Abstract
It is shown that the two strands of the chloroplast genome from Euglena gracilis are asymmetric with regards to nucleotide composition. This asymmetry switches at both the origin of replication and a location that is halfway around the circular genome from the origin. In both halves of the genome the leading strand is G+T-rich, having a bias toward G over C and T over A, and the lagging strand is A+C-rich. This asymmetry is probably the result of a difference in mutation dynamics between the leading and lagging strands. In addition to composition asymmetry, the two strands differ with regards to coding content. In both halves of the genome the vast majority of genes are coded by the leading strand. These two aspects of strand asymmetry are then applied to a statistical test for selection on codon usage. The results indicate that selection on codon usage is limited to genes on the leading strand; no gene on the A+C-rich lagging strand shows evidence for selection, suggesting that highly expressed genes are coded predominantly on the strand of DNA that is the leading strand during replication. On the basis of these observations it is proposed that the coding strand bias is generated by selection to code highly expressed genes on the leading strand to coordinate the direction of replication and transcription, thereby increasing the potential rate of both reactions.
Keywords: codon bias, nucleotide composition, selection
There is strong evidence that the codon usage of certain plastid genes is influenced by selection (1). The frequency of specific codons, referred to as major codons, is significantly increased in highly expressed genes such as some of the genes that code for products involved in the process of photosynthesis. In contrast, low-expression genes, such as those coding subunits of the plastid RNA polymerase, have a codon usage bias that is dominated by the genome composition bias, which is A+T-rich. Since the major codons match the plastid tRNA population, it has been suggested that this pattern of codon usage variation among genes is a result of selection for increased translation efficiency in highly expressed genes (1, 2). Evidence for selection has been observed in all plants and algae studied to date, but there are differences in selective intensity between lineages as indicated by variation among lineages in both the number of genes with an increased frequency of major codons and the degree of bias toward major codons in highly expressed genes. In general, codon bias is strong in the green algae, of intermediate strength in other algae, and extremely weak in the flowering plants (1).
Work on codon usage of plastid genes has focused on the major codons in two sets of codon groups. The first set is composed of the twofold-degenerate groups with a pyrimidine at the third position (particularly GAY, CAY, TAY, AAY, and TTY). For these codon groups it has been shown that the major codons of the plastid are the C-terminating codons (2). As a result, there is a bias toward C at twofold-degenerate sites in highly expressed genes, which is noticeable given the strong A+T bias in the genome overall (2). The second set is composed of the fourfold-degenerate codon groups, and for each of these codon groups, the codon with a T at the third position is the major codon, with the exception of valine, for which the major codon is GTA (3).
The single known exception to the general pattern discussed above is the green alga Euglena gracilis. In terms of anticodon sequence, the tRNA genes coded by Euglena are essentially identical to all other fully sequenced plastid genomes (4). Despite this, an increased frequency of C at twofold-degenerate sites is not observed in any chloroplast gene of Euglena, including psbA (1, 4) which codes for the most prominent translation product of the chloroplast (5). Instead, all of the genes coded by the Euglena chloroplast genome have a high A+T content at the third codon position of every codon group (1). This apparent lack of increase in major codon usage in highly expressed genes seems to suggest that selection is not adapting codon usage to the tRNA population in Euglena. However, a test comparing the codon usage of each gene to an expected distribution generated from noncoding base frequencies indicated that several highly expressed genes from Euglena have a codon usage bias that is significantly different from expectation (1). In addition, codon bias levels in Euglena genes are strongly correlated with levels from homologous genes of other algae (6). Both of these observations suggest that, although there is no obvious increase in the use of major codons for twofold-degenerate groups, selection does act on the codon usage of at least some Euglena chloroplast genes. These apparently conflicting lines of evidence make Euglena an interesting case study for codon usage bias.
In the current work, the codon usage of Euglena chloroplast genes is studied in more detail with respect to a previously unknown asymmetry in the composition of the genome. A number of genomes have been found to be asymmetrical, particularly in compositional properties (7, 8). For example, the bacterium Mycoplasma genitalium (9) and the metazoan mitochondrial genome (10) have one G+T-enriched strand and one A+C-enriched strand. In some bacterial species, the strand-specific bias switches at both the origin and terminus of replication in such a way that the leading strand has the same bias in all regions of the genome (7, 8, 9, 11). It is likely that this is the result of observed differences in the mutational dynamics of the polymerase subunits responsible for replicating the different strands of DNA (7).
