Abstract
Human telomerase consists of two essential components, telomerase RNA template (hTER) and telomerase reverse transcriptase (hTERT), and functions to synthesize telomere repeats that serve to protect the integrity of chromosomes and to prolong the replicative life span of cells. Telomerase activity is expressed selectively in germ-line and malignant tumor cells but not in most normal human somatic cells. As a notable exception, telomerase is expressed in human lymphocytes during development, differentiation, and activation. Recent studies have suggested that regulation of telomerase is determined by transcription of hTERT but not hTER. The highly regulated expression of telomerase in lymphocytes provides an opportunity to analyze the contribution of transcriptional regulation of hTERT and hTER. We report here an analysis of hTERT expression by Northern and in situ hybridization. It was found that hTERT mRNA is expressed at detectable levels in all subsets of human lymphocytes isolated from thymus, tonsil, and peripheral blood, regardless of the status of telomerase activity. hTERT expression is regulated as a function of lineage development, differentiation, and activation. Strikingly, however, telomerase activity in these cells is not correlated strictly with the levels of hTERT and hTER transcripts. The absence of correlation between telomerase activity and hTERT mRNA could not be attributed to the presence of hTERT splice variants or to detectable inhibitors of telomerase activity. Thus, transcriptional regulation of hTERT is not sufficient to account for telomerase activity in human lymphocytes, indicating a likely role of posttranscriptional factors in the control of enzyme function.
Telomerase, a specialized reverse transcriptase, seems to play a key role in maintaining telomere length and in cellular replicative life span (1–3). In humans, telomerase activity is detected in germ cells, in most malignant tumor cells, and in a few somatic cells such as lymphocytes but not in the majority of somatic cells (4–6). The mechanisms that control telomerase activity are not completely understood. The two core components of human telomerase, hTER and hTERT, have been identified recently (7–11). hTERT and hTER are both necessary and sufficient for telomerase activity in vitro (12, 13) and have been reported to have different expression profiles in normal somatic cells (7–11). The expression of hTER is ubiquitous in all types of human cells, regardless of the status of telomerase activity. In contrast, it has been reported recently that hTERT is expressed only in cells and tissues positive for telomerase activity, i.e., tumor and fetal cells, but that hTERT is not detected in normal somatic cells that lack telomerase activity (8, 9, 11, 14, 15). Furthermore, introducing hTERT into normal somatic cells that do not normally express hTERT or telomerase activity is sufficient to reconstitute telomerase activity in these cells (16–19). Thus, it has been concluded that regulation of telomerase activity, at least in human fibroblasts and some epithelial cells, is controlled at the level of hTERT transcription (7, 8).
B and T lymphocytes, the major antigen-specific lineages of the immune system, require extensive cell division and clonal expansion for their functions. Recently, telomerase and its regulation of telomere length have drawn considerable attention for their potential roles in controlling cellular replicative life span (20–22). Interestingly, it has been found that telomerase activity is expressed and regulated stringently during lymphocyte development and differentiation (23, 24) and also that telomerase activity is induced dramatically in mature resting lymphocytes on antigenic activation (25–30). Furthermore, elevated telomerase activity was correlated with apparent telomere lengthening in tonsil germinal-center (GC) B cells (24), suggesting that telomerase may play an important role in the control of human lymphocyte replicative life span. Because telomerase activity is highly regulated in lymphocytes, defining the molecular basis for this regulation is of particular interest and may suggest mechanisms for manipulation of the immune response for therapeutic benefit. Identification of the genes encoding the two essential components of telomerase allows direct analysis of transcriptional regulation of telomerase. Recent analysis of hTER in lymphocytes determined that, although ubiquitously present, expression of hTER is regulated and is correlated with overall telomerase activity at various stages during lineage development, differentiation, and activation (24, 31). However, the presence of hTER in cells that express no detectable telomerase activity suggested that normally hTER is not a determining factor for telomerase activity (6, 31).
The expression of hTERT in lymphocytes has not been assessed previously; therefore, the role of hTERT in the regulation of telomerase activity in lymphoid cells is not known. Here, we report an analysis of the expression of hTERT, the more recently identified telomerase catalytic component, in human lymphocytes and show that hTERT mRNA, like hTER, is present in all B and T lineage cells analyzed. As was observed for hTER, hTERT expression is regulated during lymphocyte development, differentiation, and activation. However, hTERT as well as hTER transcripts were detected in lymphocytes with no detectable telomerase activity; there was no strict quantitative relationship between telomerase activity and hTERT or hTER transcript levels in lymphocyte populations, suggesting that transcription of hTERT and hTER is not sufficient for telomerase activity and indicating that other mechanisms contribute to the regulation of telomerase activity in human lymphocytes.
