experiments.
Table 1.
Correlation between leuB mRNA levels determined as described for Fig. 2 and leuB− reversion rates
| Cells | relA genotype | Condition | pg leuB mRNA/ :μg of total RNA | leuB− reversion rate × 10−9 |
|---|---|---|---|---|
| CP79 | relA | unstarved | 29 ± 5 | * |
| CP78 | relA+ | unstarved | 30 ± 5 | * |
| CP79 | relA | −arg then − leu | 17 ± 6 | 0.15 ± 0.02 |
| CP79 | relA | −leu | 17 ± 5 | 0.22 ± 0.08† |
| CP79pyrD | relA | −arg then − leu | 28 ± 5 | 0.25 ± 0.06‡ |
| CP79pyrD | relA | −leu | 35 ± 12 | 0.44 ± 0.12‡ |
| CP79ΔspoT207 | relA | −leu | 126 ± 11 | 0.60 ± 0.11§ |
| CP78pyrD | relA+ | −leu | 285 ± 103 | 1.26 ± 0.26‡ |
| CP78 | relA+ | −leu | 269 ± 60 | 1.47 ± 0.48† |
| CP78 | relA+ | −arg then − leu | 335 ± 53 | 1.80 ± 0.30 |
| CP78pyrD | relA+ | −arg then − leu | 347 ± 136 | 2.30 ± 0.70‡ |
| CP79spoT203 | relA | −leu | 378 ± 149 | 2.50 ± 0.62§ |
Table 2.
Comparisons between leuB− reversion rates and leuB mRNA levels in CP78, CP79, and 78AL
| Cells | Condition | leuB mRNA* | MR × 10−9† |
|---|---|---|---|
| CP79 relA− | log | 29 ± 5 | Not applicable |
| CP79 relA− | Leu-starved | 17 ± 5 | 0.22 ± 0.1 |
| CP78 relA+ | log | 30 ± 5 | Not applicable |
| CP78 relA+ | Leu-starved | 269 ± 60 | 1.5 ± 0.5 |
| 78AL‡ | log | 213 ± 4 | 1.0 ± 0.1 |
| 78AL | log + IPTG | 1,275 ± 175 | 2.7 ± 0.5 |
| 78AL | SP − Leu | 63 ± 4 | 0.9 ± 0.1 |
| 78AL | SP − Leu + IPTG | 333 ± 32 | 1.6 ± 0.5 |
SP, stationary phase.
pg leuB mRNA/μg total RNA.
leuB− mutation rates (MR) ×10−9 (see Materials and Methods).
Lacks attenuation and ppGpp-binding sequences.
RNA Isolation.
Cells were collected and lysed according to Emory and Belasco (28). The lysate was extracted twice with phenol/chloroform/isoamyl alcohol (50:49:1) and once with chloroform. The RNA was precipitated with 0.5 M ammonium acetate and 2.5 volumes ethanol overnight at −20°C, was pelleted and washed with 70% ethanol, and was resuspended in diethyl pyrocarbonate water. It was reprecipitated with 2.5 M LiCl at −20°C for 1 h and was pelleted. After this, the pellet was washed with ethanol and was resuspended in diethyl pyrocarbonate water, and the RNA was treated with 10 units of RNase-free DNase (Promega). Phenol/chloroform/isoamyl alcohol extraction was used to remove Dnase, and the final RNA was precipitated with ammonium acetate and ethanol as above. RNA concentration was determined by reading the OD at 260 nm.
RNA Probe Preparation.
The E. coli leuB, pyrD, and glpK genes were PCR amplified and cloned into a vector containing opposable T3 and T7 promoter sites (pBluescript II SK(+), Statagene). For each gene, plasmid DNA was isolated (QuantumPrep mini-prep kit, Bio-Rad) and cut with a restriction enzyme to produce a 100- to 600-bp section of the gene with either the T3 or T7 promoter at one end. Antisense RNA probes were produced by in vitro transcription (Ampliscribe T3 and T7 in vitro transcription kit, Epicentre Technologies, Madison, WI). The probes were labeled with biotin (BrightStar Psoralen-Biotin kit, Ambion) and were purified by using denaturing polyacrylamide gel electrophoresis. The probes contained some vector sequence at the 5′ end that was nonhomologous to the target mRNA, producing protected hybrid fragments that were shorter than the full-length probe and separable by gel electrophoresis. This difference in protected vs. full length probe is evidence of a working assay. As a standard, positive-sense mRNA was generated for each gene from the same plasmid but using the other promoter.