In the case of the Euglena chloroplast genome, one asymmetrical feature has been noted previously: the circular chromosome is strongly biased in terms of which strand is the coding strand (4). The entire genome consists of 143,172 bp, with a total of 62 protein-coding genes, including ORFs, and 41 RNA-coding genes. Moving outwards from the origin of replication (site 1) in one direction, almost all genes from site 1 to about 70000 are coded on the strand designated as strand A. In the other direction from the origin of replication, almost all genes from site 143172 to 70000 are coded on strand B (ref. 4, also see Fig. 1).
Figure 1.
Plot of (T − A)/(T + A) (solid line) and (C − G)/(C + G) (dashed line) for strand A of the chloroplast genome of Euglena gracilis. The plot is performed over a sliding window of length 5,000 and a shift of 500, starting at the origin of replication. Above the plot is a representation of the coding strand, either strand A or B, for known genes and ORFs (4). Each box represents the number of genes indicated but does not imply anything about operon structure or spacing of the genes in that region. The shaded box represents the region where the three repeated rDNA operons are coded. The approximate location of the atpA gene is also indicated.
In this study it is shown that the two strands of the Euglena chloroplast genome also differ in compositional properties. Strand A has a bias toward G over C and T over A from nucleotide 1 to approximately 70000 but a bias toward A and C from about nucleotide 70000 to the last base of the circular genome. Therefore, the strand that codes the majority of genes in each half of the genome also has a bias toward G and T. Since the statistical test of codon usage applied previously was based on an expectation generated from noncoding base frequencies (1), the strand asymmetry demonstrated in this work requires that the test utilize the base frequencies for a specific strand and genome location. When these factors are controlled for it is found that many more genes than previously thought have a codon bias that is significantly greater than expected. In addition, selection appears to be limited to genes on the G+T-rich strand, which is the leading strand in both directions from the origin of replication. Therefore, coding strand bias is correlated with gene expression level. It is proposed that this bias is due to selection to coordinate transcription and DNA replication.
MATERIALS AND METHODS
All sequences were extracted directly from the complete genome sequence of Euglena gracilis (ref. 4, GenBank accession no. X70810). Genes greater than 350 nucleotides in length, to avoid sampling bias in codon usage calculations, and noncoding regions greater than 30 bases in length were used for analyses. Coding sequence locations were determined from information in the GenBank file, and the definition of strands A and B are taken from ref. 4. The codon adaptation index (CAI; ref. 12) of each gene was calculated as described previously (1).
To test CAI relative to the composition of noncoding regions, the circular genome was divided into two sections which were separated by sites 0 and 69075 of the genome (the numbering is that used in the GenBank file). This division is based on the analysis presented below. For both halves of the genome, cumulative dinucleotide frequencies from all noncoding regions were calculated. Dinucleotide frequencies for the opposite strand were determined from the complement so that four tables of dinucleotide frequencies were obtained; one for each strand in each half of the genome. These tables were used to perform a test on each individual gene as described previously (1). For 500 replicates, a random codon table was generated with the same amino acid usage as the actual gene. Codon frequencies were assigned at random for each codon group on the basis of the composition of the second codon position of that codon group as well as the dinucleotide frequency table for that region and strand in which the gene is coded. The CAI value was then calculated for the random codon table. From the 500 replicates for the gene a distribution was calculated and compared with the observed value. Any gene with a CAI value greater than 2 standard deviations above the mean has a codon usage bias that is significantly different than expected on the basis of the composition of noncoding regions, which is regarded as evidence for selection on codon use, assuming that noncoding dinucleotide frequencies are an accurate reflection of mutation bias (1).
Strand asymmetry was calculated for the genome overall and for noncoding regions by using the method described by Lobry (9, 11). For noncoding regions, the value (T − A)/(T + A), where A and T represent the number of occurrences of the two nucleotides, respectively, was calculated separately for each region. For the entire genome, the values (T − A)/(T + A) and (C − G)/(C + G) were calculated for windows of length 5000, starting with nucleotides 1 to 5000 and continuing by shifting 500 nucleotides downstream along strand A for each new window.
RESULTS AND DISCUSSION
Strand Asymmetry in Euglena.