MATERIALS AND METHODS
Isolation of B and T Cells from Tonsil, Thymus, and Peripheral Blood.
Tonsils were obtained from tonsillectomies of children and adults with chronic tonsillitis (Holy Cross Hospital, Silver Spring, MD), thymuses from children undergoing pediatric cardiac surgery (Fairfax County Hospital, Fairfax, VA), and peripheral blood of normal donors from National Institutes of Health blood bank (Bethesda, MD). Subsets of B and T cells were isolated from tonsil, thymus, and peripheral blood as described (23, 24). In brief, mononuclear cells were isolated by lymphocyte separation medium (Organon Teknika–Cappel) from minced tonsil, thymus, or buffy coat blood diluted in Hanks’ solution (Biofluids, Walkersville, MD). Subsets of B and T cells were selected further by immunomagnetic separation procedures (Dynal, Great Neck, NY). The purity of the isolated subsets of B and T cells was analyzed by flow cytometry (FACScan, Becton Dickinson) according to the manufacturer’s instructions and was generally 90–95%.
Activation of T Lymphocytes in Vitro.
Activation of peripheral-blood CD4+ T cells was conducted as described (32). Anti-CD3 (OKT3) was obtained from Ortho Biotech (Raritan, NJ), and anti-CD28 mAb 9.3 (IgG2a) was purified on protein A-Sepharose. Anti-CD3 and anti-CD28 antibodies were immobilized on magnetic beads (Dynal) and used to stimulate CD4+ T cells.
PCR-Based Telomerase Assay.
A modified telomeric repeat amplification protocol (TRAP) was used as described (33). Generally, the telomerase assay was carried out in two steps: telomere synthesis and PCR amplification. Telomere synthesis was carried out in an Eppendorf tube containing cell extracts from 5 × 104 cells, followed by PCR consisting of 28 cycles of amplification in a DNA Engine (PTC-225, Tetrad MJ Research, Watertown, MA); one-third of PCR products (≈3,000 cell equivalents) from each sample was loaded per well of a 12% polyacrylamide gel, stained with a sensitive DNA dye (SYBR Green I, Molecular Probes), and visualized by the Storm system (Molecular Dynamics, Sunnyvale, CA), a dual imaging system for phosphorimages and fluorescent images. The activity of telomerase was quantitated by a 2-fold serial dilution of cell lysates and expressed as the maximal dilution of lysate that generated a clear DNA ladder. In experiments involving of mixing of lysates, cell lysate from thymocytes or GC B cells was mixed with cell lysates from peripheral-blood resting CD8+ T and CD19+ B lymphocytes or with lysis buffer at 1:1 or 1:3 ratios; telomerase activity was then measured as described.
Northern Hybridization.
Total RNA was extracted from purified cells by using STAT-60 RNA solution (Tel-Test, Friendswood, TX). For hTERT analysis, messenger RNA was isolated from the total RNA by using an mRNA isolation kit (Dynal). From each sample, 0.4 μg of mRNA was used. For hTER analysis, 10 μg of total RNA was used for each sample. RNA was separated on 1.2% agarose/formaldehyde gels and transferred to nylon membranes (Hybond-N+, Amersham Pharmacia) by using Turboblotter (Schleicher & Schuell). The hTERT probe was a 1,261-bp fragment (from nucleotides 1,895–3,155) generated by reverse transcription–PCR (RT-PCR) from tonsil B cell mRNA and cloned into PCR II vector (Invitrogen). The PCR primers were hTERT-1950S (5′-CGATTGTGAACATGGACTACGTCGTGGG-3′) and hTERT-3210A (5′-GCGTTCTTGGCTTTCAGGATGGAGTAGC-3′). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was generated by PCR of the entire GAPDH coding sequence and cloned into PCR II vector. The cloned DNA fragments were confirmed by DNA sequencing. The probes were generated by in vitro transcription by using a Strip-EZ T3/T7 Kit (Ambion, Austin, TX) in the presence of [α-32P]UTP (New England Nuclear). Prehybridization and hybridization were carried out at 65°C for 16 h, following by two washes with 2× SSC/0.1% SDS at room temperature for 5 min, one wash with 1× SSC/0.1% SDS at 68°C for 30 min, and two washes with 0.1× SSC/0.1% SDS at 68°C for 30 min. The hTER and 7SK probes and hybridization conditions have been described (31). The membranes were exposed to phosphorimager screens, and the results were analyzed by using imagequant (Molecular Dynamics). When levels of mRNA were assayed on a cell-equivalent basis, it was observed consistently that the levels of GAPDH as well as levels of hTERT varied as a function of lymphocyte development or activation. hTERT levels therefore are expressed here normalized to cell equivalents.
