Abstract
On the basis of the B lymphotropic Epstein–Barr virus (EBV), we have constructed a virus-free packaging cell line that allows encapsidation of plasmids into herpesvirus particles. This cell line harbors an EBV mutant whose packaging signals have been deleted. The gene vectors, which can encompass very large, contiguous pieces of foreign DNA, carry all cis-acting elements involved in amplification and encapsidation into virus-like particles as well as those essential for extrachromosomal maintenance in the recipient cell. Although this first-generation packaging cell line suffers from unwanted recombination between the helper virus genome and gene vector DNAs, this approach opens the way to delivery and stable maintenance of any transgene in human B cells.
Viruses can efficiently introduce foreign genetic information into eukaryotic cells, sometimes even without adverse affects. For this reason, most of the gene vectors currently in use are of viral origin. These vectors usually consist of a viral origin of replication and additional cis-acting elements needed for encapsidation into virus-like particles. Among viral vectors, those based on Epstein–Barr virus (EBV), a member of the herpes virus family, are especially promising because EBV has a number of unique, advantageous features (1). They include efficient gene delivery into B cells that cannot be transduced easily by any other means, and suitable vectors can accommodate up to 150 kb of foreign DNA. Consequently, complete genomic loci encompassing all their regulatory regions can be transferred into target cells. Moreover, these vectors are maintained extrachromosomally, as is wild-type EBV, avoiding the problems that follow random integration in the host cell chromosome. Finally, the ability of EBV to latently infect its target cells sustains a long-term persistence of the introduced genes.
With the exception of adenoviruses, no packaging system free of wild-type virus is available for large DNA viruses. Vectors derived from herpes viruses or other large DNA viruses (which are not merely viruses with a hitchhiking gene) require a helper cell line for vector encapsidation to provide the necessary factors in trans. In the case of large DNA viruses, the packaging cells are infected with wild-type virus or a conditionally mutant virus that complements for those functions the gene transfer vector lacks (1). As a consequence, both the encapsidated vectors and the helper virus are released, which is a major drawback. To overcome this intrinsic problem we have constructed a first-generation helper cell line for encapsidation of EBV-derived vectors in which supernatants stay free of helper virus.
METHODS
Cells.
293 is a transformed epithelial cell line whose differentiation stage is not fully characterized (2). Raji is an EBV-positive human Burkitt’s cell line (3). B95.8 is a lymphoblastoid cell line obtained by infection of marmoset monkey peripheral blood leukocytes with EBV (4). B lymphocytes from peripheral blood buffy coats, adenoids, and tonsils were purified on a Ficoll cushion after T cell rosetting by using sheep erythrocytes, as described (5). All cell lines were grown in RPMI 1640 medium/10% FCS (Life Technologies, Paisley, Scotland).
Recombinant DNA Plasmids.
pMBO131 is an F factor-based prokaryotic replicon that carries the F factor origin of replication, the chloramphenicol-resistance gene, and partitioning genes (6). The mini-EBV consists of all EBV latent genes cloned onto an F plasmid (7). The EBV reporter plasmids consist of the latent and lytic replication origins (oriP, oriLyt), the green fluorescent protein (GFP) gene, and the hygromycin-resistance gene, which were cloned onto a pBR322 plasmid backbone (p2122). The p2124 plasmid additionally contains the terminal repeats. p588 is identical to p2124, except that it lacks the GFP gene (8). The MDR1 gene was cloned in front of the simian virus 40 early promoter and introduced into a plasmid that encodes for the EBNA1 gene, both EBV replication origins and the terminal repeat (TR), to give p2194. All plasmids were propagated in Escherichia coli strain DH5α. Allelic exchange was performed in E. coli strain BJ5183 (9).
DNA Transfections.
DNA transfections into 293 cells were performed by using Lipofectamine in Optimem minimal medium (Life Technologies).
Infections.
Raji cells were infected overnight with filtered (0.45-μm pore size) supernatants from TR−2/293 cells in which the lytic cycle had been induced by transfecting an expression plasmid encoding BZLF1 (10). For evaluation of the virus titers, 1 × 105 Raji cells were infected with 0.5 ml supernatant. When selection was required, 1 × 106 cells were incubated with 1 ml supernatant.