The nucleotide composition of strand A from the Euglena chloroplast genome is shown in Fig. 1, starting and ending at the origin of replication. The values (T − A)/(T + A) and (C − G)/(C + G) are plotted over a sliding window, so that the plot represents regional biases toward pyrimidines. Above the plot of compositional bias is a representation of the coding strand for all genes. As noted previously, there is a bias in the coding strand of the Euglena chloroplast genome. Over the first half of the genome, most genes (38 of 51) are coded by strand A, whereas over the second half most genes (45 of 52, including the 3 rDNA operons) are coded by strand B (4).
It can be seen in Fig. 1 that strand A has a bias toward T over A and G over C over most of the first half of the genome, whereas in the second half of the genome, strand A has a complementary bias toward A and C. Any strand with a bias toward T over A and G over C will be referred to here as G+T-rich, although there is actually a higher representation of A than G because of the strong A+T bias of the genome. Therefore, strand A is G+T-rich over the half of the genome between sites 1 and roughly 70000, whereas strand B is G+T-rich over the other half. The main exception to this general trend is the small bias in strand A toward T around position 125 kb from the origin. This is the location of the three repeats of the rDNA operon on strand B, indicated by the shaded box above the plot. This deviation may simply reflect selection on the structural properties of the rRNA products. There are also two short sections that are an exception to the strand bias. One is where strand A has a slight bias toward C over G roughly 30 kb from the origin, and the other where it shows a slight bias toward T over A about 90 kb from the origin. Because the first correlates with a region where 7 genes are coded by strand B and the second with a region where strand B codes 12 genes, these small exceptions may reflect selection for amino acid composition of these genes.
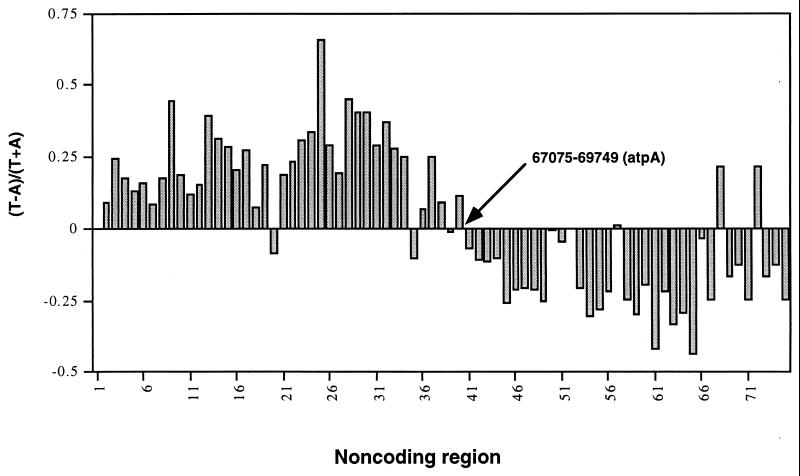
The strand asymmetry in base composition and the difference between the two halves of the genome is most apparent in noncoding DNA (Fig. 2). The position of the atpA gene, which lies at the approximate point in Fig. 1 where the strand shift occurs, is indicated in the figure. Almost all sequences to the left (upstream) of atpA have a bias of T over A, whereas most sequences downstream have a bias toward A over T. The bias toward C over G was not measured for individual noncoding regions because many are under 100 nucleotides in length and strongly biased toward A and T. However, the bias is observed; the noncoding regions up to nucleotide 69075 have a cumulative composition of 2,694 A vs. 3,771 T and 804 G vs. 712 C, whereas noncoding regions downstream of nucleotide 69075 have a composition of 4,041 A vs. 3,344 T and 871 G vs. 1,180 C. The few regions that are exceptions to the rule are quite short, and the two regions downstream of the atpA gene with a strong T bias are identical 86-bp spacer sequences within the rDNA repeat, that is, within the region around 125 kb in Fig. 1 that is noted above. Overall, noncoding regions are asymmetrical, and the two halves of the genome display different strand-specific biases. Although this pattern of asymmetry is very similar to what has been observed in some other organisms (8, 9, 10), none of the other fully sequenced plastid genomes has a composition asymmetry that is similar to what is observed in Euglena (data not shown).
Figure 2.