RT-PCR Analysis of hTERT Expression.
The first strand cDNA was synthesized by reverse transcription from mRNA by using the SuperScript Choice System (Life Technologies, Gaithersburg, MD). The cDNA samples were amplified in a 20-μl reaction mixture containing 200 μM each of the four deoxynucleoside triphosphates, 2 units AmpliTaq DNA polymerase (Perkin–Elmer), and 0.2 μM primers. Primers hTERT-1680S (5′-CCAAGTTCCTGCACTGGCTGATG-3′) and hTERT-2951A (5′-TTTGCGACGCATGTTCCTCCCAGC-3′) were used to amplify hTERT coding region from nucleotide 1,680 to nucleotide 2,951 to generate a 1,272-bp fragment. Primers hTERT-2164S (5′-GCCTGAGCTGTACTTTGTCAA-3′) and hTERT-2620A (5′-CGCAAACAGCTTGTTCTCCATGTC-3′) were used to amplify the hTERT coding region from nucleotide 2,164 to nucleotide 2,620 to generate a 457-bp fragment (35). The reaction mixtures were heated at 94°C for 60 s, followed by 28 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s.
RNA in Situ Hybridization.
Sections (10 μm thick) were cut from frozen tonsil tissues. In situ hybridization was carried out by using a digoxigenin nucleic acid detection kit (Boehringer Mannheim) according to the manufacturer’s instructions. In brief, the sense and antisense hTERT probes were synthesized by in vitro transcription with an AmpliScribe T7 transcription kit (Epicentre Technologies, Madison, WI) and labeled with digoxigenin-UTP. The prehybridization and hybridization were carried out at 43°C in a Hybaid OmniSlide humidity incubator (National Labnet, Woodbridge, NJ) for 16 h. The sections were washed sequentially, once with 2× SSC at 43°C for 5 min, thrice with 60% formamide in 0.2× SSC for 5 min at 43°C, and twice with 2× SSC for 5 min at room temperature. Signal amplification was carried out with anti-digoxigenin antibody alkaline phosphatase-conjugate (1:200 dilution). Detection was accomplished with the detection buffer [100 mM Tris⋅HCl, pH 9.5/100 mM NaCl/50 mM MgCl2/0.18 mg/ml 5-bromo-4-chloro-3-indolyl phosphate/0.34 mg/ml nitro blue tetrazolium chloride/0.24 mg/ml levamisole (Sigma)].
RESULTS
hTERT Expression and Telomerase Activity in Tonsil and Peripheral-Blood B Cells.
