Abstract
Darwin’s finches comprise a group of passerine birds first collected by Charles Darwin during his visit to the Galápagos Archipelago. The group, a textbook example of adaptive radiation (the diversification of a founding population into an array of species differentially adapted to diverse environmental niches), encompasses 14 currently recognized species, of which 13 live on the Galápagos Islands and one on the Cocos Island in the Pacific Ocean. Although Darwin’s finches have been studied extensively by morphologists, ecologists, and ethologists, their phylogenetic relationships remain uncertain. Here, sequences of two mtDNA segments, the cytochrome b and the control region, have been used to infer the evolutionary history of the group. The data reveal the Darwin’s finches to be a monophyletic group with the warbler finch being the species closest to the founding stock, followed by the vegetarian finch, and then by two sister groups, the ground and the tree finches. The Cocos finch is related to the tree finches of the Galápagos Islands. The traditional classification of ground finches into six species and tree finches into five species is not reflected in the molecular data. In these two groups, ancestral polymorphisms have not, as yet, been sorted out among the cross-hybridizing species.
Keywords: birds, Galápagos Islands
When Charles Darwin returned in 1836 from his 5-year round-the-world voyage on board the H.M.S. Beagle, he brought with him a rich collection of geological, zoological, and botanical material, which he then distributed to various experts for analysis (1). In his zoological collection were skins of finch-like birds from the Galápagos Islands, nearly 1,000 km west of the coast of Ecuador. The skins were analyzed by the ornithologist John Gould and identified as representing nine new species of four genera, Geospiza, Cactospiza, Camarhynchus, and Certhidea (2). Later, five additional species and two genera (Platyspiza, Pinaroloxias) were collected by other expeditions to the Galápagos Archipelago and the Cocos Island 720 km northeast of the archipelago (3, 4). The group, referred to as Darwin’s finches, subsequently became one of the best known and the most studied cases of adaptive radiation.
Based on morphological, behavioral, and ecological data, Darwin’s finches currently are divided into three lineages comprising six genera and 14 species (3, 4). The first lineage is the ground finches, so named because they feed on seeds on the ground in the arid regions of the archipelago. Of all the Darwin’s finches, they are the most finch-like in appearance. They are classified into a single genus, Geospiza, with six species. Three of these species differ mainly in body size and in the size of their heavy, finch-like beaks; they are the large, medium, and small ground finches (G. magnirostris, G. fortis, and G. fuliginosa, respectively). The other three ground finch species have longer beaks and supplement their seed diet with cactus flowers and soft pulp (the large and small cactus ground finches, G. conirostris and G. scandens, respectively) or the eggs and blood of other birds and reptile ticks (the sharp-beaked ground finch, G. difficilis).
The second lineage is the tree finches, which spend most of their time in foliage and vegetation and only occasionally forage on the ground. The six species of this group are classified into three genera, Cactospiza, Camarhynchus, and Platyspiza. All, except the vegetarian finch, P. crassirostris, are insect eaters. One of the two Cactospiza species, the woodpecker finch, C. pallida, ferrets insects out of cracks with cactus spines or twigs, which it holds in its chisel-shaped beak. The other, the mangrove finch, C. heliobates, uses its stout, straight beak to catch insects in mangrove swamps. The three species of Camarhynchus differ in body and beak size, the latter adapted to the size of the insects they feed on. They are the large, medium, and small tree finch, C. psittacula, C. pauper, and C. parvulus, respectively. The vegetarian finch eats buds, young leaves, blossoms, and fruits with its thick, short, slightly decurved beak.
True to their name, the two species in the third lineage, the warbler-like finches, resemble warblers in their small size and slender beaks. The warbler finch, Certhidea olivacea, even uses a similar method of catching insects to the warblers. The second species, the Cocos finch, Pinaroloxias inornata, is the only Darwin finch species that lives outside the Galápagos Archipelago. It feeds predominantly on insects, both on the ground and in the trees.