Plot of (T − A)/(T + A) of strand A for all noncoding regions greater than 30 nucleotides in length. The noncoding regions are numbered from 1 to 75, starting at the origin of replication and proceeding downstream. These numbers represent the order of the regions but not genome location. The relative location and nucleotides of the atpA gene, which is located between noncoding regions 40 and 41, are indicated to show the genome location at which the symmetry changes in Fig. 1.
Figs. 1 and 2 demonstrate that there are two asymmetrical features of the genome, coding strand bias and compositional bias. Interestingly, both of these biases switch strands at essentially the same genomic location, roughly 69 kb from the origin, just downstream from the atpA gene (Fig. 1). As a result of this correlated change in biases the G+T-rich strand codes for the majority of genes over both halves of the genome. Strand A is defined as the strand that runs 5′ to 3′ as base numbering increases (4). Since the switch in asymmetry occurs at the origin and again at the point roughly halfway around the genome from the origin where replication terminates (4, 13), it is the leading strand of replication that is G+T-rich in both halves of the genome. Because the composition asymmetry is most apparent in noncoding sequences, it is probably the result of an asymmetrical mutation process. A difference between the leading and lagging strand polymerases (or subunits), either in misincorporation properties or proofreading functions, could account for the asymmetry. This possibility is supported by the observation that the two replication strands in Escherichia coli have significantly different mutational dynamics because of differences in the polymerase subunits responsible for replicating the different strands (7).
Strand Asymmetry and Codon Bias.
In the current study we are interested in the codon usage of Euglena chloroplast genes: specifically, whether there is any evidence for selective constraints and on what genes those constraints act. In addressing this issue I studied each gene separately, comparing the level of codon bias of that gene to an expected value. Because composition bias contributes to codon bias (14, 15), the compositional asymmetry demonstrated in Fig. 2 requires that both the strand and the genomic location of the gene be taken into consideration. To accomplish this, genes were divided into two groups, those coded by a G+T-rich strand (strand A from 0 to 70 kb and strand B from 70 kb to 143 kb) and those coded by an A+C-rich strand. The level of codon bias was measured by using CAI. CAI measures, in essence, the relative frequency of major codons in different genes and is positively correlated with expression level in plastid genomes (1), probably because of selection acting on highly expressed genes to match codon usage to the tRNA population (1, 16).
As expected, the two sets of genes have different average compositional properties. Genes coded by the G+T-rich strand have a much higher T content at fourfold-degenerate sites and higher CAI values than genes coded on the A+C-rich strand, and there is almost no overlap between the ranges of CAI values for G+T-rich strand genes and A+C-rich strand genes (Table 1). The two exceptions are in region 1; the ccsA gene, coded on strand B from nucleotides 2171–3549, which has a CAI of 0.380, and rps3, coded on strand A from nucleotides 52501–53668, which has a CAI of 0.242. Without these two genes the CAI ranges are 0.246–0.338 for strand B and 0.346–0.436 for strand A. However, variation in CAI will necessarily follow from variation in composition properties. As discussed above, selection in the plastid favors T at the third position of most fourfold-degenerate codon groups. Because CAI measures the frequency of major codons, genes on the G+T-rich strand could have higher CAI values because of the higher frequency of T at fourfold-degenerate sites resulting from the composition properties of that strand. Therefore, to make inferences about selection we have to determine whether the differences in codon usage between genes are due simply to the asymmetrical composition bias in the genome.
Table 1.
Compositional properties of genes in different genomic regions and on different strands
| Position | Strand | Genes | Mean CAI (Range) | Mean % T* (Range) |
|---|---|---|---|---|
| Entire | Both | 34 | 0.355 | 0.484 |
| genome | A | 20 | 0.358 | 0.518 |
| B | 14 | 0.352 | 0.435 | |
| 0–70 kb | Both | 23 | 0.351 | 0.475 |
| A (G+T-rich) | 17 | 0.363 | 0.555 | |
| (0.242–0.436) | (0.383–0.696) | |||
| B (A+C-rich) | 6 | 0.316 | 0.248 | |
| (0.246–0.380) | (0.147–0.312) | |||
| 70–143 kb | Both | 11 | 0.365 | 0.503 |
| A (A+C-rich) | 2 | 0.300 | 0.322 | |
| (0.295–0.305) | (0.281–0.361) | |||
| B (G+T-rich) | 9 | 0.379 | 0.543 | |
| (0.310–0.509) | (0.458–0.667) |
T composition at fourfold-degenerate sites.