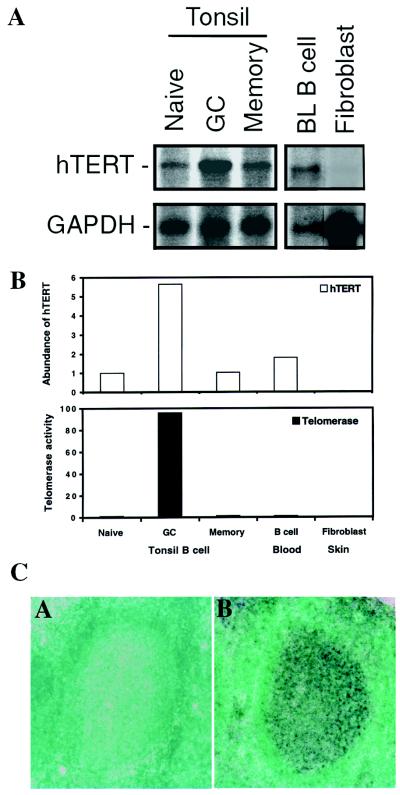
Tonsil B cells consist of three major subsets, which have been reported to express widely different levels of telomerase activity as well as hTER (24, 29, 34). To analyze hTERT expression in B cell subsets, cell lysate and mRNA were prepared from purified tonsil naïve, GC, and memory B cells, as well as from peripheral-blood resting B cells, and analyzed by TRAP assay and Northern hybridization. Telomerase activity was detected at high levels in GC B cells and was low or undetectable in naïve and memory tonsil B cells and peripheral-blood resting B cells (Fig. 1B). Interestingly, hTERT mRNA was detected in all subsets, and the levels of hTERT mRNA were 4- to 6-fold higher in GC B cells than in naïve, memory, or peripheral-blood resting B cells (Fig. 1 A and B). Although the high level of telomerase activity in GC B cells correlated with elevated hTERT mRNA in these cells, it was notable that hTERT was expressed also in tonsil naïve, memory, and peripheral-blood B cells that express low to undetectable levels of telomerase activity. The expression of high levels of hTERT mRNA in GC B cells was confirmed further by in situ hybridization of tonsil tissue with a ribonucleotide hTERT probe (Fig. 1C). High levels of hTERT mRNA were detected in GC B cells and much lower quantities were detected in the surrounding non-GC B cells. Thus, hTERT mRNA is present in all B cell subsets isolated from both tonsil and blood with the highest level in GC B cells. The presence of hTERT mRNA in peripheral-blood B cells that express little to no telomerase activity raises the question of whether hTERT mRNA is the predominant determinant of telomerase activity in lymphocytes, as has been reported for other cell types.
Figure 1.
hTERT mRNA expression and telomerase activity in tonsil and peripheral-blood B cells. (A) Representative Northern blots of hTERT expression in naïve, GC, and memory B cells isolated from human tonsils, resting B cells isolated from peripheral blood, and cultured fibroblasts from neonatal foreskin are shown. The blots were probed sequentially with hTERT and GAPDH probes. (B) The relative abundance of hTERT mRNA and telomerase activity in B cell subsets. The hTERT level from each cell subset shown in A was analyzed by phosphorimager and imagequant software. The relative abundance of hTERT mRNA was normalized on the basis of cell equivalents. The open bars represent the relative abundance of hTERT expression for each cell subset. The level of hTERT in naïve cells was arbitrarily set at 1. Telomerase activity was measured by a modified TRAP assay from ≈104 cells, and the results were normalized on the basis of cell equivalents. The black bars represent the relative levels of telomerase activity. The level of telomerase in naïve cells was arbitrarily set at 1. (C) hTERT mRNA expression in tonsil section by in situ hybridization. The tonsil sections were probed with sense (C, A) and antisense probe (C, B). Strong expression of hTERT is observed in GC cells but not in surrounding naïve and memory B cells.
hTERT Expression in Thymocytes, Tonsil T Cells, and Blood T Cells.
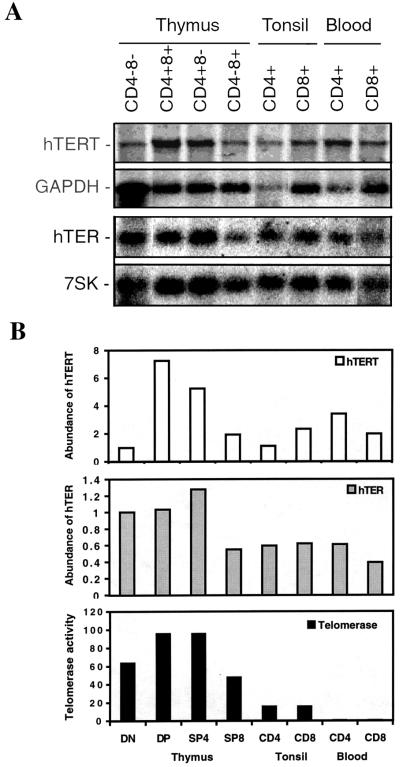
