Abstract
Genomic imprinting plays a fundamental role in cancer and some hereditary diseases, including Beckwith–Wiedemann syndrome (BWS), a disorder of prenatal overgrowth and predisposition to embryonal malignancies such as Wilms tumor. We have previously shown that the KVLQT1 gene on chromosomal band 11p15 is imprinted, with expression of the maternal allele, and that the maternal allele is disrupted in rare BWS patients with balanced germ-line chromosomal rearrangements. We now show that an antisense orientation transcript within KVLQT1, termed LIT1 (long QT intronic transcript 1) is expressed normally from the paternal allele, from which KVLQT1 transcription is silent, and that in the majority of patients with BWS, LIT1 is abnormally expressed from both the paternal and maternal alleles. Eight of sixteen informative BWS patients (50%) showed biallelic expression, i.e., loss of imprinting (LOI) of LIT1. Similarly, 21 of 36 (58%) BWS patients showed loss of maternal allele-specific methylation of a CpG island upstream of LIT1. Surprisingly, LOI of LIT1 was not linked to LOI of insulin-like growth factor II (IGF2), which was found in 2 of 10 (20%) BWS patients, even though LOI of IGF2 occurs frequently in Wilms and other tumors, and in some patients with BWS. Thus, LOI of LIT1 is the most common genetic alteration in BWS. We propose that 11p15 harbors two imprinted gene domains—a more centromeric domain including KVLQT1 and p57KIP2, alterations in which are more common in BWS, and a more telomeric domain including IGF2, alterations in which are more common in cancer.
Keywords: genomic imprinting, chromosomal domain, cancer, insulator
Genomic imprinting is an epigenetic modification of a gene or the chromosome on which it resides that leads to preferential expression of a specific parental allele of the gene. Imprinting is normally potentially reversible in that a maternal allele in one generation can become a paternal allele in the next. Abnormal imprinting is thought to play a role in several human diseases. In particular, loss of imprinting (LOI) is one of the most frequent genetic alterations in cancer, and by definition can involve both abnormal activation of the normally silent allele of a growth-promoting gene, such as insulin-like growth factor II (IGF2), or silencing of the normally expressed allele of a growth inhibitory gene, such as p57KIP2 (reviewed in ref. 1).
Beckwith–Wiedemann syndrome (BWS), which causes prenatal overgrowth and predisposition to cancer, has been a beacon for the investigation of genomic imprinting in human disease. We and others first mapped BWS in families to 11p15.5 by using genetic linkage analysis (2, 3), determined that a group of BWS-associated balanced germ-line chromosomal rearrangements (termed BWSCR1) are clustered in this band (4) and that they are all of maternal origin (4, 5). These parental origin-specific chromosomal rearrangements, as well as paternal uniparental disomy in other BWS patients (6), suggested that BWS involves one or more imprinted genes on 11p15.5. We later found that all of the BWSCR1 breakpoints disrupt the KVLQT1 gene (7), which encodes a voltage-gated potassium channel, and that KVLQT1 is imprinted and expressed from the maternal allele, consistent with a critical role for this gene in BWS (7). However, the genetics of BWS are complicated by two other observations: some BWS patients show LOI of IGF2 (8–10), which maps 350 kb telomeric to KVLQT1 (7); and 5% of BWS patients show germ-line mutations in p57KIP2 (11–13), which maps 40 kb centromeric to KVLQT1 (4, 7).
Our laboratory has been engaged in identifying imprinting of known and novel genes from 11p15, including IGF2, H19, KVLQT1, p57KIP2, TSSC3, and TSSC5. With the exception of IGF2, all of these genes are preferentially expressed from the maternal allele (7, 14–20). We have now identified a paternally expressed transcript in antisense orientation within KVLQT1. More importantly, analysis of this transcript reveals abnormal imprinting (LOI) of LIT1 (long QT intronic transcript 1) in a majority of BWS patients and no association of LOI of LIT1 with LOI of IGF2. Based on these observations, we offer a unifying hypothesis involving two distinct imprinted gene domains of 11p15: the centromeric imprinted domain that includes p57KIP2 and KVLQT1 and the telomeric imprinted domain including IGF2. By this hypothesis, BWS predominantly involves inactivation of maternally expressed genes within the centromeric domain, and cancer predominantly involves activation of a maternally repressed gene within the more telomeric imprinted domain.