To control for variation in the composition properties of different genome locations, we applied a previously developed test for selection on codon usage. This test utilizes noncoding base compositions to generate an expected distribution of CAI for each gene. The distribution is then used to determine whether the actual gene has a codon usage (CAI) that is significantly different from expectation (1). A significant difference means that composition properties of the genome, or region, cannot account for codon usage of that gene. This deviation is then taken as evidence that selection acts on that particular gene (1), based on an assumption that noncoding DNA is an accurate reflection of composition bias. The asymmetry of the Euglena genome means that the test for a specific gene must utilize the base composition of the noncoding regions from the same strand and genome location that codes the gene. Therefore, four sets of noncoding dinucleotide frequencies were used, one for each of strands A and B over the first half of the genome, and one for each of strands A and B over the second half of the genome. For each gene the appropriate frequency table was applied to generate the distribution. This will test whether the codon usage of a specific gene is within the expected range, given the genomic location of that gene, allowing us to assess selection with regards to the differences between the sets of genes in Table 1.
The results of this test are presented in Table 2. Two main conclusions can be reached. First, many more genes give a significant result than when the test was applied previously. The initial test, which did not account for strand asymmetry, gave evidence for selection on only 6 of 36 chloroplast genes (including two ORFs) from Euglena (1). The test accounting for asymmetry gives a significant result for 23 of 34 genes. Second, there is a clear asymmetry in the distribution of significant results. All of the genes that have a significant CAI value are on the G+T-rich strand; none of the 8 protein-coding genes greater than 350 nucleotides in length that are coded by an A+C-rich strand have a significant value. Therefore, the higher CAI values of genes on the G+T-rich strand (Table 1) are not simply due to compositional properties; selection appears to contribute to codon bias to at least some degree. It should be noted, however, that the intensity of selection is not as strong in Euglena as in other green algae. Although the results presented here show that CAI values of a number of genes are significantly greater than expectation, they are still low relative to homologous genes from other algae (1). Regardless, the results from Table 2 strongly suggest that selection does influence codon usage of a significant number of genes in Euglena.
Table 2.
Results from the codon bias test
| Genome location* | Gene | CAI† |
|---|---|---|
| Region 1 | psbD | P < 0.001 |
| strand a (G+T-rich) | ycf13 | P < 0.001 |
| psaA | P < 0.0001 | |
| psaB | P < 0.001 | |
| rpl2 | P < 0.01 | |
| rpl22 | P < 0.001 | |
| rps3 | NS | |
| rpl16 | P < 0.05 | |
| rpl14 | NS | |
| rpl5 | P < 0.001 | |
| rps8 | P < 0.01 | |
| rps2 | P < 0.001 | |
| atpI | P < 0.001 | |
| atpF | P < 0.001 | |
| psbC | P < 0.0001 | |
| psbA | P < 0.0001 | |
| atpA | P < 0.001 | |
| Region 1 | ycf4 | NS |
| strand B (A+C-rich) | ccsA | NS |
| tufA | NS | |
| rps7 | NS | |
| rps12 | NS | |
| rpl20 | NS | |
| Region 2 | rps4 | NS |
| strand A (A+C-rich) | rps11 | NS |
| Region 2 | rpl12 | P < 0.001 |
| strand B (G+T-rich) | rps9 | NS |
| atpB | P < 0.001 | |
| petB | P < 0.001 | |
| psbB | P < 0.0001 | |
| rpoC2 | P < 0.001 | |
| rpoC1 | P < 0.001 | |
| rpoB | P < 0.001 | |
| rbcL | P < 0.0001 |
The fact that significant results were obtained only for genes on the G+T-rich strand indicates that there is a correlation between codon usage and genome organization in Euglena. On the basis of the model developed for Borrelia burgdorferi (17) it is proposed that this correlation is a secondary effect of two different correlations: first, the established correlation between expression level and CAI (1), and second, a correlation between gene expression and genome organization. The proposal is that the second correlation exists and that it is due to selective pressure to code most genes, and in particular highly expressed genes, on the leading strand. This organization would increase the likelihood that the RNA and DNA polymerases are moving in the same direction during replication, reducing the number of “head-on collisions” between the two enzymes. Such collisions have been shown to significantly decrease the rate of replication (17, 18), so the basis of the selective pressure would be to increase the potential rate of DNA replication and, possibly, the rate of transcription during the replication cycle (17). Under such a model, the selective pressure to code any particular gene on the leading strand would increase with the expression level of that gene, leading indirectly to the observed correlation between CAI and strand. The secondary nature of this correlation is supported by the observation that the nontranslated rRNA and tRNA genes are almost exclusively coded on the leading strand (4). It should be noted that this type of correlation is not limited to Euglena; a similar correlation between CAI and coding strand exists in E. coli, with high CAI genes being coded at a significantly higher frequency on the leading strand (7). In addition, a recent analysis of bacterial genomes showed that in every case the majority of genes are coded by the leading strand of replication (8).