Members of the assemblage resemble one another in nest architecture, egg pattern, and courtship displays, yet differ in morphology, plumage to some extent, and song, and they hold overlapping territories (3, 4). In a widely reproduced diagram (3, 4) of the presumed relationships among the 14 species, drawn on the basis of phenotypic similarities before the advent of rigorous methods of phylogenetic analysis, the warbler-like finches are depicted as being closest to the ancestral form, followed by the tree and ground finches. Roughly similar relationships also were inferred from allozyme frequency data (5). The objective of the present study was to use well-established molecular markers to answer the following questions: First, are the Galápagos finches truly monophyletic? Second, is the Cocos finch directly related to the Galápagos finches? Third, did the warbler finch descend from the same ancestor as the rest of the group? Fourth, is the vegetarian finch really a member of the tree finch group? And fifth, can the species classification of the ground and tree finches be corroborated by molecular data?
MATERIALS AND METHODS
DNA Isolation, Cloning, and Sequencing.
Blood samples were collected from the wing veins and genomic DNA was extracted from the samples by using the Qiagen Blood-Kit. The DNA was amplified by PCR, cloned, and sequenced by methods described elsewhere (6). The primer pairs used for the amplification of the control region (cr) were: M1 (sense) 5′-CATCAGACAGTCCATGAAATGTAGG-3′ (nucleotides 100–124 of Fringilla; ref. 7) and H1261 (antisense; ref. 8) 5′-AGGTACCATCTTGGCATCTTC-3′ (nucleotides 1261–1282 in tRNAPhe; annealing temperature 58°C). The cytochrome b (cytb) amplification primers were CB1 (sense) 5′-CCAACATCTCHKCHTGATGAAAYTT-3′ (nucleotides 78–101), and CB2 (antisense) 5′-GATGAAKGGGTVTTCTACTGGTTG-3′ (nucleotides 1024–1047; annealing temperature 56°C). Amplifications of the entire mtDNA circle were performed by using the PTC-200 Programmable Thermal Controller and the Expand Long PCR System Kit (Boehringer Mannheim). Sequencing reactions were processed by the LI-COR Long READIR 4200 DNA Sequencer (MWG Biotec, Ebersberg, Germany) and the Automated Laser Fluorescent Sequencer (Pharmacia Biotech). Each clone was sequenced in both directions, and at least two independently derived clones were sequenced from each individual.
Sequence Analysis.
Bias in base composition was calculated by using the formula in ref. 9. Transitions (Ts)/transversions (Tv) ratios were estimated from expected substitution frequencies in neighbor-joining (NJ) trees by using the pamp program in the paml package (10). Saturation of substitutions was assessed by plotting the number of Ts against the mean number of Tv. Genetic distances in pairwise comparisons were calculated by the Tamura-Nei (11) method. Phylogenetic relationships were assessed by using the three forms of the optimization principle to choose the most probable tree: the minimum number of mutational changes in the maximum parsimony method, the smallest sum of branch lengths in the minimum evolution method, and the highest likelihood of observing the data set in the maximum likelihood method. The maximum parsimony trees were drawn by using paup version 3.1.1 (12) with 10 independent heuristic searches and random taxon addition. The minimum evolution trees were assessed by using the NJ method (13) in the 3.1.1 version of paup followed by a branch swapping procedure. The α parameter of the γ distribution used to correct the Tamura-Nei (11) distances was estimated by using the gz-gamma program (14). Maximum likelihood trees were drawn with the dnaml program in the phylip 3.5c package (15), by using the estimated Ts/Tv ratios and the empirical base frequencies. Statistical support of the phylogenies was sought through the bootstrapping test (seqboot in phylip) and bootstrap confidence level test (bcl in mega; ref. 16). Heterogeneity of substitution rates along different lineages was tested by the two-cluster and branch-length methods (17) by using the lintree program.