MATERIALS AND METHODS
Isolation of DNA and RNA from Tissues.
All BWS patients were part of a self-referred BWS registry. Only patients having a diagnosis of BWS confirmed by clinical examination were studied. Fibroblasts were obtained by skin biopsy, with informed consent approved by an institutional review board, and cells were cultured in DMEM supplemented with 10% fetal bovine serum. DNA was isolated by digestion with proteinase K (0.2 mg/ml) in the presence of 1% SDS in buffer TE9 (0.5 M Tris⋅HCl, pH 9.0/20 mM EDTA/10 mM NaCl) at 50°C overnight, followed by phenol/chloroform extraction and ethanol precipitation. RNA was isolated by using RNAzol B (Tel-Test, Friendswood, TX), following the manufacturer’s protocol.
Identification of Polymorphisms in LIT1.
Ten sets of primers were used to identify polymorphisms, and genomic DNA of 16 fetuses was directly sequenced by using PCR. The following three transcribed polymorphic sites were identified: single-nucleotide polymorphism 1 (SNP1) in expressed sequence tag (EST) AA331124, SNP2 in EST H88273, and SNP3 in q91 cDNA. The primers that identify these polymorphisms are as follows: for SNP1, Lit105 5′-GATCCTNTCCAGGCAGCTTCTTCCACA-3′ and Lit205 5′-CATAAGGTAGGTAAGTTTGTGTCCCTG-3′; for SNP2, Lit102 5′-TCTGGTTCATGTCACTCTGTGGAGCAG-3′ and Lit202 5′-CTCCCAAAAGCAGAGTTTTGGCAATAT-3′; and for SNP3, Lit111 5′-CAGCCACCTCTGTGGCGTGAATGTTCT-3′ and Lit211 5′-GCTCAAACCCGTCTCTGAAATGCACGG-3′. PCR products were purified by using QIAquick (Qiagen, Chatsworth, CA) and directly sequenced by using an ABI377 automated sequencer.
Analysis of Allele-Specific Expression of LIT1.
RNA was treated with RNase-free DNase (Boehringer Mannheim) and reverse transcription–PCR (RT-PCR; avian myeloblastosis virus reverse transcriptase, Boehringer Mannheim), and sequencing of PCR products was used to analyze allele-specific gene expression. RT was carried out by using either primers CRT1, CRT2, or CRT3, which are centromeric to Lit105/Lit205, Lit102/Lit202, and Lit111/Lit211, respectively, or TRT1, TRT2, or TRT3, which are telomeric to Lit105/Lit205, Lit102/Lit202, and Lit111/Lit211, respectively. The primer sequences are as follows: CRT1, 5′-TTCCATGCAGATGGGGATCCTCTCCAG-3′; CRT2, 5′-ACAGTGCTGTGAGTCTCTGGTTCATGT-3′; CRT3, 5′-TGTCTCCATGACAATATCAGTGCAGTT-3′; TRT2, 5′-GAAGATAACTCACCTCTCCCAAAAGCA-3′; TRT1, 5′-CCTTTAT CAAGA- CATTGTTTAGAGTAA-3′; TRT3, 5′-ACAGGGCTGCATGAGTCATAGGATCCC-3′. RT-PCR was always carried out in parallel in the presence and absence of reverse transcriptase, to rule out any DNA contamination.
Analysis of Allele-Specific Expression of IGF2.
The genotyping of a SNP corresponding to the known ApaI polymorphism was performed by direct sequencing of the PCR product amplified by using primers IGF102 (5′-CTTGGACTTTGAGTCAAATTGGCCTGG-3′) and IGF201 (5′-TTTGGTCTTACTGGGTCCCTCTGACTG-3′). The RT-PCR reactions were carried out by using the following primers that cross an intron–exon boundary to exclude any possibility of genomic DNA contamination: IGF101 in exon 8 (5′-TGGCCCTCCTGGAGACGTACTGTGCTA-3′) and IGF201 in exon 9 (see above). The RT-PCR products were isolated from 1% agarose gels and directly sequenced. That the product corresponded to cDNA was confirmed by sequencing from exon 8 through exon 9 by using IGF101.