Nuclease Protection Assay and Densitometry.
The biotinylated antisense RNA probe of defined length was hybridized overnight to the target mRNA in solution at 50°C. After digestion of single strand nucleic acids by T2 ribonuclease, the hybrids were precipitated, were separated by denaturing polyacrylamide gel electrophoresis, and were transferred to positively charged nylon membrane by semidry electroblotting. The biotinylated probe was conjugated to streptavidin-alkaline phosphatase and CDP-Star by using Ambion’s Bright-Star BioDetect kit. Chemiluminescent probes were detected with x-ray film. Standards were run with each sample, and relative intensities of different mRNA species within a single RNA extract were compared by densitometry using an Astra (Sodertalje, Sweden) 1200S document scanner with transparency adapter and Scanalytics (Bellerica, MA) one-dscan software. The probe molecules were present in 4- to 10-fold molar excess of the target mRNA to demonstrate linearity between the amount of RNA present and the amount of binding to the probe.
Replacing the leu with the tac Promoter (Strain 78AL).
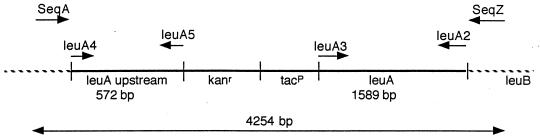
A suicide vector, pBanz2, containing the tac promoter (tacP), a kanamycin resistance cassette, and ends that are homologous to the E. coli chromosome flanking the leu promoter was generated (see Fig. 1) and then was used to transform E. coli CP79 (relA2, leuB−). Double crossover homologous recombination between the chromosome and the plasmid replaced the chromosomal leu promoter with the tac promoter and kanamycin resistance cassette (Fig. 1). The construct was generated as follows: (i) leu upstream and leuA regions were PCR amplified, with specific restriction sites in the primers from the CP78 chromosome, and were cloned into T-Vectors (29); (ii) the leu upstream and the leuA fragments were recovered from the T-Vectors by restriction digestion; (iii) the kanamycin cassette was acquired by restriction digest of plasmid pBSL15 (29); (iv) the fragments were sequentially cloned into plasmid pKK223–3 (tacp, ampr; Amersham Pharmacia); and (v) the 3.6-kilobase construct was recovered by PCR amplification by using the Expand High Fidelity PCR System of Boehringer-Mannheim with primers LeuA4 and LeuA2 (Fig. 1; see Primers) and was cloned into suicide vector pWM91 [oriR6K, ampr, sucroses due to sacB (30)] to generate plasmid pBanz2, which was introduced into CP79 by electroporation.
Figure 1.
Construct map of 78AL and 79AL. Recombination with the suicide vector pBanz2, which contained the above construct, resulted in the arrangement of genes into the genomes of CP78 and CP79 as shown. The PCR primers described are depicted in their respective approximate positions and in their direction of polymerization. The recombination event introduced a kanamycin resistance gene cassette and the tac promoter, replacing the endogenous leu promoter.
Plasmid pBanz2 is propagated only in E. coli strains that produce the λ Pir protein and will not replicate in CP78 or CP79, therefore rendering pBanz2 a suicide vector for these strains. When introduced into CP79 by electroporation, single-crossover homologous recombination resulted in integration of the suicide vector into the leu operon. An integrant (CP79-integrant) was selected by resistance to kanamycin and ampicillin. Double-crossover homologous recombinants that maintained the 3.6-kilobase fragment (kanr) but that lost the remainder of the suicide vector containing the toxic sacB gene were selected by sucrose and kanamycin resistance and were screened for ampicillin sensitivity. P1vir transduction was used to transfer the recombinant promoter to CP78 (relAwt, leuB−), resulting in the recombinant 78AL. A PCR fragment, using a primer set that flanked the construct sequences in the E. coli chromosome (see Primers), was amplified from 78AL and was sequenced to confirm replacement of the leuP with the tacP.