RESULTS
Because the radiation of Darwin’s finches was expected to have occurred relatively recently, two segments of the rapidly evolving mtDNA (18) were chosen to answer the questions posed above—the gene encoding the cytb protein and the noncoding cr. During the study, it was realized that some of the primers also amplified copies of nuclear mtDNA (numt; ref. 19). Thus far, four distinct numts have been identified in the Darwin’s finches and related species (A.S., unpublished work), but space limitations do not permit their description here. Suffice it to say that the results obtained by using the four numts are fully consistent with those presented in this paper, which is restricted to the use of mtDNA. The main evidence that the sequences presented here are of mitochondrial, rather than of nuclear, origin is that amplification under long PCR conditions with properly placed and oriented (head-to-head) primers yields a fragment corresponding in length to avian mtDNA and containing cytb and cr sequences on which the present study is based (not shown).
The study involved 13 of the 14 currently recognized Darwin’s finch species (samples of C. heliobates, which has become very rare, were not available). As an outgroup, we included the dull-colored grassquit, Tiaris obscura, which we have identified in another study as the most likely nearest relative of the Darwin’s finches (A.S., unpublished work). Genomic DNA isolated from these samples was amplified by PCR and the amplification products, which encompassed 922 bp of the cytb and 1,127 bp of the cr segments, were cloned and sequenced; the sequences have been deposited in the GenBank database. In total, 41 and 55 individuals were tested for the cytb and cr segments, respectively. The number of individuals per species ranged from two to five. Samples from different islands were included when available. The sequences were aligned by eye by using the published Fringilline finch sequences as a reference (7).
The Darwin’s finch sequences reveal two characteristic features of avian and other mtDNAs—biased base composition and skewed substitutional Ts to Tv ratios (19–21). In both segments, cytosine is the most frequently occurring base of the L strand, followed by adenine, thymine, and guanine (36% C, 26% A, 23% T, 13% G in cytb; 30% C, 29% A, 13% T, 27% G in cr). The bias is particularly pronounced at the third codon position in the cytb gene (0.54 as compared with 0.06 and 0.22 at the first and second positions, respectively, expressed by using the formula in ref. 9). Similarly, Ts are preponderant over Tv in both the cytb and cr segments. In the cytb gene, the majority (75%) of the substitutions in the Darwin’s finch sequences occurred at the third codon position and 90% of these are Ts (Fig. 1). In the cr segment, substitutions are distributed unevenly over the sequence. In both the cytb and cr Darwin’s finch segments, however, divergences were below the saturation level, so that all sites, Ts and Tv, could be used for phylogeny reconstruction. The cr can be subdivided into individual segments that show different evolutionary rates. Heterogeneity tests (see Materials and Methods) were conducted on three segments of 278, 471, and 378 bp and revealed that the 3′ part of the cr designated as domain III by Marshall and Baker (7) evolves 2.4 times as fast as the rest of the region (domains I and II; the cr evolves 1.5 times as fast as the cytb gene). Trees based on domain III sequences have the same topologies as those based on the entire region (not shown). Trees based on domains I or II, on the other hand, have somewhat different topologies, presumably because of the small number of informative sites. Rate heterogeneity also was observed for some lineages, but their removal did not change the topology of the group (not shown).
Figure 1.
Relationships between Ts (%) and Tv (%) per site in the cytb (●) and cr (○) full-length nucleotide sequences of the Darwin’s finches and the dull-colored grassquit. Ts and Tv were obtained by the Tamura-Nei (11) method in mean pairwise comparison of all 14 tested species. The low degree of sequence divergence (<2% at either segment within Darwin’s finches) results in little or no substitutional saturation. A 10:1 ratio of Ts to Tv is found in comparisons within Darwin’s finches; transitional saturation is indicated only in comparisons involving the dull-colored grassquit, for which the ratio falls to 5:1. All per site Tv levels above 0.5% are those between Darwin’s finches and the grassquit.