Southern Hybridization of LIT1.
Five micrograms of genomic DNA was digested with 10 units of BamHI or BamHI plus NotI at 37°C overnight. The digested DNA was phenol/chloroform-extracted, ethanol-precipitated, and electrophoresed on 0.8% agarose gels, transferred to Hybond-N+ filters, and fixed by UV crosslinking. Filters were hybridized with an EST 592241 probe, labeled by random priming (21). Hybridizations were carried out at 65°C overnight as described (22). The filters were washed in 0.1× SSC/0.1% SDS at 65°C.
RESULTS
Identification of an Antisense-Orientation Transcript in KVLQT1 and Its Expression from the Paternal Allele.
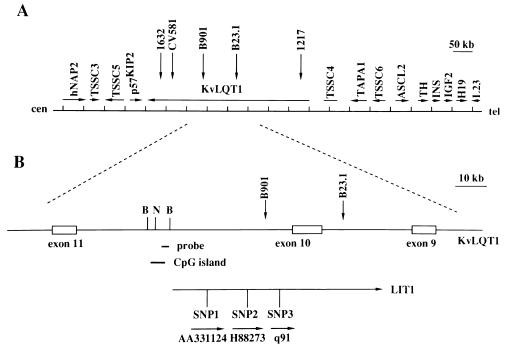
We previously showed that a cluster of balanced germ-line chromosomal rearrangement breakpoints in BWS patients spans 350 kb, and all five of these breakpoints are located within the KVLQT1 gene (refs. 4, 7; Fig. 1A). We also identified several transcripts in the same region (4). As a first step toward identifying a potential role for these transcripts, we sought to determine whether any of them were imprinted. Genomic DNA from 16 fetuses was sequenced by using primers designed to analyze 10 ESTs located within introns of KVLQT1 and near BWS chromosomal rearrangement breakpoints (7) to identify any frequent polymorphisms. In this manner, in ESTs AA331124, H88273, and q91 (GenBank and ref. 4), three frequent SNPs were identified (Fig. 1B).
Figure 1.
Location of paternally expressed transcript LIT1. (A) Transcriptional map of genes from human chromosome 11p15.5. The imprinted genes in the centromeric domain include TSSC3, TSSC5, p57KIP2, and KVLQT1. The imprinted genes in the telomeric domain include ASCL2, IGF2, and H19. TSSC4 and TSSC6 are not imprinted and are located between these two imprinted gene domains. (B) Location of three transcribed polymorphisms within LIT1. Lit1 contains at least 15 EST clones and spans at least 60 kb. It is transcribed from centromere to telomere, and is transcribed in antisense orientation to KVLQT1 (see Fig. 2 and text). SNP1, SNP2, and SNP3 are three transcribed SNPs located within EST clones AA331124, H88273, and q91, respectively. B and N, BamHI and NotI sites, respectively. Probe refers to EST clone 592241, used as a probe. 1632, CV581, B901, B23.1, and 1217 correspond to germ-line balanced chromosomal rearrangement breakpoints in BWS patients (4).