Primers.
The 572-bp leu upstream region was amplified with forward primer LeuA4 (5′AAGGCGCATGCTCAGTAACGGGC-3′), including a SphI site (underlined) and reverse primer LeuA5 (5′-CAACGGCTAGCACGCATGGTCGA-3′) with a NheI site (underlined). The 1,589-bp leuA gene was amplified with forward primer LeuA3 (5′-GCCGGAATTCATGAGCCAGC-3′, including the leuA start codon (underlined and bold) and an EcoRI site (underlined), and reverse primer LeuA2 (5′-ATCCTGCAGCACACGGTTTC-3′) with a PstI site (underlined).
Primers used to amplify the regions surrounding the integration site (see Fig. 1) were as follows: The forward primer, from upstream of the leu upstream region, was SeqA (5′-CACGCTAAGTACGCTCATC-3′), and the reverse primer, originating within the leuB gene downstream of the leuA gene, was SeqZ (5′-GGCGATACGTTCGATCTCA-3′). The size of the amplified fragment from this set of primers was designed to allow parents and recombinants to be distinguished: The parental product was 3,683 bp, and the recombinant product was 4,254 bp (see Fig. 1).
RESULTS
The present study concerns two isogenic E. coli multiple auxotrophs (leuB−, argH−, thr−, his−) differing only in relA (and therefore ppGpp levels). Investigations have concentrated on the leuB− point mutation, which is a C-to-T transition resulting in a serine-to-leucine substitution at amino acid residue 286 of the LeuB protein (26). In a typical mutation rate experiment, 80% of the colonies counted proved to be true revertants (26). Most mutations occurred immediately when starvation was imposed (25), coinciding with the burst in ppGpp levels (16); a positive correlation was established between leuB− reversion rates and the concentration of ppGpp (26). Reversion rates (25, 26) of the leuB−, argH−, and thr− mutant alleles in response to amino acid starvation were higher in the relA+ strain (CP78) than in the relA− strain (CP79), in contrast to mutations in genes regulating maltose catabolism or pyrimidine biosynthesis (32). Stringent response mutations, like transcription-induced lac− reversions (33), are recA-independent (data not shown), distinguishing them from prolonged stress-induced “adaptive” mutations (8–14) and from DNA damage-induced “SOS” mutagenesis, both of which require recA (34, 35).
Derepression of the leu Operon Is Specific to Leucine Starvation.
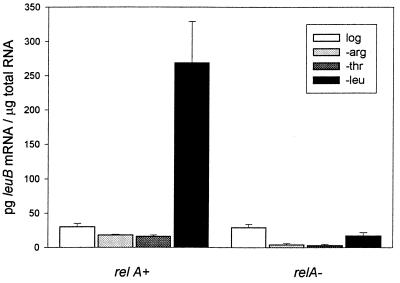
Fig. 2 illustrates the specificity of leuB gene expression resulting from starvation for leucine, arginine, or threonine. The difference between leuB mRNA levels in the leucine-starved relA+ compared with the relA− strain may be attributed to derepression (36) plus ppGpp activation in the case of the relA+ strain and to continued repression in the relA− strain. Although starvation for either arginine or threonine evokes the stringent response and high ppGpp levels (16), it does not activate the leu operon because of the presence of leucine. The data suggest that, in the absence of ppGpp (i.e., the relA− mutant), removal of inhibition by attenuation alone (i.e., leucine starvation) does not allow leu operon expression at 15 min starvation because transcript levels stay the same.
Figure 2.
leuB mRNA abundance in E. coli CP78 (relA+, ppGpp+) and CP79 (relA−, ppGpp−). Cells were grown to log phase (log) in minimal medium or were grown to log phase and were washed and transferred to minimal medium without arginine (−arg), threonine (−thr), or leucine (−leu) for 15 min. Total RNA was recovered, and the level of leuB transcript was determined by nuclease protection assay and densitometry (see Materials and Methods).