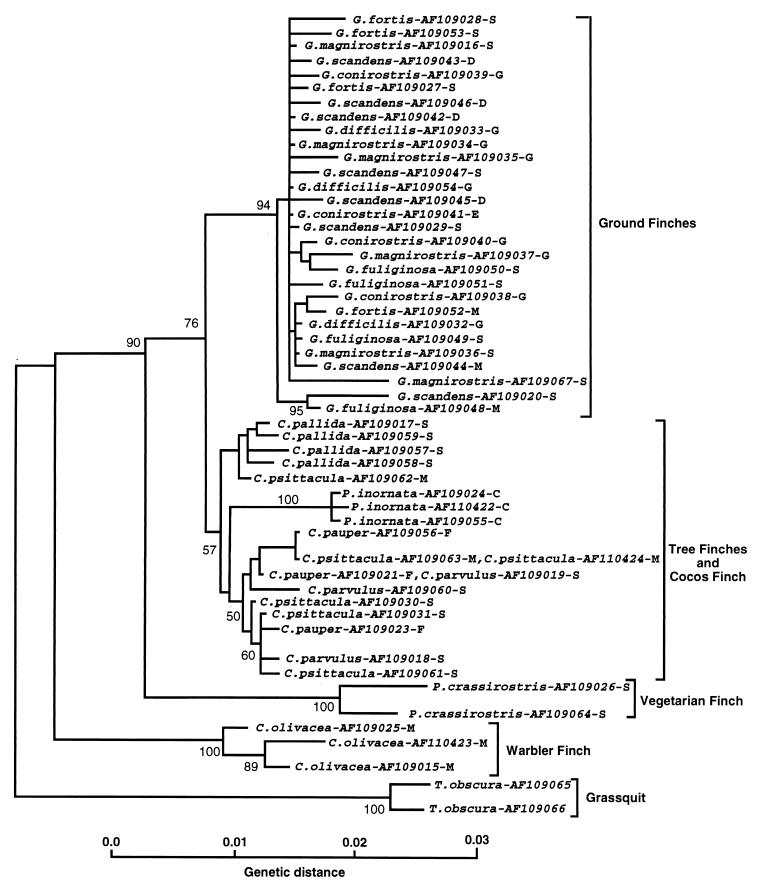
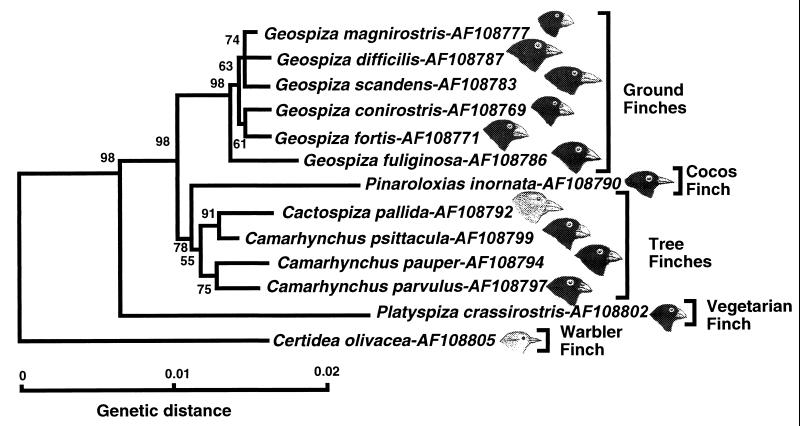
The alignment of the Darwin’s finch cytb sequences is gap free; for the optimal alignment of the 53 cr sequences only a few minor gaps had to be introduced. This fact alone suggests that the Darwin’s finches are an assemblage of closely related species, and the phylogenetic analysis of the sequences bears out this suggestion. Phylogenetic relationships among the Darwin’s finches were deduced from NJ, maximum parsimony, and maximum likelihood trees produced by using the cytb and cr sequences separately or combined. The basic topology of all of these trees was the same (Fig. 2) and did not change when different outgroup species were used (not shown). In all of them, four groups of sequences could be distinguished, arranged in the same branching order, the warbler finch sequences branching out first after the outgroup, the vegetarian finch sequences next, and the tree finch together with the Cocos finch sequences (which in some trees intermingle with the tree finch, whereas in others they are outside of the group; compare Figs. 2 and 4), diverging from the ground finch sequences last. In the tree-robustness tests, these groups were recovered with high bootstrap values. The tree topology was also congruent with that of the trees based on the numt sequences (A.S., unpublished work) and on nuclear microsatellite DNA variation (22). It is thus indicated by several independent molecular markers, both mitochondrial and nuclear.