Telomerase activity also is regulated tightly during T cell lineage development and activation (23, 25–28). To determine the relationship between telomerase activity and hTERT expression during these processes, we analyzed T cell subsets isolated from thymus, tonsil, and peripheral blood. In agreement with previously reported findings, levels of telomerase activity were high in thymocytes, intermediate in tonsil T cells, and generally undetectable in peripheral-blood T cells isolated from the majority of normal adults (Fig. 2B). Low levels of telomerase activity were detected more frequently in peripheral-blood T cells isolated from cord blood and from children under age 10 (data not shown). hTER was present in all subsets of T cells and was 1.5- to 2-fold higher in thymocytes than in peripheral-blood T cells (Fig. 2 A and B). hTERT mRNA was detected in all subsets of T cells regardless of the levels of telomerase activity in these subsets (Fig. 2 A and B). In parallel with high levels of telomerase activity and hTER transcripts, high levels of hTERT mRNA were found in DP (CD4+8+) and SP (CD4+8−) CD4 thymocytes (Fig. 2B). However, despite an ≈64-fold higher level of telomerase activity in DN (CD4−8−) and single positive (CD4−8+) CD8 thymocytes (high telomerase populations) relative to telomerase in peripheral-blood CD4+ and CD8+ T cells (low telomerase populations), the levels of hTERT mRNA in peripheral-blood T cells were as high as or higher than levels in thymus DN and SP CD8 cells (Fig. 2 A and B). Thus, in contrast to previously reported findings that hTERT expression is correlated closely with telomerase activity in fibroblasts and other nonlymphoid normal somatic cells, we found that there is not a strict correlation between telomerase activity and hTERT mRNA in T cell subsets. Furthermore, the presence of hTER and hTERT in resting adult peripheral-blood T cells is not always sufficient to constitute detectable telomerase activity.
Figure 2.
hTERT mRNA expression and telomerase activity in normal human T cells. (A) Representative Northern blot of hTERT and hTER expression in thymocytes, tonsil T cells, and peripheral-blood T cells is shown. The blots were probed sequentially with either hTERT and GAPDH or hTER and 7SK probes. (B) Comparison of relative abundance of hTERT and hTER expression and telomerase activity. The levels of hTERT and hTER from each cell subset were determined by imagequant software. The relative abundance of hTERT mRNA and hTER were calculated by normalizing on the basis of cell equivalents. The bars represent relative abundance of hTERT (open bars) and hTER (gray bars) transcripts for each cell subset. The level of hTERT and hTER in double-negative (DN) thymocyte was arbitrarily set at 1. Telomerase activity was measured by a modified TRAP assay as described (33). The black bars represent the relative telomerase activity. The level of telomerase enzymatic activity in the CD4−CD8− subset was arbitrarily set at 1. DP, double-positive; SP, single-positive.
The presence of hTERT and hTER transcripts in resting lymphocytes with undetectable levels of telomerase activity suggested the possibility that telomerase activity might be repressed by an inhibitory mechanism in these cells. To test for factor(s) in peripheral-blood resting lymphocytes that might actively inhibit telomerase, we measured telomerase activity from cell lysates of thymocytes or GC B cells that had been mixed with lysate from telomerase-negative peripheral-blood resting T or B lymphocytes or with control lysis buffer. No obvious inhibition of telomerase activity was detected in these mixtures. A typical result is shown in Fig. 3.
Figure 3.
Absence of detectable telomerase inhibitory effect in mixtures of cell lysates from telomerase-negative and telomerase-positive cells. Cell lysate from telomerase-positive GC B cells was mixed with either cell lysis buffer or cell lysates from telomerase-negative blood CD8+ T cells at ratios of 1:1 or 1:3, respectively. The specificity of the assay was controlled by RNase treatment of cell lysates and by inclusion of a 36-bp internal control for PCR amplification. GC, GC B cells; B, peripheral-blood resting CD8+ T cells; LB, cell lysis buffer; IC, internal control.
hTERT Expression in CD4+ T Cells Activated in Vitro.