Primary B cells (2 × 107) were infected with 5 ml of filtered (0.45-μm pore size) supernatants from TR−2/293 cells into which the mini-EBV and an expression plasmid encoding BZLF1 had been transfected. B cells then were plated in 96-well cluster plates (2 × 105/well) and fed once a week.
Hygromycin and Colchicine Selection.
Selection of Raji cells in 96-well cluster plates with 100 μg/ml of hygromycin (Calbiochem) was done as described (11). Colchicine selection was done at a concentration of 3 ng/ml of culture medium. Colchicine-resistant clones were propagated under the same conditions.
Southern Blot Analysis.
DNA preparation and hybridization were performed as described (11).
Immunostaining.
Detection of viral capsid antigens in lytically induced TR−2/293 cells was done as described previously (11).
RESULTS
Cloning of a TR-Negative EBV Mutant.
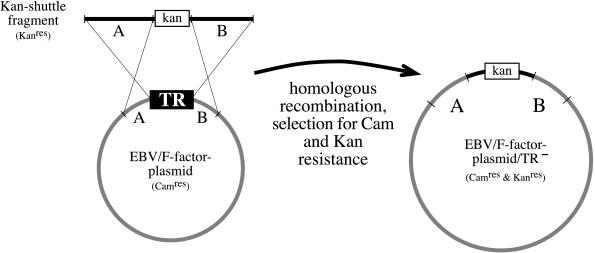
Incorporation of recombinant DNA into an EBV particle depends on two cis-acting elements, the lytic origin of DNA replication and the TRs (10, 12). The TRs are located at both ends of the EBV DNA in its linear conformation and constitute packaging-signal sequences absolutely required for encapsidation (8, 12, 13). A further constraint is the size of the DNA to be encapsidated. The lytic replication of herpes virus DNA follows, at least partially, a “rolling circle” mechanism, such that integral numbers of vector copies can be encapsidated only if their total size is about 165 kb (14). As a consequence, an EBV genome with its TRs deleted cannot be incorporated into EBV particles but is expected to provide the factors in trans required for encapsidation. We recently have cloned the entire EBV genome in E. coli with the aid of an F plasmid (11). It also carries the genes for hygromycin resistance in eukaryotic cells, chloramphenicol resistance in E. coli, and the gene that encodes the enhanced GFP (eGFP). In E. coli, the cloned EBV genome is amenable to any genetic modification, including generation of mutant EBV genomes, which are crippled for otherwise indispensable viral functions, i.e., packaging-signal sequences. To delete the TRs, they were replaced in E. coli for the kanamycin-resistance gene by targeted allelic exchange (Fig. 1). A recA-positive, recBC-negative E. coli strain (BJ5183) (9) harboring the complete EBV genome as an F factor plasmid was transformed with a linear DNA fragment encompassing the kanamycin-resistance gene flanked by those EBV sequences that constitute the left and right bordering regions adjacent to the TRs. Thus, homologous recombination events between the TRs’ bordering regions and the EBV/F factor DNA result in a single plasmid that encodes resistance against kanamycin and chloramphenicol and lacks the TRs. Plasmid DNAs prepared from bacterial clones were analyzed by Southern blot analysis by using a TR-specific probe confirming the absence of the TRs (data not shown). Further examination with numerous restriction enzymes revealed a perfect recombination between the parental EBV/F factor plasmid and the kanamycin-resistance gene, which resulted in deletion of the TRs (Fig. 2).
Figure 1.
Generation of B95.8/F factor DNA mutant lacking its packaging signals. The TRs were replaced by the kanamycin-resistance gene via homologous recombination. The sequences that flank the TR in the wild-type virus DNA were cloned to the left and right of the kanamycin-resistance gene to provide regions of homology that permit homologous recombination in E. coli. This plasmid then was linearized and introduced into the BJ5183 bacterial strain, and cells carrying recombinant molecules were selected for resistance against kanamycin and chloramphenicol.