Figure 2.
Phylogenetic tree of Darwin’s finches and the dull-colored grassquit based on 55 cr sequences and drawn by the NJ algorithm of the mega program (16) using Tamura-Nei (11) distances. The numbers on the nodes indicate the percent recovery of these nodes in 500 bootstrap replications. The species names are followed by GenBank accession codes of the sequences and an abbreviation of the island where the blood samples were collected. C, Cocos Island; D, Daphne Major; E, Espanola; F, Floreana; G, Genovesa; M, Marchena; S, Santa Cruz.
Figure 4.
Maximum likelihood tree of the tree, Cocos, and vegetarian finch cr sequences. A Ts/Tv ratio of 10:1 and empirical base frequencies were used. Ts/Tv ratios of 8:1 and 5:1 give identical topologies to that shown. Numbers on nodes indicate bootstrap recovery of that node in 500 bootstrap replications. For symbol explanations see Fig. 2.
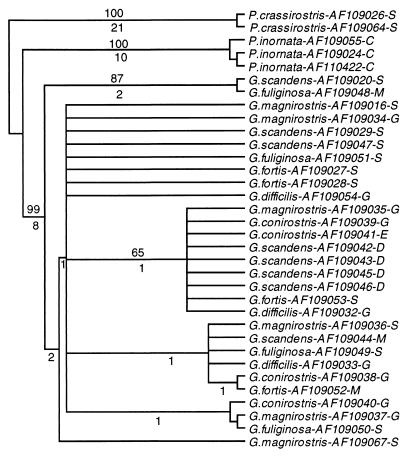
A definitive arrangement of the individual sequences within the two polytypic groups, one encompassing all ground finch sequences, and the other the tree finch sequences, could not be obtained, however. Not only did the branching order of individual sequences within each group differ among the various trees and the individual nodes could be only poorly reproduced by bootstrap resampling, but also sequences from different morphologically identified species were intermingled. Attempts to resolve the intra-group branching pattern and make it consistent with morphological species differentiation by using different tree-drawing methods failed (Figs. 3 and 4). Nor could any obvious correlation between sequences and the origin of the samples from the various islands be ascertained, although the data set was not large enough to exclude the existence of such a correlation definitively. Consistent with the failure to differentiate the species in the trees was the observation that the ranges of intra- and inter-species genetic distances within the ground finch group were 0–0.9% and 0–1.2% substitutions per site, respectively; within the tree finch group the ranges were 0.2–1.4% and 0.2–1.3% substitutions per site, respectively. Hence, the intra- and interspecific distances overlapped within each group. Moreover, in the cytb data set, three instances were found of identical sequences shared by different species: one G. conirostris sequence also was found to be present in some individuals of G. fortis, G. magnirostris, and G. fuliginosa. In the cr, a C. parvulus sequence was found to be present in C. pauper.
Figure 3.
Maximum parsimony tree of the ground, Cocos, and vegetarian finch cr sequences. The tree drawn by using the paup program is a consensus of eight equally parsimonious trees of 125 steps based on the 50% majority rule. Consistency and retention indices are 0.90 and 0.89, respectively. Numbers above the branches indicate percent bootstrap recovery in 500 replications. Numbers below internal branches indicate the number of substitutions that are common to all eight trees. For symbol explanations see Fig. 2.
DISCUSSION
These results, summarized in Fig. 5, provide answers to the questions posed at the onset of the study. First, because in all the phylogenetic trees based on the mtDNA sequences, the dull-colored grassquit of Ecuador does indeed assume the outgroup position and because a related study identifies the Tiaris group as the nearest relative of Darwin’s finches (A.S., unpublished work), it follows that the finches constitute a monophyletic group. This interpretation is further supported by the distribution and sequences of the numts (A.S., unpublished work), microsatellite DNA data (22), as well as by morphological (3, 4), behavioral (23), biochemical (5), and karyological (24) data. The mtDNA and microsatellite data are consistent with the origin of Darwin’s finches from a single ancestral species, possibly from a single founding population (25), which reached the Galápagos Archipelago from Central or South America.