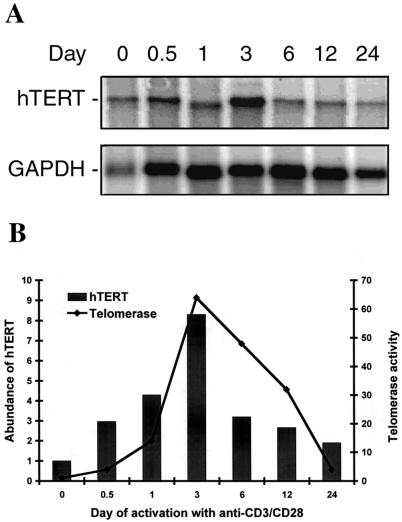
Activation of peripheral-blood CD4+ T cells through the engagement of T cell receptor and costimulatory receptor in vitro results in a significant induction of telomerase activity (23, 25–28). In agreement with previous findings, telomerase activity was detectable as early as 1 day after stimulation with anti-CD3 plus anti-CD28- mAbs conjugated magnetic beads, peaked between day 3 and day 6 (≈96-fold increase), and declined to basal levels between day 24 and day 30 (Fig. 4B). hTERT mRNA was increased at 12 h after stimulation, peaked at around day 3 with an ≈8-fold increase of hTERT mRNA, and decreased progressively from day 6 to day 24 after stimulation (Fig. 4 A and B). Thus, the kinetics of hTERT induction generally is correlated with the induction of telomerase activity in CD4+ T cells activated in vitro.
Figure 4.
hTERT mRNA expression in peripheral-blood CD4+ T cells during in vitro activation. (A) Representative Northern blot of hTERT mRNA expression in peripheral-blood CD4+ T cells after in vitro stimulation with anti-CD3 and anti-CD28 antibodies. Peripheral-blood CD4+ T cells were isolated from healthy donors, stimulated with anti-CD3 plus anti-CD28, and cultured for various time intervals up to 24 days. The Northern blots were probed sequentially with hTERT and GAPDH probes. (B) Comparison of relative abundance of hTERT mRNA and telomerase activity. The hTERT mRNA levels at each time point were determined by phosphorimager and imagequant software. The relative abundance of hTERT mRNA was normalized to the number of cell equivalents used to isolate mRNA. The bars represent relative abundance of hTERT expression at each given time point; the level before the stimulation (time 0) was set at 1. Telomerase activity was measured by a modified TRAP assay as described (33). The line represents the relative telomerase activity; the level before stimulation was set at 1.
No Alternate Splicing of hTERT Was Detected in Resting B and T Cells.
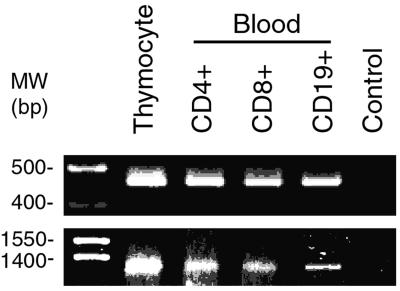
Recently, it has been reported that variants of hTERT transcripts are expressed in human fetal tissues and tumor cell lines (11, 35). Because these variant transcripts either resulted in deletion of critical reverse-transcriptase motifs of hTERT (e.g., α-splice deletion results in a loss of 36 bp in reverse-transcriptase motif A) or caused a frameshift mutation (e.g., β-splice deletion results in a loss of 182 bp and a reading-frame truncation; ref. 11), it was hypothesized that these variant transcripts may serve as a mechanism for regulating telomerase activity. The presence of such splicing variants might contribute to a lack of correlation between levels of hTERT mRNA and telomerase activity. Because these variants might not be resolved adequately by conventional Northern hybridization, we applied RT-PCR with two sets of oligonucleotides covering the regions of reported deletions to characterize further hTERT mRNA detected in normal peripheral-blood B and T lymphocytes. Analysis of mRNA from thymocytes and peripheral-blood lymphocytes identified no evidence for the existence of splicing transcripts of hTERT that delete α- and/or β-sites between A and B motifs or other detectable sites in the reverse-transcriptase region (Fig. 5).
Figure 5.
RT-PCR analysis of hTERT expression in B and T lymphocytes. (Upper) Representative result of the RT-PCR analysis of B and T lymphocyte hTERT expression with primers hTERT-2164S and hTERT-2620A (35). The amplified products were separated on a 2.0% agarose gel and visualized by ethidium bromide staining. The α- and β-deletions of hTERT result in a loss of 36 bp and 182 bp, respectively. (Lower) Representative result of RT-PCR analysis of B and T lymphocyte hTERT expression with primers hTERT-1680S and hTERT-2974A, which cover the entire region of reverse transcriptase. The amplified products were separated on a 1.2% agarose gel and visualized by ethidium bromide staining. The DNA molecular weight markers were run alongside the samples and indicated at the left.