Mutation Rates Are Correlated with leuB mRNA Abundance.
As shown in Table 1, a quantitative correlation exists between leuB− reversion rates and transcript abundance [the Pearson correlation = 0.967, significant at the 0.01 level (2-tailed)]. This correlation is particularly meaningful in that the mutation rates were determined in the past (25, 26, 32) and, in effect, predicted the mRNA levels that have now been found. Preliminary data suggest that the leuB mRNA accumulation pattern follows that of ppGpp; that is, the peak is very sharp and occurs within the first 10 min (16). To obtain the data for Table 1, cells were starved for 15 min before measuring leuB mRNA levels, which were too variable at earlier time points to be useful for comparative purposes.
In fluctuation tests, to determine leuB− reversion rates in cells grown with limiting levels of leucine (25, 32), cells first experience starvation in the liquid minicultures; therefore, leu operon derepression and leuB− reversions presumably occur before plating (it is not possible to distinguish between mutations that occur at the end of growth and mutations that occur within a few hours after plating). If cells are grown with limiting levels of threonine or arginine, the stringent response also occurs in the liquid culture at the end of growth, but derepression of the leu operon does not; leu operon expression and leuB− reversions must then occur after plating to the selective medium lacking leucine. It is not possible to measure reversion rates without derepressing the operon (16, 25, 32).
Derepression of the leu Operon Is Specific Within the Genome.
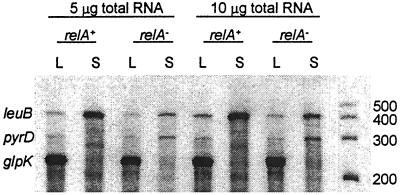
Three biotin-labeled probes were prepared to quantify mRNA from genes involved in different metabolic pathways: leuB (β-isopropylmalate dehydrogenase, EC 1.1.1.85), pyrD (dihydroorotate oxidase, EC 1.3.3.1), and glpK (glycerol kinase, EC 2.7.1.30). Fig. 3 illustrates the results of hybridizing these probes to RNA isolated from log and from 60 min leucine-starved relA+ and relA− cells. As anticipated, based on previous mutation rate studies (32), leucine-starved relA+ cells had higher leuB mRNA levels than starved relA− cells whereas the opposite was true for pyrD mRNA levels. Thus, both pyrD mRNA levels and reversion rates of pyrD− (an A-to-G transition) were higher in the relA− than in the relA+ strain (37). The pyrD promoter has a GC-rich discriminator sequence (38), as does pyrB1, which is inhibited by ppGpp (39, 40). As an internal control, the gene encoding glycerol kinase (glpK) was examined and found to be down-regulated in the absence of growth to the same extent in the two strains when amino acid starvation was imposed (37). (In the experiment summarized in Fig. 3, glpK mRNA levels were saturating.) Thus, relA+ strains starved for leucine (but not arginine or threonine; Fig. 2) derepressed the leuB but not the pyrD or glpK genes.
Figure 3.
Ribonuclease protection assay of E. coli CP78 (relA+, ppGpp+) and CP79 (relA−, ppGpp−). Total RNA was recovered, and the levels of specific transcripts were determined by nuclease protection assay and densitometry (see Materials and Methods). Total RNA [5 μg (lanes 1–4) and 10 μg (lanes 5–8)] was hybridized simultaneously with probes for leuB, pyrD, and glpK mRNA. E. coli CP78 and CP79 cells were grown to log phase (L) and then were washed and starved for leucine for 60 min (S). RNA markers 200–500 nt in length were run in the last lane. The leuB hybridized fragment is 436 nt, pyrD is 313 nt, and glpK is 244 nt.
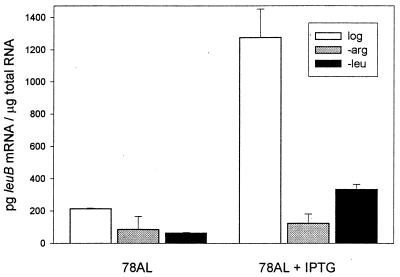
The leu Operon Can Be Activated by an Inducible tac Promoter.