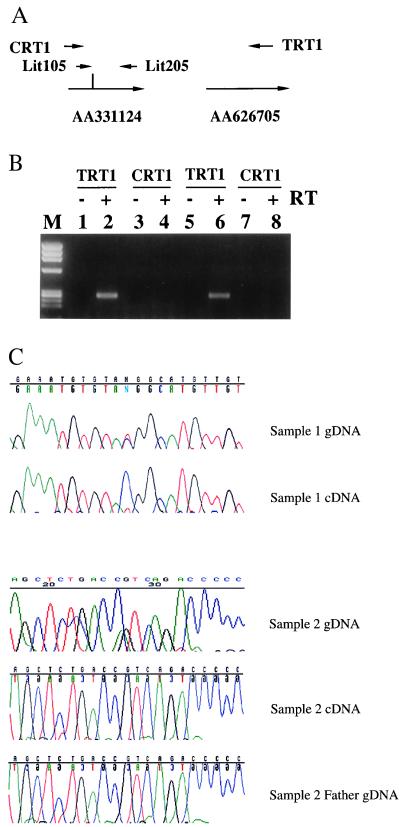
To determine the direction of transcription, RT-PCR was performed by using RT primers at both centromeric and telomeric ends of all three ESTs in both the presence and absence of reverse transcriptase (Fig. 2A). RT-PCR products were obtained only for RT products generated from the telomeric end of each EST and only in the presence of reverse transcriptase (Fig. 2B), indicating that they represented genuine transcripts oriented from centromere to telomere. This is opposite in orientation to KVLQT1 (Fig. 1), which is transcribed from telomere to centromere and expressed from maternal chromosome (7). Twelve additional ESTs were identified in the region, including AA359588, U77321, AA155694, AA533100, AA155639, AA701413, AA626705, AA889050, R66208, AA693940, AA029517, and AA622687. Sequence analysis of the 15 total EST clones indicated that they are all colinear with genomic sequence, indicating the absence of introns. PCR amplification of cDNA by using an RT primer derived from EST AA626705 and PCR primer pairs within EST AA331124 indicated that these two ESTs are part of the same contiguous transcript (Fig. 2). Similar experiments described elsewhere showed exon connection among nine other ESTs spanning 60 kb (23). Consistent with the idea of a large transcript corresponding to these separate ESTs, we previously observed a coinciding band at the upper limit of resolution by Northern blot analysis using three cDNA clones spaced over 80 kb (4), which we now believe corresponds to the same antisense orientation transcript, which we term LIT1 (long QT intron transcript 1; Fig. 1).
Figure 2.
LIT1 is transcribed from centromere to telomere and is expressed from the paternal chromosome. (A) Primers and polymorphisms used. CRT1 and TRT1 are gene-specific primers used in the reverse transcription reaction and are located centromeric and telomeric, respectively, to the PCR fragment amplified by using Lit105/Lit205 spanning SNP1. The vertical line indicates SNP1. EST AA331124 (Fig. 1) is 1 kb centromeric to EST AA626705. (B) LIT1 transcription is from centromere to telomere. RT-PCR reactions were carried out on RNA isolated from patient 41 (lanes 1–4) and patient 3 (lanes 5–8). Because only RT-PCR amplification was obtained from the +RT reaction using the telomeric RT primer, AA331124 and AA626705 are transcribed from centromere to telomere. In addition, because TRT1 is located on AA626705 and Lit 105/Lit 205 is on AA331124, these two EST clones are both part of LIT1. Similar experiments were carried out for the other ESTs in this region. Lane M contains a φX174/HaeIII size marker. The size of the PCR product of Lit105/Lit205 is 269 bp. (C) Examples showing imprinting of LIT1. Samples 1 and 2 are heterozygous for SNP2, GAAATGTGTA(C/T)GGCATGTTGT, and SNP1, AGCTCTGACC(G/A)TCAGACCCCC, respectively (gDNA). Both samples show monoallelic expression on RT-PCR analysis, C for sample 1 and G for sample 2. RT-PCR was always performed in parallel in the presence and absence of reverse transcriptase, and only those reactions with no products in the −RT lane were sequenced (cDNA). The expressed allele in sample 2 was of paternal origin (father gDNA).
These 3 transcribed polymorphisms were then used to analyze tissue samples from 11 separate heterozygous (informative) normal individuals, including 2 fibroblast cultures, 8 kidney samples, and 4 fetal tissues (kidney, heart, adrenal, placenta). All 14 tissues from these 11 individuals showed monoallelic expression. For example, Fig. 2C shows genomic DNA from a kidney heterozygous for SNP2, with expression of only the C allele of LIT1. Similarly, a second normal individual displayed heterozygosity for SNP1, and again monoallelic expression was observed (Fig. 2C). Genotyping of parental DNA indicated that the expressed G allele was derived from the paternal chromosome (Fig. 2C). The parental origin of two additional individuals also was determined, and in these cases, as well, the paternal allele was expressed (data not shown). Finally, in experiments described elsewhere, two paternal and two maternal chromosomes from separate individuals were segregated in somatic cell hybrids, and LIT1 was only expressed from the paternal chromosome (23).