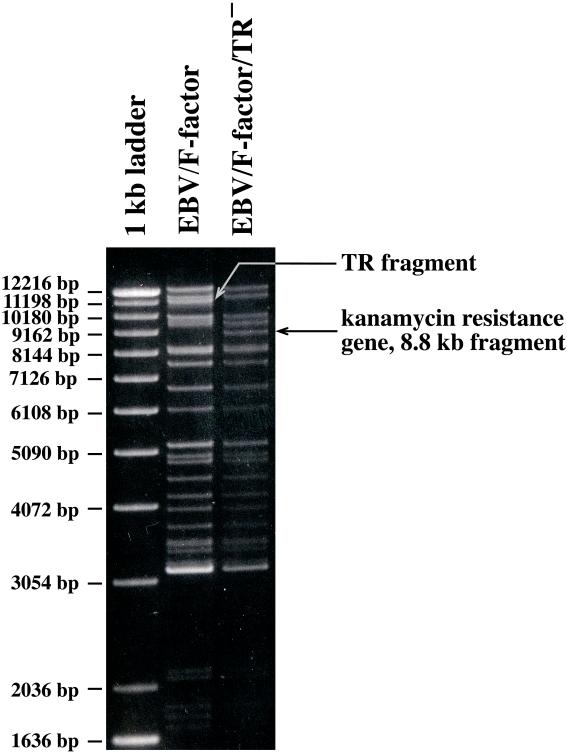
Figure 2.
Restriction fragment analysis of TR− mutant DNA in comparison with the wild-type B95.8/F factor DNA. DNA was purified from a chloramphenicol- and kanamycin-resistant E. coli clone and digested with the BamHI restriction enzyme. The restriction pattern of the modified B95.8/F factor DNA was compared with the one obtained with wild-type B95.8/F factor DNA. Both restriction fragment patterns were identical with one exception. A new fragment was generated by the exchange of the BamHI fragment that carries the TRs (disappearance of the 11.2-kb fragment in EBV/F factor) against the kanamycin-resistance gene (appearance of an 8.8-kb fragment in EBV/F factor/TR− DNA).
Generation of an EBV Helper Cell Line That Encapsidates TR-Positive Reporter Plasmids.
Experiments with the parental, wild-type B95.8/F factor plasmid had shown previously the ability of 293 cells to maintain the latent status of EBV and to support the EBV lytic cycle after transfection with a BZLF1 expression plasmid (11, 15). To establish an EBV helper cell line, the TR-negative EBV DNA was prepared from E. coli and transfected into 293 cells. After hygromycin selection, 12 293 cell clones grew out that carried the TR-less EBV/F factor plasmid as revealed by Southern blot analysis (data not shown). Cell clones were transfected with a BZLF1 expression plasmid together with the EBV reporter plasmid p2124 that carries the TRs, the latent and lytic EBV replication origins, the hygromycin-resistance gene, and the GFP gene (Fig. 3). Three days after transfection, cells were analyzed for viral capsid antigen (VCA) expression to monitor the induction of the lytic cycle, and Raji cells were infected with supernatants obtained from these cultures. All cell clones showed expression of VCA (Fig. 4). Incubation of Raji cells with the supernatants resulted in successful infection as indicated by green fluorescence of the Raji cells after exposure to UV light (Fig. 4). Moreover, addition of hygromycin to the culture medium gave rise to resistant Raji clones in all seeded wells after 5 weeks of selection, confirming the data provided by GFP fluorescence shortly after infection (Table 1). Control experiments with a reporter plasmid that differed from p2124 only by the absence of TRs (p2122, Fig. 3) yielded neither green cells when exposed to UV light nor hygromycin-resistant clones after selection. For unknown reasons, one of the 293 cell clones, TR−2/293, proved to be much more efficient in producing infectious particles and was used in further experiments. We estimated the titer of encapsidated p2124 vectors in serial dilutions to be at about 6 × 103 infectious particles per ml by counting the number of GFP-positive Raji cells after infection with the TR-positive EBV reporter plasmids, which is one to two orders of magnitude lower than what is observed with the 293 cell line stably transfected with the wild-type B95.8/F factor plasmid (11).
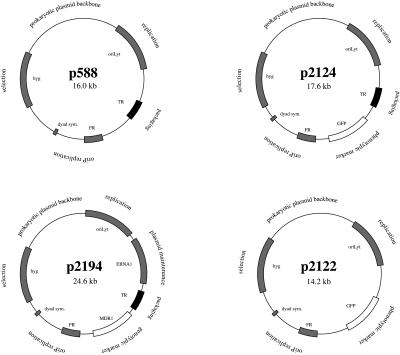
Figure 3.