On the basis of results in Table 2, then, it is proposed that the asymmetrical organization of the Euglena chloroplast genome is caused by a selective pressure to coordinate replication and transcription similar to what has been suggested previously for B. burgdorferi (17). This coordination would be independent of the compositional asymmetry, which is probably generated by different misincorporation biases of the enzymes that replicate the leading and lagging strands. The fact that the two features switch strands at roughly the same genomic location (Fig. 1) is probably due to the shared dependence on replication, not to any real influence of one on the other.
Asymmetry and Amino Acid Composition.
The strand asymmetry demonstrated in Figs. 1 and 2 has been shown above to be an important factor in the analysis of codon bias. In addition, because genomic base composition is correlated with amino acid composition in a number of organisms (19–22), it is possible that the asymmetry in Euglena gives rise to variation in amino acid composition among genes in different genomic sections. The amino acids that would be expected to be influenced are those coded by codons that are either G+T-rich or A+C-rich. There are 8 that fall into this category: Phe (TTY), Cys (TGY), Trp (TGG), Gly (GGN), Val (GTN), Lys (AAR), Asn (AAY), and Pro (CCN). The first five are expected to be present in an increased frequency on the leading strand in both halves of the genome and the last three to be present in a decreased frequency.
The number of occurrences of for each of these 8 amino acids is given in Table 3 for both the G+T-rich and A+C-rich genome regions. As expected, the five amino acids coded by G+T-rich codons occur at a significantly higher frequency on the leading strand (χ2 = 7.05, P < 0.01). The only exception is glycine (GGN), which is present in higher frequency on the A+C-rich strand. When this amino acid is excluded the difference between the two strands is strongly significant (χ2 = 33.9, P < 0.001). In addition, the three amino acids coded by A+C-rich codons are present at a significantly lower frequency on the leading strand (χ2 = 13.3, P < 0.01), primarily because of a difference in the representation of Lys (AAR). These results are consistent with strand bias having an influence on amino acid composition of the proteins coded by each strand, although differences in coding properties will have to be tested in future studies to evaluate the significance of the differences observed between the strands.
Table 3.
Differences in amino acid composition of genes coded on different strands
| Codons | G+T-rich strand
|
A+C-rich strand
|
||
|---|---|---|---|---|
| Number | Frequency | Number | Frequency | |
| All | 9,778 | — | 1,854 | — |
| TTY | 635 | 0.065 | 55 | 0.030 |
| TGY | 110 | 0.011 | 13 | 0.007 |
| TGG | 198 | 0.020 | 8 | 0.004 |
| GGN | 729 | 0.075 | 185 | 0.100 |
| GTN | 632 | 0.065 | 116 | 0.063 |
| Total | 2,304 | 0.236 | 377 | 0.203 |
| AAY | 505 | 0.052 | 101 | 0.054 |
| AAR | 697 | 0.071 | 196 | 0.106 |
| CCN | 379 | 0.039 | 73 | 0.039 |
| Total | 1,581 | 0.162 | 370 | 0.200 |
Concluding Remarks.