Biallelic Expression of LIT1 in Patients with BWS.
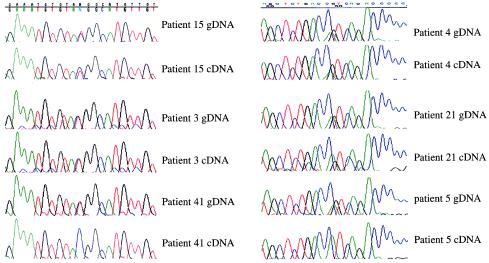
SNP1 and SNP2 were used to type normal fibroblasts or peripheral blood lymphocytes from 37 patients with BWS. Sixteen BWS patients were found to be heterozygous for at least one of the two polymorphic sites, allowing analysis of imprinting of LIT1. Eight of the 16 informative BWS patients (50%) showed biallelic expression of LIT1, indicating relaxation of imprinting of the normally silent maternal allele of LIT1 in these patients (Fig. 3, Table 1). By convention, relaxation of imprinting is also termed LOI (loss of imprinting), and this term will be used here for convenience. In contrast to the BWS patients, 14 tissues from 11 control patients without BWS showed normal imprinting of LIT1 (Fig. 2C and data not shown). Thus, LOI of LIT1 was frequent and specific for BWS.
Figure 3.
Biallelic expression of LIT1 occurs frequently in BWS patients. Samples from patients 15, 3, and 41 are heterozygous for SNP2 (gDNA), and patients 4, 21, and 5 are heterozygous for SNP1 and were analyzed in the same manner as in Fig. 2. Examples of biallelic (patients 15, 3, 4, and 21) and monoallelic (patients 41 and 5) expression for each polymorphism are shown. The results for all 16 informative BWS patients analyzed are shown in Table 1.
Table 1.
Analysis of Imprinting of LIT1 and IGF2 in BWS Patients
| Patient | LIT1
|
IGF2
|
|
|---|---|---|---|
| Allele-Specific Expression | CpG Island Methylation | Allele-Specific Expression | |
| 1 | LOI | I | |
| 2 | LOI | ||
| 3 | LOI | LOI | I |
| 4 | LOI | LOI | I |
| 5 | I | I | |
| 6 | I | ||
| 7 | LOI | ||
| 8 | LOI | LOI | |
| 9 | I | ||
| 10 | LOI | ||
| 11 | I | I | |
| 12 | I | I | |
| 13 | LOI | ||
| 14 | I | ||
| 15 | LOI | LOI | |
| 16 | I | ||
| 17 | I | I | |
| 18 | LOI | ||
| 19 | LOI | ||
| 20 | LOI | ||
| 21 | LOI | LOI | I |
| 22 | LOI | LOI | |
| 23 | I | ||
| 24 | I | I | |
| 25 | I | I | |
| 26 | I | ||
| 27 | LOI | ||
| 28 | I | ||
| 29 | LOI | ||
| 30 | I | ||
| 31 | LOI | ||
| 32 | I | ||
| 33 | LOI | ||
| 34 | LOI | ||
| 35 | LOI | ||
| 36 | LOI | ||
| 37 | LOI | ||
| 38 | LOI | LOI | |
| 39 | I | I | |
| 40 | I | ||
| 41 | I | LOI | |
| 42 | I | ||
| 43 | I | ||
LOI, loss of imprinting; I, normal imprinting.
Frequent Hypomethylation of a Maternally Methylated CpG Island in BWS Patients with LOI of LIT1.
We have analyzed the genomic sequence near and within LIT1 and have identified a CpG island 200 bp upstream of the 5′ end of EST AA359588, which is at the 5′-most end of LIT1. The CpG island is located on a 6.0-kb BamHI fragment, and there is a methylation-sensitive NotI site in this fragment that is methylated on the maternal chromosome. While the parental origin of the allele-specific LIT1 transcripts is described here, the parental origin of the LIT1 CpG island methylation is described in a separate publication (23). Briefly, this was shown by the establishment of four somatic cell hybrids carrying chromosome 11 of known parental origin (two paternal, two maternal), and on only the maternal chromosome was the CpG island methylated (23). We have also confirmed parental origin-specific methylation by examination of parthenogenetic ovarian teratomas (methylated) and androgenetic hydatidiform moles (unmethylated) (data not shown).