DISCUSSION
Regulation of telomerase activity is a highly controlled process as evidenced by its selective expression in germ-line cells, in limited types of normal somatic cells, and in a majority of tumor cells. The results presented in this paper provide insights into this regulated process. First, expression of hTERT is highly regulated during human lymphocyte lineage development, differentiation, and activation. However, in contrast to the recent proposal that transcription of hTERT determines telomerase activity in normal somatic cells, human lymphocytes express hTERT independently of the presence, absence, or quantitative level of detectable telomerase activity. Together, these results suggest that regulation of telomerase activity is cell-type-specific and is controlled at multiple levels, including both transcriptional and posttranscriptional events.
In the present study and in previous studies (23, 24, 31), we have analyzed telomerase activity and expression of hTER and hTERT in lymphocytes in several systems: (i) B cell differentiation in tonsil—from naïve to GC to memory B cells; (ii) T cell lineage development—thymocyte subsets (DN, DP, CD4 SP, and CD8 SP), tonsil T cell subsets, and peripheral-blood T cell subsets; and (iii) peripheral-blood CD4+ T cells in the course of in vitro stimulation. Telomerase activity and its two essential components, hTER and hTERT, are regulated during tonsil B cell differentiation, T cell intrathymic development, and activation of peripheral-blood CD4+ T cells. Parallel regulated expression of hTER and hTERT is found during tonsil B cell differentiation and peripheral-blood CD4+ T cell activation, and the abundance of hTER and hTERT generally correlates with the levels of telomerase activity in these cells. However, the high levels of telomerase activity detected in all subsets of thymocytes do not correlate well with the levels of hTERT mRNA during T cell lineage development. Although all subsets of thymocytes express much higher (48- to 96-fold greater) levels of telomerase activity than do resting peripheral-blood T cell subsets, hTERT mRNA levels are increased only moderately in DP and SP CD4 subsets and are not increased at all in DN and SP CD8 subsets, compared with the hTERT mRNA levels in resting peripheral-blood T cells. It is possible that CD8 SP thymocytes have decreased or terminated transcription of hTERT in the course of differentiation, while the previously translated telomerase protein remains active. However, it is more difficult to explain the low levels of hTERT in DN thymocytes on the basis of a similar argument. Taken together, these data suggest that the regulation of telomerase activity during intrathymic T cell development is controlled at levels beyond transcription of hTER and hTERT. Further study is needed to delineate the mechanism underlying this regulation and particularly to develop reagents/assays for analysis of the hTERT protein and its interaction with other telomerase components.
Two molecular mechanisms have been described that may contribute to the regulation of telomerase activity in humans: transcriptional regulation of hTER and hTERT and posttranscriptional alternative splicing of hTERT (8, 9, 11, 35). It has been reported that regulation of telomerase activity in human fibroblasts occurs at the level of transcription of hTERT. There are two lines of evidence that support this idea: first, it has been observed that there is no detectable hTERT mRNA in human fibroblasts, which correlates with a lack of telomerase activity in these cells; and second, it has been shown that reconstitution of telomerase activity could be achieved in human fibroblasts by introducing exogenous hTERT expression. Ulnaer et al. (35) recently reported another mode of regulation of telomerase by alternatively spliced forms of hTERT transcripts during human fetal kidney development. Both telomerase activity and full-length hTERT transcripts were detected in fetal kidney at an early gestational phase. However, after ≈15 weeks of gestation, telomerase activity disappeared, and, although hTERT mRNA persisted at similar levels, only a β-deletion transcript of hTERT, resulting in a truncated protein that lacks several essential motifs of reverse transcriptase for enzymatic activity, was expressed. The data presented here indicate that additional modes of telomerase regulation act at as yet undefined levels, which may include translational and/or posttranslational mechanisms. The failure to identify telomerase inhibitor(s) from telomerase-negative cells by mixing of cell lysates does not rule out the possibility that inhibitory factor(s) are active in vivo but not detectable under these conditions. The question of whether telomerase activity is controlled by hTERT protein and hTER alone and/or by additional interacting proteins remains to be answered.
It seems that the regulation of telomerase activity may be complex. It is conceivable that controlling telomerase gene expression at the transcriptional level offers an economic and effective mode of regulation for cells that do not require rapidly available telomerase activity. In contrast, for cells such as lymphocytes that may require a mechanism for rapid induction and regulation of telomerase activity, there could be an advantage to maintaining basal levels of hTER transcript and hTERT transcripts (or protein), with posttranscriptional regulation of enzymatic activity. The understandings of telomerase regulation will not only satisfy the curiosity surrounding this complex and important enzyme but will also provide a foundation for the pursuit of telomerase-based therapeutic intervention.