The data presented here demonstrate that there is a strong asymmetry in the chloroplast genome of Euglena gracilis. Two features are found to be asymmetrical: nucleotide composition bias and which strand acts as the coding strand for genes. The strand-specific biases of both features are correlated in that they change at both the origin and the putative terminus of DNA replication. It is suggested that these are two independent features that are correlated because of a common reliance on replication. Composition asymmetry is proposed to arise from different mutational dynamics of the leading and lagging strand. When these biases are accounted for there is statistical evidence that selection influences the codon usage of a large number of genes, but apparently only genes coded on the leading strand of replication. It is proposed that this arises from a correlation between expression level and selection for codon usage coupled with selection to code genes, and in particular highly expressed genes, on the leading strand. This organization would limit “collisions” between RNA and DNA polymerase and, therefore, increase the speed of replication. The result is the coding strand asymmetry that is observed. Finally, it is also shown that amino acid frequencies vary between genes coded on each strand in a manner consistent with the mutation bias.
If this model concerning selection and coding strand asymmetry for Euglena is correct, then it raises questions about the plastid genome of other algae as well as plants. No other plastid genome for which we have a complete sequence available—which includes a diatom, a red alga, the cyanelle of Cyanophora paradoxa, a green alga, and several land plants—has such an obvious organization in terms of coding strand bias. If the organization in Euglena does in fact reflect selection to coordinate replication and transcription, we need to determine whether there is any evidence that other plastid genomes have a similar organization. Finally, the evolutionary relationship of the Euglena chloroplast to other plastids has been problematic for some time (23). Both the asymmetry described here and the unusual codon usage bias (6) could influence sequence analyses, and an accurate assessment of its relation to other algae will require a serious consideration of the unusual composition properties of the Euglena chloroplast genome.
Acknowledgments
I thank Sean Graham for reading and commenting on this manuscript and two anonymous reviewers for helpful comments. This work was supported in part by National Science Foundation Grant MCB-9727906.
ABBREVIATIONS
- R
purine
- Y
pyrimidine
- CAI
codon adaptation index
References
- 1.Morton B R. J Mol Evol. 1998;46:449–459. doi: 10.1007/pl00006325. [DOI] [PubMed] [Google Scholar]
- 2.Morton B R. J Mol Evol. 1993;37:273–280. doi: 10.1007/BF00175504. [DOI] [PubMed] [Google Scholar]
- 3.Morton B R. J Mol Evol. 1996;43:28–31. doi: 10.1007/BF02352296. [DOI] [PubMed] [Google Scholar]
- 4.Hallick R B, Hong L, Drager R G, Favreau M R, Montfort A, Orsat B, Spielman A, Stutz E. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullet J E, Klein R R. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morton B R. In: Evolutionary Biology. Hecht M, MacIntyre R, Clegg M, editors. New York: Plenum; 1999. , in press. [Google Scholar]
- 7.Francino M P, Ochman H. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 8.McLean M J, Wolfe K H, Devine K M. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 9.Lobry J R. Science. 1996;272:745–746. doi: 10.1126/science.272.5262.745. [DOI] [PubMed] [Google Scholar]
- 10.Jermiin L S, Graur D, Crozier R H. Mol Biol Evol. 1995;12:558–563. [Google Scholar]
- 11.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 12.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koller B, Delius H. EMBO J. 1982;1:995–998. doi: 10.1002/j.1460-2075.1982.tb01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp P M. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 15.Sharp P M, Stenico M J, Peden F, Lloyd A T. Biochem Soc Trans. 1993;21:835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- 16.Ikemura T. Mol Biol Evol. 1985;2:13–35. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 17.McInerney J O. Proc Natl Acad Sci USA. 1998;95:10698–10703. doi: 10.1073/pnas.95.18.10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French S. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 19.D’Onofrio G, Mouchiroud D, Aissani B, Gautier C, Bernardi G. J Mol Evol. 1991;32:504–510. doi: 10.1007/BF02102652. [DOI] [PubMed] [Google Scholar]
- 20.Berkhout B, van Hemert F J. Nucleic Acids Res. 1994;22:1705–1711. doi: 10.1093/nar/22.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter T D. Biochim Biophys Acta. 1995;1261:394–400. doi: 10.1016/0167-4781(95)00037-h. [DOI] [PubMed] [Google Scholar]
- 22.Foster P G, Jermiin L S, Hickey D A. J Mol Evol. 1997;44:282–288. doi: 10.1007/pl00006145. [DOI] [PubMed] [Google Scholar]
- 23.Martin W, Somerville C C, Loiseaux-de Goer S. J Mol Evol. 1992;35:385–404. doi: 10.1007/BF00162973. [DOI] [PubMed] [Google Scholar]