To determine the methylation status of this CpG island in BWS, genomic Southern blotting was used to analyze the methylation-sensitive NotI site within the CpG island. A single 6.0-kb or 4.2-kb band indicates that either both chromosomes are methylated or both are unmethylated, respectively. The presence of equal intensity of both bands indicates differential methylation.
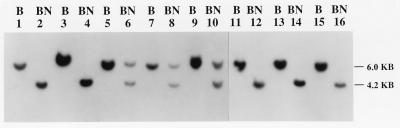
To determine the methylation status of the LIT1 CpG island in patients with BWS, we isolated genomic DNA from several different tissues, including fibroblasts, peripheral blood lymphocytes, tongue, and kidney, and digested the DNA either with BamHI or with BamHI plus NotI. Of 36 BWS patients analyzed by genomic Southern hybridization at this site, 15 showed two bands (4.2 kb and 6 kb) of equal intensity (Fig. 4, Table 1), indicating that the DNA was normally methylated on the maternal chromosome in those patients, which was the same result observed in 8 samples from non-BWS individuals (data not shown). However, 21 of the 36 BWS patients (58%) showed a single band of 4.2 kb, indicating hypomethylation of the normally methylated maternal allele (Fig. 4, Table 1), suggesting abnormal imprinting in these patients. Consistent with this hypothesis, hypomethylation of the CpG island in these patients was found in all cell types examined, including fibroblasts, peripheral blood lymphocytes, and tongue, indicating that the altered DNA methylation occurred in the germ line or early in development.
Figure 4.
Frequent loss of methylation of a CpG island NotI site on the maternal chromosome in BWS. BamHI digestion (labeled B) yields a 6.0-kb fragment containing the NotI site, and BamHI plus NotI digestion (labeled BN) yields a 6.0- and 4.2-kb fragment, corresponding to the methylated and unmethylated maternal and paternal alleles in normal individuals. Patients 21 (lanes 1 and 2), 22 (lanes 3 and 4), 1 (lanes 11 and 12), 2 (lanes 13 and 14), and 3 (lanes 15 and 16) show only a single 4.2-kb band after BamHI plus NotI digestion, indicating loss of methylation of the NotI site. Patients 23 (lanes 5 and 6), 24 (lanes 7 and 8), and 25 (lanes 9 and 10) show a normal pattern of methylation. Genomic Southern blots were hybridized by using EST clone 592241 as a probe.
Finally, to confirm that hypomethylation of the LIT1 CpG island was caused by abnormal imprinting, we compared methylation to allele-specific expression of LIT1 in nine BWS patients for whom there was sufficient high molecular weight DNA for methylation analysis and that were informative for one or more polymorphisms in LIT1. This analysis revealed a complete correlation between hypomethylation and LOI, and a similar complete correlation between normal methylation and normal imprinting of LIT1. Thus, all six BWS patients with biallelic expression of LIT1 also showed hypomethylation of the LIT1 CpG island (Figs. 3 and 4, and Table 1, patients 3, 4, 8, 15, 21, and 22). In contrast, three patients with normal imprinting of LIT1 showed normal differential methylation of the NotI CpG island (Table 1, patients 5, 11, and 17). This result was statistically significant [P = (0.58)6(0.42)3 = 0.0028]. Therefore, BWS patients frequently exhibit LOI of LIT1, manifested both by abnormal expression of the normally silent maternal allele and by hypomethylation of the normally methylated maternal allele.
Discordant Regulation of LIT1 and IGF2 Imprinting in BWS.