Figure 5.
NJ tree of combined cytb and cr sequences. For simplicity’s sake, a single representative sequence is displayed for each species of Darwin’s finches, and the outgroup has been left out. The drawings point out differences in one of the distinguishing morphological characters of Darwin’s finch species—the shape of the beak. (Bird drawings based in part on ref. 4.)
Second, the molecular data place the Cocos finch within the Darwin’s finch assemblage and in some trees (Fig. 2) within the tree finch group. The species may have arisen relatively recently as part of the tree finch radiation. Its relatively large genetic distance from the other tree finches as revealed by the rate constancy test (data not shown) and by the branch lengths on the NJ tree (Fig. 2) can be explained as being the result of an adaptation to the diverse conditions on the Cocos Island. The mtDNA, as well as the microsatellite data (22), support the hypothesis that the ancestors of the Cocos finch migrated from the Galápagos Archipelago to the Cocos Island relatively recently.
Third, the molecular data corroborate the placement of the warbler finch at the base of the Darwin’s finch tree as suggested by some taxonomists (3) and by the allozyme frequency data (5). This species is apparently the oldest in the Darwin’s finch assemblage, presumably the result of an early radiation of the ancestors. It has long been debated whether the warbler finch should or should not be included in the Darwin’s finch assemblage. Although the majority of taxonomists have regarded the warbler finch as being derived from finch-like ancestors (3), since Darwin’s time some uncertainty has persisted about this issue (e.g., refs. 26 and 27). Darwin himself included this species in the family Parulidae, the American warblers (28). According to this view, Darwin’s finches then would have been founded by at least two different species—the finch-like and the warbler-like ancestors. Other taxonomists have argued, however, that the warbler-like appearance of the species is the result of morphological convergence (3), and this view now is upheld by the molecular data: the warbler finch is part of the monophyletic Darwin’s finch group and Parulidae were clearly not among its ancestors (A.S., unpublished work). The relatively large genetic distances among the individual warbler finches tested (Fig. 2) hint at the existence of taxonomical diversity within this species. Taxonomists indeed have divided the warbler finch, which inhabits all the major and a few minor islands of the archipelago (3), into eight species distinguished by plumage color (3, 29). Heterogeneity of the species has been demonstrated by the microsatellite data to be large enough to justify splitting the species into two (22). A more definitive characterization of mtDNA heterogeneity will require testing of a large sample set collected from the different islands (all of the warbler finch samples tested in the present study were from Marchena).
Fourth, contrary to the traditional view (3, 5), the vegetarian finch is not a member of the tree finch group, but rather a separate branch that diverged from the ancestral stock after the divergence of the warbler finch but before the divergence of the tree finches (Fig. 2). The inclusion of the vegetarian finch in the tree finch group was based primarily on similar ecology, but it also seemed supported by biochemical data (5). The molecular data, however, clearly and consistently separate it, with high bootstrap support, from the Camarhynchus and Cactospiza species (Fig. 2; see also ref. 22). Fifth, the traditional classification of ground finches into six species and of tree finches into five species (excluding the vegetarian finch) is not reflected in the molecular data.