In strain 78AL, the chromosomal leu promoter (including the attenuator and ppGpp discriminator sequences) has been replaced by the tac promoter (see Materials and Methods). Fig. 4 summarizes the effect of 1 mM IPTG on leuB mRNA levels during growth and during 30 min of starvation for either arginine or leucine. As might be expected in this construct, IPTG enhances leuB mRNA levels maximally during growth (in the presence of leucine), in contrast to the conditions found optimal for derepression in the parental strain CP78 (Fig. 2). Reversion rates in the construct also were enhanced maximally by IPTG during growth (Table 2) and were recA-independent. These results are consistent with those of Herman and Dworkin (33), which showed that recA-independent reversions of lac− were enhanced ≈2-fold by IPTG induction of the lac operon.
Figure 4.
Construct 78AL was grown to log phase in the absence or presence of 1 mM IPTG (log) or was grown to log phase and was washed and transferred to minimal medium without arginine (−arg) or leucine (−leu), in the presence or absence of IPTG. Total RNA was recovered, and the level of leuB transcript was determined by nuclease protection assay and densitometry (see Materials and Methods).
DISCUSSION
Transcription Is Known to Enhance Mutation Rates.
Various kinds of evidence document an effect of transcription on mutation rates; the nontranscribed ssDNA strand is more vulnerable to mutations than double-stranded DNA. Fix and Glickman (41) observed that 77% of C to T transitions originated in the nontranscribed strand; this strand bias has been confirmed in other systems (42, 43). In fact, it is known that cytosines deaminate to uracils in ssDNA at >100× the rate seen in double-stranded DNA (3, 4, 44). The first investigators to propose transcriptional activation as a mechanism for increasing mutation rates in specific operons were Brock (45) and Herman and Dworkin (33). Their investigations demonstrated that recA-independent lac− reversion rates of frameshift and point mutations were higher when transcription was induced by IPTG and that the effect was specific to that gene; that is, lys− reversions were not affected, and mutants associated with uninducible regulatory systems (i−o+) were not differentially influenced by IPTG. Transcription-enhanced mutations also have been shown for reversions of a lys− frameshift allele in yeast (46); reversion rates of an uninducible lys allele and mutations to canavanine resistance at an unrelated locus were not affected. The mutability of the nontranscribed strand also was demonstrated (47) in a plasmid system in which a 4-fold increase in the frequency of transitions occurred selectively in the nontranscribed strand when transcription was induced; for a number of reasons, this strand bias could not be ascribed to transcription-coupled repair (48). In our view, these studies demonstrating enhanced mutation rates as a result of artificially induced transcription are all examples of specifically induced mutations. However, they have not been recognized as such, nor have they been related to transcription induced by starvation (derepression), which is directly relevant to environmental stress and evolution.
Up-Regulation of the leu Operon Requires ppGpp Activation.
When bacteria are starved for an amino acid, the biosynthetic operon is derepressed; that is, it is no longer down-regulated by attenuation (thr, his, ilv, and leu), repression (arg), or both (trp and phe). However, in addition to derepression, our data indicate that the operon is always activated by ppGpp. At 15 min starvation, leu operon expression apparently depends on ppGpp activation because leucine starvation of the relA+ strain, but not the relA− strain, enhanced leuB mRNA levels (Fig. 2). Although leuB mRNA levels increased somewhat after 60 min of starvation in the ppGpp-deficient strain (Fig. 3), this may be related to the fact that the relA− allele is somewhat “leaky” (17).
leu Operon Activation by IPTG in the Construct Is Less Effective than ppGpp Activation in the Parental Strain.