Schematic representation of the various plasmids used in this study. All plasmids are derived from the p588 plasmid (8) that consists of the two replication origins, oriP and oriLyt, the TR packaging-signal sequences, the EBNA1 gene, and the hygromycin-resistance gene (hyg). The other constructs carry a phenotypic marker (GFP in p2124 and MDR1 in p2194) or lack the TR packaging-signal sequences, as in p2122.
Figure 4.
(Top) GFP production in cells incubated with supernatants from induced 293 cell lines carrying the TR-less B95.8/F factor DNA. Supernatant (0.5 ml) from p2124 encapsidated in the TR−2/293 cell line was mixed with 105 Raji cells in 0.5 ml culture medium. About 3% of the cells were GFP-positive, indicating a virus titer of at least 6 × 103 infectious viruses per ml. GFP fluorescence was investigated 48 h after infection. (Middle) Corresponding phase-contrast light microscopy. (Bottom) Production of the VCA in TR−2/293 after induction of the lytic cycle. Fixed cells were incubated with a mAb against VCA and a second anti-mouse antibody coupled to the Cy5 fluorochrome. Stained cells were visualized in UV light.
Table 1.
Frequencies of gene transfer and recombinations between helper virus and gene vectors
| Type of experiment
|
Gene transfer, no. of positive wells/wells plated | Recombination frequency (accidental GFP transfer), no. of positive wells/wells plated | |
|---|---|---|---|
| Cells infected | Supernatant with packaged plasmid | ||
| Raji | p2124 | 288/288 (100%)*† | ND |
| Raji | p2194 | 72/96 (75%)†‡ | ND |
| Raji | p588 | 96/96 (100%)* | 32/96 (33%) |
| Raji | p1495 | 192/192 (100%)* | 5/192 (2.6%) |
| Primary B cells | p1495 | 96/96 (100%)§ | 0/96 (0%) |
ND, not done.
Proliferation of Raji cells under selection with 200 μg/ml of hygromycin.
GFP-positive Raji cells.
Proliferation of Raji cells under selection with 3 ng/ml of colchicine.
Proliferation of immortalized B cell clones.
EBV Helper Cell Line Encapsidates Mini-EBV Plasmids Efficiently.
Mini-EBVs are recombinant E. coli F factor plasmids that carry all EBV genes necessary for in vitro B cell immortalization but lack most genes expressed during the lytic phase of EBV’s life cycle (7). As a consequence, the encapsidation of mini-EBVs depends on a helper virus or on a cell line that provides all necessary viral proteins in trans. We tested the cell line TR−2/293 for packaging of mini-EBVs. TR−2/293 was transiently cotransfected with mini-EBV plasmid DNA and the BZLF1 expression plasmid (7). After 3 days, supernatant was harvested, filtered, and used to infect primary B lymphocytes as described (8). Immortalized B cells grew out in all wells of 96-well cluster plates after 3 weeks (Table 1). Sixty of these B cell lines were analyzed further. DNA was extracted, cleaved with appropriate restriction enzymes, blotted according to Southern, and hybridized with the F factor DNA as a probe (Fig. 5). Because restriction enzyme cleavage yields characteristic F plasmid fragments distinct for mini-EBV or EBV/F factor genomes (16.7 or 10.4 kb), their DNAs can be readily distinguished. All cell lines were found to carry the characteristic F factor fragment derived from the mini-EBV plasmid only, whereas an infection with the helper virus could not be assessed (Fig. 5). In line with these results, none of the immortalized B cell clones was found to produce GFP, which can be provided by the TR-less B95.8/F factor genome only (Table 1).
Figure 5.
Southern blot analysis of immortalized B cell lines that were established with virus stocks derived from the TR−2/293 packaging cell line after transfection with mini-EBV plasmid DNA p1495.4 (24). Total DNA from the immortalized B cell lines was digested with BamHI, separated by electrophoresis, blotted, and hybridized with a probe specific for the pMBO131 plasmid (6). DNAs extracted from the wild-type B95.8 cell line served as a negative control. The positive control consisted of 10 μg of a 293 cell clone that stably carries the B95.8/F factor plasmid (11). The number of copies of this plasmid was estimated to lie between one and two copies per cell. All tested immortalized B cell lines were found to carry the intact mini-EBV F-factor DNA and no helper virus DNA.