The six ground finch species are distinguished by body size, as well as beak size and shape (3, 4). The species identification based on these traits is, however, not always easy. The variation of each of the characters is contiguous and often overlapping between species; even bivariate plots (e.g., beak length and beak depth) cannot always separate the species (4, 30). Biochemical studies using allozyme variation also failed to differentiate the individual species (5). The identification of the tree finches is not without problems either (3, 4). The molecular data fail to distinguish the morphologically defined ground and tree finch species altogether. The inter- and intraspecies genetic distances overlap and on the phylogenetic trees, individuals representing different morphologically identified species are intermingled (Fig. 2). The situation resembles that of the Lake Victoria haplochromine flock, in which at least some species are morphologically well differentiated, but cannot be distinguished at the molecular level (31, 32). Although molecular polymorphism does exist in the fish flock in both mtDNA (32) and nuclear DNA (33) segments, it is widely shared by different species (33). In the ground and tree finches, too, identical or closely related alleles (haplotypes) of mtDNA segments (34) and of allozyme-encoding nuclear genes (5) are shared by different species. This sharing can be explained by either interspecies hybridization or incomplete sorting of haplotypes (34, 35). In the case of ground and tree finches, both explanations may apply. Hybridization with the production of fertile offspring occurs with an estimated frequency of up to 5% in the ground finches (30, 36, 37). In the tree finches, interspecies hybridization also has been documented (3, 4, 38), but its frequency and the fertility of the hybrids have not been established. Trans-species polymorphism of neutral markers (33) and of alleles under balancing selection (39) is known to occur in the haplochromine flock and is presumed to occur, with or without hybridization, in many other situations (40). It therefore can be expected to exist also in the ground and tree finches, although it will be difficult to distinguish it from introgression as a result of hybridization. The adaptive radiation of the Lake Victoria haplochromine flock is of a young age: it may have taken place within the last 12,000 years (41). Analogously, the incomplete species differentiation within the ground and tree finch groups is probably a sign of an adaptive radiation in progress.
Acknowledgments
We thank Jane Kraushaar for editorial assistance and Gudrun Labib and Ljubica Sanader for technical assistance. We also thank Dr. Peter T. Boag, Queen’s University, Kingston, Ontario, Canada, and Dr. G. Seutin, Florida Museum of Natural History, for help in collecting DNA samples; Dr. Dagmar Klein for help with sample shipments; the Galápagos and Costa Rica National Parks Services and the Charles Darwin Research Station (Galápagos), the staff on Cocos Island, and the Smithsonian Tropical Research Institute for administrative and logistical support; and the National Science Foundation for financial support to B.R.G. and P.R.G.
ABBREVIATIONS
- cr
control region
- cytb
cytochrome b
- NJ
neighbor-joining
- numt
nuclear mtDNA
- Ts
transitions
- Tv
transversions
Footnotes
References
- 1.Sulloway F J. Bull Brit Mus Nat Hist Zool Ser. 1982;43:49–94. [Google Scholar]
- 2.Gould J. Proc Zool Soc London. 1837;5:4–7. [Google Scholar]
- 3.Lack D. Darwin’s Finches. Cambridge: Cambridge Univ. Press; 1947. [Google Scholar]
- 4.Grant P R. Ecology and Evolution of Darwin’s Finches. Princeton, NJ: Princeton Univ. Press; 1986. [Google Scholar]
- 5.Yang S Y, Patton J L. The Auk. 1981;98:230–242. [Google Scholar]
- 6.Sato A, Figueroa F, Wedekind C, Klein J. Mol Marine Biol Biotechnol. 1998;7:221–231. [PubMed] [Google Scholar]
- 7.Marshall H D, Baker A J. Mol Biol Evol. 1997;14:173–184. doi: 10.1093/oxfordjournals.molbev.a025750. [DOI] [PubMed] [Google Scholar]
- 8.Baker A J, Marshall H D. In: Avian Molecular Evolution and Systematics. Mindell D P, editor. San Diego: Academic; 1997. pp. 51–82. [Google Scholar]