LOI of IGF2 has been proposed to play a major role in the etiology of BWS (8, 10). The frequency of LOI in BWS has been reported to range from 67% to 81% (8, 10). We therefore sought to determine the frequency of LOI of IGF2 in BWS patients and whether there was any relationship between LOI of IGF2 and LOI of LIT1. To obviate any potential problems with restriction enzyme digestion or heteroduplex formation, analysis at a polymorphic ApaI site in IGF2 was performed by direct sequencing of PCR-amplified genomic DNA or cDNA, the latter across an exon–intron boundary to avoid any possible genomic DNA contamination. Of 37 BWS patients examined, 10 were found to be heterozygous at the ApaI site in their genomic DNA (Fig. 5, Table 1). Of these, only 2 (20%) showed LOI of IGF2 (Fig. 5, Table 1). These results also were confirmed by direct analysis by ApaI digestion (data not shown). There was no association between LOI of LIT1 and LOI of IGF2, as 4 of the 8 patients with normal imprinting of IGF2 exhibited LOI of LIT1, and 1 of the 2 patients with LOI of IGF2 exhibited LOI of LIT1 (Figs. 3 and 5, Table 1). Thus, LOI of LIT1 occurs much more frequently in BWS than does LOI of IGF2, and LOI of LIT1 is independent of LOI of IGF2.
Figure 5.
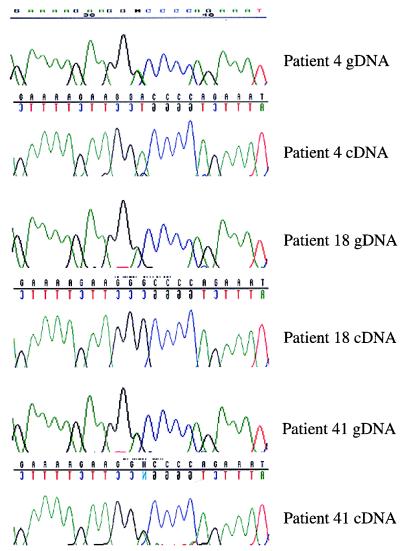
Normal imprinting of IGF2 in most BWS patients. Patients 4, 18, and 41 are heterozygous for a SNP, GAAAAGAAGG(A/G)CCCCAGAAAT (gDNA). Patients 4 and 18 show monoallelic expression (cDNA, A and G, respectively), and patient 41 shows LOI of IGF2. The results for all 10 BWS patients informative for IGF2 are listed in Table 1.
DISCUSSION
We have shown that LIT1, a paternally expressed transcript within and in antisense orientation to KVLQT1, was abnormally expressed from the maternal allele in half of patients with BWS (8 of 16 informative, 50%). In addition, biallelic expression of LIT1 in BWS was linked to loss of methylation on the maternal chromosome of a CpG island NotI site upstream of LIT1. When assayed by DNA methylation of LIT1, 21 of 36 patients (58%) showed LOI of LIT1. This switching of the maternal chromosome to a paternal pattern of expression and methylation, termed LOI of LIT1 in keeping with the nomenclature for other genes, is the most frequent genetic change in BWS (combining the allele-specific expression and methylation data, 23 of 43 patients, 53%). We have also shown that LOI of another gene previously implicated in BWS, IGF2 (8, 10), was uncommon in these patients (2 of 10 patients, 20%). Moreover, we found that LOI of IGF2 is not linked to LOI of LIT1. Thus, we believe that LOI of LIT1 is crucial to the pathogenesis of BWS, and that LOI of LIT1 is unrelated to LOI of IGF2, even though IGF2 may also be important in BWS.
We propose a model in which 11p15 harbors two separate imprinted domains. The more centromeric imprinted domain includes KVLQT1, p57KIP2, TSSC5, and TSSC3 and spans 500 kb. The telomeric imprinted domain includes IGF2, H19, and ASCL2 and spans 200 kb. These domains are separated by ≈300 kb. According to our model, hypomethylation of the LIT1 CpG island is linked to LIT1 expression, and LIT1 expression is antagonistic to expression of other maternally expressed genes in the same region, such as KVLQT1 and p57KIP2. Consistent with this idea, three other imprinted genes, IGF2R, IGF2, and UBE3A have also been shown to overlap antisense transcripts (24–26), the expression of which has been proposed to serve a regulatory role in silencing the sense orientation transcript (25). Thus, abnormal expression of LIT1 on the maternal chromosome could be linked to silencing of nearby maternally expressed genes on the same chromosome.