Infection of CD21-Positive Cells with EBV Vectors That Contain the MDR1 Gene Confers Resistance Against Colchicine.
Genes mediating resistance to cytotoxic anticancer drugs can be used to dominantly select gene-modified hematopoietic cells in vitro and in vivo (16). The human MDR1 gene encodes a membrane-located drug effluent pump that decreases the intracellular concentration of a number of important antitumor drugs (17). Colchicine is a highly toxic cytostatic drug that blocks cells during mitosis, but expression of the MDR1 gene provides resistance against it (17, 18). To test whether MDR1 can be transferred efficiently into B cells, we cloned the MDR1 gene under the control of a constitutively active promoter onto an EBV-derived vector plasmid that includes all elements necessary for packaging and maintenance in the infected cell (p2194) (Fig. 3). This plasmid was transfected into the 293 TR−2 helper cell line, and supernatants were used to infect Raji cells, which were selected with colchicine in the culture medium. Two to 3 weeks after infection, colchicine-resistant cell clones grew in the infected cell population only (Table 1), indicating a successful gene transfer and a functional MDR1 gene.
Frequency of Recombination Between Helper Virus Genome and Gene Vector DNA.
One major problem by using viral packaging cell lines is recombination between helper virus DNA and the gene vector to be encapsidated. To estimate the frequency of recombination, we used a packageable EBV vector plasmid that, in contrast to the helper virus genome, lacks the GFP gene but carries the hygromycin-resistance gene as a selectable marker in the recipient cells (p588, Fig. 3). Accidental transfer of the GFP gene onto the reporter plasmid after recombination with the helper virus can be observed readily in the infected cells, providing a simple and accurate means to evaluate the number of recombinants. Hygromycin resistance, on the other hand, provides a reliable measure of the frequency of gene transfer via infection. Small gene vector plasmids, such as p588, that contain the origins of DNA replication of EBV and the TRs showed a high percentage of recombination, with roughly one-third of the clones GFP-positive after selection with hygromycin (Table 1). Using the much larger mini-EBV p1495 (about 83 kb in size) as reporter plasmid for packaging and recombination frequency, only about 3% of the hygromycin-resistant Raji cell clones were found to harbor the GFP gene after recombination with the helper virus (Table 1).
DISCUSSION
Here, we describe a novel packaging cell line that is suitable for encapsidation of EBV-based gene transfer vectors. This cell line carries an EBV mutant that is devoid of its TRs, which prevents packaging of its DNA into virions. The EBV mutant genome supports the lytic phase of EBV’s life cycle and is capable of encapsidating foreign DNA that carries the TR-packaging signals. This novel helper cell line encapsidates mini-EBV plasmids with an efficiency roughly 10 times higher than the previously used HH514 cell line (7, 19). Moreover, HH514 supernatants contain a high proportion of helper virus, as documented previously (7). Attempts to package EBV vectors devoid of TRs did not produce any infectious particles, confirming that the TRs are absolutely required for encapsidation. Exogenous genes present on EBV-derived gene vector plasmid were expressed readily after infection of B cells, as exemplified by the MDR1 gene that also can be used as an in vivo selectable marker. The MDR protein is a paradigm for other dominant selection markers applicable to control expansion and elimination of transduced cells in vivo. Because of EBV’s host range, the MDR1 gene is transferred efficiently to B cells, but a global, protective effect against cytopenia induced by cytostatic drugs can be achieved only by infecting hematopoietic stem cells. It remains to be shown that they can be targeted with EBV-derived vectors with an altered cell specificity.