Acknowledgments
We thank Stephen Shaw for providing valuable antibodies; Dan Longo, Karen Hathcock, and Shannon Murray for critical reading of the manuscript; and the anonymous reviewers for comments and suggestions. We thank Holy Cross Hospital, Fairfax County Hospital, and the National Institutes of Health blood bank for assistance in obtaining tonsil, thymus, and blood. This work was supported in part by Army contract DAMD17-93-V-3004, by the Naval Medical Research and Development Command, and by the Henry M. Jackson Foundation.
ABBREVIATIONS
- hTER
human telomerase RNA template
- hTERT
human telomerase reverse transcriptase
- GC
germinal center
- TRAP
telomeric repeat amplification protocol
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- DN
double-negative
- DP
double-positive
- SP
single-positive
References
- 1.Blackburn E H. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T M, Cech T R. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Science. 1998;279:334–335. doi: 10.1126/science.279.5349.334. [DOI] [PubMed] [Google Scholar]
- 4.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 5.Greider C W. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng N P, Hathcock K S, Hodes R J. Immunity. 1998;9:151–157. doi: 10.1016/s1074-7613(00)80597-x. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C P, Adams R R, Chang E, Allsopp R C, Yu J. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 9.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 10.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 12.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 13.Beattie T L, Zhou W, Robinson M O, Harrington L. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 14.Kolquist K A, Ellisen L W, Counter C M, Meyerson M, Tan L K, Weinberg R A, Haber D A, Gerald W L. Nat Genet. 1998;19:182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan S, Eppenberger U, Mueller H, Shinkai Y, Narayanan R. Cancer Res. 1998;58:622–625. [PubMed] [Google Scholar]
- 16.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 17.Vaziri H, Benchimol S. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 18.Counter C M, Meyerson M, Eaton E N, Ellisen L W, Caddle S D, Haber D A, Weinberg R A. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 19.Wen J, Cong Y S, Bacchetti S. Hum Mol Genet. 1998;7:1137–1141. doi: 10.1093/hmg/7.7.1137. [DOI] [PubMed] [Google Scholar]
- 20.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 21.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sedivy J M. Proc Natl Acad Sci USA. 1998;95:9078–9081. doi: 10.1073/pnas.95.16.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng N, Levine B L, June C H, Hodes R J. J Exp Med. 1996;183:2471–2479. doi: 10.1084/jem.183.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng N P, Granger L, Hodes R J. Proc Natl Acad Sci USA. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiyama K, Hirai Y, Kyoizumi S, Akiyama M, Hiyama E, Piatyszek M A, Shay J W, Ishioka S, Yamakido M. J Immunol. 1995;155:3711–3715. [PubMed] [Google Scholar]
- 26.Buchkovich K J, Greider C W. Mol Biol Cell. 1996;7:1443–1454. doi: 10.1091/mbc.7.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi H, Sakaguchi N. Biochem Biophys Res Commun. 1996;219:649–655. doi: 10.1006/bbrc.1996.0288. [DOI] [PubMed] [Google Scholar]
- 28.Bodnar A G, Kim N W, Effros R B, Chiu C P. Exp Cell Res. 1996;228:58–64. doi: 10.1006/excr.1996.0299. [DOI] [PubMed] [Google Scholar]
- 29.Hu B T, Lee S C, Marin E, Ryan D H, Insel R A. J Immunol. 1997;159:1068–1071. [PubMed] [Google Scholar]
- 30.Igarashi H, Sakaguchi N. Blood. 1997;89:1299–1307. [PubMed] [Google Scholar]
- 31.Weng N, Levine B L, June C H, Hodes R J. J Immunol. 1997;158:3215–3220. [PubMed] [Google Scholar]
- 32.Levine B L, Bernstein W B, Connors M, Craighead N, Lindsten T, Thompson C B, June C H. J Immunol. 1997;159:5921–5930. [PubMed] [Google Scholar]
- 33.Kim N W, Wu F. Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norrback K F, Dahlenborg K, Carlsson R, Roos G. Blood. 1996;88:222–229. [PubMed] [Google Scholar]
- 35.Ulaner G A, Hu J F, Vu T H, Giudice L C, Hoffman A R. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]