- 9.Kornegay J R, Kocher T D, Williams L A, Wilson A C. J Mol Evol. 1993;37:367–379. doi: 10.1007/BF00178867. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 12.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Version 3.1.1. [Google Scholar]
- 13.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Gu X, Zhang J. Mol Biol Evol. 1997;14:1106–1113. doi: 10.1093/oxfordjournals.molbev.a025720. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. phylip: Phylogenetic Inference Package. Seattle: University of Washington; 1986–1993. , Version 3.5c. [Google Scholar]
- 16.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetic Analysis. University Park, PA: The Pennsylvania State University; 1993. , Version 1.01. [Google Scholar]
- 17.Takezaki K, Rzhetsky A, Nei M. Mol Biol Evol. 1995;10:512–526. [Google Scholar]
- 18.Brown W M. In: Evolution of Genes and Proteins. Nei M, Koehm R K, editors. Sunderland, MA: Sinauer; 1983. pp. 62–88. [Google Scholar]
- 19.Quinn T W. In: Avian Molecular Evolution and Systematics. Mindell D P, editor. San Diego: Academic; 1997. pp. 3–28. [Google Scholar]
- 20.Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, Garcia-de-la-Torre C, Varela P, Recio M J, Ferre S, Martínez-Laso J. Cell Mol Life Sci. 1998;54:1031–1041. doi: 10.1007/s000180050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards S V, Arctander P, Wilson A C. Proc R Soc London Ser B. 1991;243:99–107. doi: 10.1098/rspb.1991.0017. [DOI] [PubMed] [Google Scholar]
- 22.Petren K, Grant B R, Grant P R. Proc R Soc London Ser B. 1999;266:321–330. [Google Scholar]
- 23.Bowman R I. In: Patterns of Evolution in Galápagos Organisms. Bowman R I, Berson M, Leviton A E, editors. Pacific Division, San Francisco, CA: Am. Assoc. Adv. Sci.; 1983. pp. 237–537. [Google Scholar]
- 24.Jo N. In: Patterns of Evolution in Galápagos Organisms. Bowman R I, Berson M, Leviton A E, editors. Pacific Division, San Francisco, CA: Am. Assoc. Adv. Sci.; 1983. pp. 201–217. [Google Scholar]
- 25.Vincek V, O’hUigin C, Satta Y, Takahata N, Boag P T, Grant P R, Grant B R, Klein J. Proc R Soc London. 1997;264:111–118. [Google Scholar]
- 26.Sclater P L. Catalogue of the Passeriformes, or Perching Birds, in the Collection of the British Museum. London: Order of the Trustees; 1886. [Google Scholar]
- 27.Sushkin, P. P. (1929) Verh. VI Int. Orn. Kongr. (Berlin), 375–378.
- 28.Darwin C R. The Zoology of the Voyage of H.M.S. Beagle, Under the Command of Captain FitzRoy, R.N., During the Years 1832–1836: Part III, Birds. London: Smith Elder; 1841. [Google Scholar]
- 29.Lack D. Evolution. 1969;23:252–263. doi: 10.1111/j.1558-5646.1969.tb03509.x. [DOI] [PubMed] [Google Scholar]
- 30.Grant P R. Philos Trans R Soc London Ser B. 1993;340:127–139. [Google Scholar]
- 31.Greenwood P H. The Haplochromine Fishes of the East African Lakes. Ithaca, NY: Cornell Univ. Press; 1981. [Google Scholar]
- 32.Meyer A, Kocher T D, Basasibwaki P, Wilson A C. Nature (London) 1990;347:550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 33.Nagl S, Tichy H, Mayer W E, Takahata N, Klein J. Proc Natl Acad Sci USA. 1998;95:14238–14243. doi: 10.1073/pnas.95.24.14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeland J. Ph.D. thesis. Ontario, Canada: Queen’s University; 1997. [Google Scholar]
- 35.Klicka J, Zink R M. Science. 1997;277:1666–1669. [Google Scholar]
- 36.Grant P R, Grant B R. Evolution. 1994;48:297–316. doi: 10.1111/j.1558-5646.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- 37.Grant P R. Evol Ecol. 1994;8:598–617. [Google Scholar]
- 38.Bowman R I. Morphological Differentiation and Adaptation in the Galápagos Finches. Berkeley: Univ. of California Press; 1961. [Google Scholar]
- 39.Klein D, Ono H, O’hUigin C, Vincek V, Goldschmidt T, Klein J. Nature (London) 1993;364:330–334. doi: 10.1038/364330a0. [DOI] [PubMed] [Google Scholar]
- 40.Klein J, Sato A, Nagl S, O’hUigin C. Annu Rev Ecol System. 1998;29:1–21. [Google Scholar]
- 41.Johnson T C, Scholz C A, Talbot M R, Kelts K, Ricketts R D, Ngobi G, Beuning K, Ssemmanda I, McGill J W. Science. 1996;273:1091–1093. doi: 10.1126/science.273.5278.1091. [DOI] [PubMed] [Google Scholar]