Table 2 compares the construct 78AL and the parental strains with respect to leuB mRNA levels and leuB− reversion rates. The high basal transcription level of the uninduced tac promoter in 78AL log cells is consistent with higher mRNA levels compared with log cells of the parental strains. In growing cells, a 6-fold increase in mRNA level induced by IPTG is apparently “required” for a 2- to 3-fold increase in mutation rate in 78AL. During leucine starvation, IPTG also stimulates leuB mRNA levels. However, this is not sufficient to have a significant affect on reversion rates under these conditions. IPTG induction previously has been observed to enhance lac− reversion rates in growing (33) but not starving cells (49). With respect to affecting mutation rates, the normal starvation mechanism is apparently more efficient than IPTG induction in the construct (which lacks the ppGpp discriminator sequence). This suggests that ppGpp may be affecting the concentration of ssDNA by some mechanism related to and in addition to its effect on transcription. Work in progress concerns leuB mRNA turnover, as well as investigations into the effect of ppGpp on pausing of RNA polymerase (21), which could increase the concentration of ssDNA and mutation rates at specific pause sites. It is therefore interesting that an analysis of predicted mRNA folding (Mfold, GC Gene, University of Wisconsin) indicates that both the leuB and pyrD mutations occurred at possible pause sites (data not shown). Somatic hypermutation in mature B lymphocytes depends on transcription, and RNA polymerase pausing is thought to be a mechanism that could determine the site of mutagenesis (50).
How Are Mutations Expressed in Nongrowing Cells?
When mutations occur at the onset of starvation, reversions on the transcribed strand would immediately be immortalized by the synthesis of functional enzyme and leucine, allowing cell division. If most mutations occur on the nontranscribed strand and are never expressed, then the actual mutation rates are higher than calculated. Mutations on the nontranscribed strand could be expressed, for example, by the stimulation of DNA repair activities that attempt to repair the complementary position on the transcribed strand or by the induction of “cross-strand deamination” (51). It is also possible that a damaged base on the nontranscribed strand will cause the complementary base to become altered at a higher frequency than paired bases (52). Mutants defective in a variety of DNA repair functions should provide further insights into the mechanism of transcription-enhanced mutations.
Are Stringent Response Mutations Specifically Enhanced?
The extent to which starvation regimen-dependent mutation rates are exclusively increased by derepression and ppGpp activation of a targeted operon appears to justify use of the word “specific”. The effect of leucine starvation in up-regulating leu operon transcription is specific in that starvation for other amino acids is not effective; in the arg operon, starvation for arginine but not histidine increases argH mRNA abundance (S. McAllister, personal communication). Genes down-regulated or not regulated during the stringent response include pyrD and glpK (Fig. 3), “housekeeping” genes (53), and starvation regimen-independent genes involved in DNA, rRNA, tRNA, nucleotide, phospholipid, and cell wall synthesis (17). Whether hypermutation specific to the leu operon is described as being induced, enhanced, or directed by leucine starvation is arbitrary. The specificity of stringent response mutations and their dependence on derepression (relA) recently has been observed by others in a Bacillus subtilus system (54). Mutation rates in amino acid biosynthetic genes, but not drug resistance genes, were enhanced in relA+ compared with relA− strains under conditions that evoke the stringent response.
In our investigations, an increase in reversion rate of leuB− is taken as evidence of increased mutation rates also in the other (normal) genes of that operon. The reversion of an auxotroph to prototrophy represents the restoration of a previously existing ability and therefore only serves as a model for the analysis of mechanisms underlying “forward” mutations required in the evolution of complex metabolic networks (55, 56). We suggest that the stringent response played, and continues to play, a role in optimizing functional genes in organisms confronted with novel stress conditions and in derepressing and mobilizing genes to serve in new biochemical pathways. Genetic derepression frequently is a prerequisite for further mutations that modify existing genes, enabling them to use new substrates during acquisitive evolution (57).
Unaware of the evidence supporting his hypothesis, Fitch (58) speculated that transcribing genes might have higher mutation rates than other genes. The growth-dependent, artificially induced, specifically enhanced mutations discussed earlier occurred under nonselective conditions and are therefore not relevant to environmental stress and evolution. However, stringent response mutations occur in response to impending starvation. In nature, bacteria likely starve simultaneously for many required metabolites; nutritional stress, gene derepression, and hypermutation must indeed be very widespread.
The current paradigm of neo-Darwinism as formulated by Weisman (59) rejects any influence of the environment on the direction of variation. However, prolonged nutritional stress results in a general increase in mutation rates; the introduction of environmental effects on specific mutation rates is a reasonable extension of what is known, especially because mechanisms by which starvation can immediately and specifically affect rates of transcription and mutation are consistent with accepted principles of molecular biology. The proposed mechanism of derepression-induced hypermutation provides the critical link between mutations and the metabolic activities evoked by specific conditions of environmental stress, increasing the availability of variants most likely to evolve in that environment.