Recombination between helper virus and reporter plasmid is a recurrent pitfall of all currently available gene therapy systems that make use of viral vectors. The sequence homology between EBV-derived vectors and the helper virus, e.g., the lytic and the latent origins or DNA replication, constitute targets for homologous recombination. Herpes virus genetics relied on such recombination events to generate viral mutants, although newer methods might supersede this technique shortly (8, 11, 20–22). The rate of recombination between the vector plasmid and the helper virus DNAs varied drastically with the size of the plasmid to be encapsidated. Comparatively small plasmids showed a high rate of recombination (nearly 30%) whereas in experiments by using large plasmids such as mini-EBVs only 3% of the infected cells were found to carry recombined vector DNA (Table 1). This reduction in the recombination rate probably reflects the fact that simple cointegrates between the helper virus and a large gene transfer vector such as the mini-EBV plasmids vastly exceed the maximum size of the DNA that can be incorporated into EBV capsids. Thus, only cointegrates that have undergone several consecutive recombination events can give rise to DNA that fulfills the requirements for encapsidation. The probability that a fully functional and replication-competent viral genome will arise after recombination is low even in this rather primitive system. This provides an explanation for the observation that none of the immortalized cells that grew out after infection of primary B cells with encapsidated mini-EBV was found to carry the helper virus genome.
Vectors derived from herpes viruses provide one of the most attractive systems for gene delivery because they can accommodate large DNA inserts (1). A number of very important therapeutic genes are too large to be cloned in adenoviruses or retroviruses, and the incorporation of the gene’s own regulatory elements would be impossible in most cases. As an example, expression of the factor VIII gene is hampered severely in the absence of some of its introns (23). Herpes virus vectors also may prove to be extremely helpful in cases in which promoter and enhancer structures are indispensable for accurate, differentiation- and cell-specific gene expression.
In conclusion, this first-generation packaging cell line can be used to deliver any gene of interest into primary or established B cells in vitro, as exemplified here with the MDR1 and GFP genes. It will facilitate further the use of the mini-EBV system for genetic analysis of genes known to be involved in B cell immortalization by EBV. To adopt this system to gene therapy standards, further steps are necessary to optimize the packaging of the gene transfer vectors and to minimize DNA recombination.
Acknowledgments
Part of this work was supported by Public Health Service Grant CA70723, Grant Ha 1354/3 from the Deutsche Forschungsgemeinschaft, and Grant 10–2016-Ze from the Deutsche Krebshilfe.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- TR
terminal repeat
- GFP
green fluorescent protein
- VCA
viral capsid antigen
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Glorioso J C, DeLuca N A, Fink D J. Annu Rev Microbiol. 1995;49:675–710. doi: 10.1146/annurev.mi.49.100195.003331. [DOI] [PubMed] [Google Scholar]
- 2.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 3.Pulvertaft R J V. Lancet. 1964;i:238–240. [Google Scholar]
- 4.Miller G, Shope T, Hermann L, Stitt D, Lipman M. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeidler R, Meissner P, Eissner G, Lazis S, Hammerschmidt W. Cancer Res. 1996;56:5610–5614. [PubMed] [Google Scholar]
- 6.O’Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 7.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerschmidt W, Sugden B. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Hammerschmidt W, Sugden B. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 11.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kintner C R, Sugden B. Cell. 1979;17:661–671. doi: 10.1016/0092-8674(79)90273-3. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann J, Hammerschmidt W. J Virol. 1995;69:3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloss T A, Sugden B. J Virol. 1994;68:8217–8222. doi: 10.1128/jvi.68.12.8217-8222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Countryman J, Miller G. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baum C, Stocking C, Wagener T, Eckert H, Ostertag W. In: Concepts in Gene Therapy. Strauss M, Barranger J, editors. Berlin: de Gruyter; 1997. pp. 233–266. [Google Scholar]
- 17.Gottesman M M, Pastan I. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 18.Baum C, Hegewisch-Becker S, Eckert H G, Stocking C, Ostertag W. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilger E, Kieser A, Baumann M, Hammerschmidt W. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiley J. Nature (London) 1980;285:333–335. doi: 10.1038/285333a0. [DOI] [PubMed] [Google Scholar]
- 21.Post L E, Roizman B. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 22.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuah M K, Vandendriessche T, Morgan R A. Hum Gene Ther. 1995;6:1363–1377. doi: 10.1089/hum.1995.6.11-1363. [DOI] [PubMed] [Google Scholar]
- 24.Kempkes B, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1995;92:5875–5879. doi: 10.1073/pnas.92.13.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]