Acknowledgments
We thank M. Cashel, J. Shapiro, R. Kolter, J. Drake, R. Schaaper, C. Yanofsky, S. McAllister, and D. Reschke for criticizing this manuscript. We thank M. Cashel for bacterial strains, B. Wanner and M. Alexeyev for plasmids pWM91 and pBL15 respectively, C. Henderson for the Pearson (parametric, bivariate) correlation analysis of data in Table 1, and J. Strange and the Murdock Molecular Biology Facility for technical support. This work was supported by the University of Montana, and by a MONTS grant from the National Science Foundation EPSCoR program.
ABBREVIATIONS
- IPTG
isopropyl-d-thiogalactoside
- ssDNA
single-stranded DNA
- ppGpp
guanosine tetraphosphate
References
- 1.Mayr E. The Growth of Biological Thought, Diversity, Evolution, and Inheritance. Cambridge, MA: Belknap; 1982. [Google Scholar]
- 2.Jablonka E, Lamb M J. Epigenetic Inheritance and Evolution: The Lamarckian Dimension. Oxford: Oxford Univ. Press; 1995. [Google Scholar]
- 3.Lindahl T, Nyberg B. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 4.Frederico L A, Kunkel T A, Shaw B R. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 5.Ripley L S. Annu Rev Genet. 1990;24:189–213. doi: 10.1146/annurev.ge.24.120190.001201. [DOI] [PubMed] [Google Scholar]
- 6.Kunz B A, Kohlami S E. Annu Rev Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 7.Jablonski D, Sepkoski J, Jr, Bottjer D J, Sheehan P M. Science. 1983;222:1112–1124. doi: 10.1126/science.222.4628.1123. [DOI] [PubMed] [Google Scholar]
- 8.Cairns J, Overbaugh J, Miller S. Nature (London) 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 9.Hall B G. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro J. Trends Genet. 1997;13:98–104. doi: 10.1016/s0168-9525(97)01058-5. [DOI] [PubMed] [Google Scholar]
- 11.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 12.Torkelson J, Harris R S, Lombardo M-J, Nagendran C, Thulin C, Rosenberg S M. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster P L. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson D I, Slechta E S, Roth J R. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- 15.New York Academy of Sciences. Molecular Strategies in Biological Evolution. New York: N.Y. Acad. Sci; 1999. , in press. [PubMed] [Google Scholar]
- 16.Donini P, Santonastaso V, Roche J, Cozzone A J. Mol Biol Rep. 1978;4:15–29. doi: 10.1007/BF00775174. [DOI] [PubMed] [Google Scholar]
- 17.Cashel M, Gentry D R, Hernandez V J, Vineela D. In: Escherichia coli and Salmonella typhimurium. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1458–1496. [Google Scholar]
- 18.Hengge-Aronis R. In: Escherichia coli and Salmonella typhimurium. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1497–1512. [Google Scholar]
- 19.Chatterji D, Fujita N, Ishihama A. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 20.Heinemann M, Wagner R. Eur J Biochem. 1997;247:990–999. doi: 10.1111/j.1432-1033.1997.00990.x. [DOI] [PubMed] [Google Scholar]
- 21.Krohn M, Wagner R. J Biol Chem. 1996;271:23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- 22.Da Costa X J, Artz S W. J Bacteriol. 1997;179:5211–5217. doi: 10.1128/jb.179.16.5211-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morse D E, Morse A N C. J Mol Biol. 1976;103:209–226. doi: 10.1016/0022-2836(76)90310-7. (1976). [DOI] [PubMed] [Google Scholar]
- 24.Klyachko E V, Bochkanov S S, Shakulov S. Biokhimiya. 1984;48:1095–1102. [PubMed] [Google Scholar]
- 25.Wright B E. Mol Microbiol. 1996;19:213–219. doi: 10.1046/j.1365-2958.1996.367892.x. [DOI] [PubMed] [Google Scholar]
- 26.Wright B E, Minnick M F. Microbiology. 1997;143:847–854. doi: 10.1099/00221287-143-3-847. [DOI] [PubMed] [Google Scholar]
- 27.Luria S E, Delbruck M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emory S A, Belasco J G. J Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchuk D, Drumm M, Saulino A, Collins F S. Nucleic Acids Res. 1991;19:1154–1162. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexeyev M F. BioTechniques. 1995;18:52–56. [PubMed] [Google Scholar]
- 31.Metcalf W W, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Plasmid. 1996;35:1–14. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 32.Wright B E. FEBS Lett. 1997;402:4–8. doi: 10.1016/s0014-5793(96)01479-2. [DOI] [PubMed] [Google Scholar]
- 33.Herman R K, Dworkin N B. J Bacteriol. 1971;106:543–550. doi: 10.1128/jb.106.2.543-550.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairns J, Foster P L. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker G C. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 36.Burns R O, Calvo J, Margolin P, Umbarger H E. J Bacteriol. 1966;91:1570–1576. doi: 10.1128/jb.91.4.1570-1576.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longacre A, Reimers J M, Wright B E. Molecular Strategies in Biological Evolution. New York: N.Y. Acad. Sci.; 1999. , in press. [Google Scholar]
- 38.Larsen J N, Jensen K F. Eur J Biochem. 1985;151:59–65. doi: 10.1111/j.1432-1033.1985.tb09068.x. [DOI] [PubMed] [Google Scholar]
- 39.Donahue J P, Turnbough C L., Jr J Biol Chem. 1990;265:19091–19099. [PubMed] [Google Scholar]
- 40.Turnbough C L., Jr J Bacteriol. 1983;153:998–1007. doi: 10.1128/jb.153.2.998-1007.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fix D F, Glickman B W. Mol Gen Genet. 1987;209:78–82. doi: 10.1007/BF00329839. [DOI] [PubMed] [Google Scholar]
- 42.Skandilis A, Ford B N, Glickman B W. Mutat Res. 1994;314:21–26. doi: 10.1016/0921-8777(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 43.Francino M P, Ochman H. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 44.Frederico L A, Kunkel T A, Shaw B A. Biochemistry. 1993;32:6523–65530. doi: 10.1021/bi00077a005. [DOI] [PubMed] [Google Scholar]
- 45.Brock R D. Mutat Res. 1971;11:181–186. [PubMed] [Google Scholar]
- 46.Datta A, Jinks-Robertson S. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 47.Beletskii A, Bhagwat A S. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanawalt P, Mellon I. Curr Biol. 1993;3:67–69. doi: 10.1016/0960-9822(93)90156-i. [DOI] [PubMed] [Google Scholar]
- 49.Davis B D. Proc Natl Acad Sci USA. 1989;96:5005–5009. doi: 10.1073/pnas.86.13.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storb U, Klotz E L, Hackett J, Jr, Kage K, Bozek G, Martin T E. J Exp Med. 1998;188:689–698. doi: 10.1084/jem.188.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams L D, Shaw B R. Proc Natl Acad Sci USA. 1987;84:1779–1783. doi: 10.1073/pnas.84.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–38. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 53.Stephens J C, Artz S W, Ames B. Proc Natl Acad Sci USA. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rudner R, Murray A, Huda N. Molecular Strategies in Biological Evolution. New York: N. Y. Acad. Sci.; 1999. , in press. [Google Scholar]
- 55.Oparin A I. The Origin of Life (Macmillan, New York); trans. Margulis, S. (1953) New York: Dover; 1938. [Google Scholar]
- 56.Horowitz N H. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortlock R P. Microorganisms as Model Systems for Studying Evolution. New York: Plenum; 1984. [Google Scholar]
- 58.Fitch W M. Evolution. 1982;36:1133–1143. doi: 10.1111/j.1558-5646.1982.tb05484.x. [DOI] [PubMed] [Google Scholar]
- 59.Weismann A. Uber die Vererbung. Jena, Germany: Fischer; 1893. [Google Scholar]