Furthermore, we believe the ultimate target of these alterations in BWS patients could be p57KIP2, although this idea is not essential to our two-domain model. However, the genetic data, that the same phenotype, BWS, arises from LOI of LIT1 (this paper), from germ-line chromosomal rearrangements within KVLQT1 (7), and from mutations in p57KIP2 (11–13), are consistent with this idea. How could LOI of LIT1 or chromosomal rearrangements affect p57KIP2? There are several possible mechanisms, including enhancer competition between LIT1 and p57KIP2, or a hypothetical repressor of p57KIP2 within KVLQT1 that is displaced by maternal KVLQT1 expression. However, the model we favor is that the LIT1 CpG island serves as an insulator between p57KIP2 and its enhancer. According to this model, methylation of the CpG island would interfere with both insulator function and LIT1 expression, as has been suggested for a CpG island near H19 regulating the expression of H19 and IGF2 (27, 28). When the CpG island is unmethylated, as it is on the paternal chromosome, it effectively blocks p57KIP2 from its enhancer, and expression of LIT1 could also compete with p57KIP2 for the enhancer on the same chromosome. Synergistic effects of both insulator and enhancer competition could effectively silence p57KIP2 expression. In BWS patients with LOI of LIT1, the CpG island is abnormally hypomethylated on the maternal chromosome, allowing LIT1 expression and permitting the insulator to function, which could synergistically silence expression of p57KIP2. This idea would predict that the BWSCR1 germ-line chromosomal rearrangements have no direct effect on imprinting of LIT1. By our model, these rearrangements would simply separate p57KIP2 from its enhancer genetically, in the same manner that the unmethylated LIT1 CpG island could separate p57KIP2 from its enhancer epigenetically. If p57KIP2 is indeed a target of these genetic changes, then its expression will be reduced in these patients.
The other element of our two-domain model is that IGF2 lies within an independently regulated, more telomeric imprinted domain. We believe that LOI of IGF2 is more specifically associated with cancer, as it occurs more frequently in embryonal and other tumors than in BWS per se, and LOI of IGF2 is also specifically associated with a clinical phenotype of overgrowth and cancer in the absence of BWS (29). Consistent with this idea, we have found that LIT1 appears to be normally imprinted in Wilms tumor (23). In addition, mouse knockout experiments disrupting the maternal allele of H19 and leading to biallelic expression of IGF2 cause prenatal overgrowth but not phenotypic features of BWS (30). Similarly, although chimeric mice overexpressing an IGF2 transgene show increased overall and organ growth, they do not exhibit characteristic histologic changes in, for example, the kidneys (31). In contrast, germ line disruption of p57KIP2 causes anatomical and histologic defects consistent with BWS but not overgrowth (32). Furthermore, disruption of IGF2 imprinting has no effect on the more centromeric group of imprinted genes (33).
By our model, BWS patients with paternal uniparental disomy would have loss of expression of maternally expressed genes within the centromeric imprinted domain, as well as biallelic expression of IGF2, simply because of the replacement of a maternal with a paternal chromosome. Uniparental disomy patients have been reported to show an increased frequency of cancer compared with other BWS patients (34, 35), consistent with the importance of IGF2 for malignancy, as we propose. However, it is important to bear in mind that BWS is also associated in some patients with LOI of IGF2. Thus, the phenotypes caused by alterations in the two domains, predominantly BWS in the centromeric domain and cancer in the telomeric domain, although largely distinct, do overlap. This is exactly the prediction of Haig’s hypothesis of antagonistic imprinted domains (36). In support of our two-domain hypothesis, we have recently identified a region that escapes genomic imprinting between these two imprinted domains (37). The identification of frequent alterations in an imprinted antisense transcript in disease, reported here, should be crucial to our understanding of the genetics of BWS, as well as the mechanism of genomic imprinting.
Acknowledgments
We thank the University of Washington fetal tissue bank for specimens, and gratefully acknowledge helpful discussions with Steve Elledge, Shirley Tilghman, Patrick Onyango, and Jason Ravenel. The Oshimura laboratory contributed the methylation analysis of LIT1 in somatic cell hybrids to this paper. This work was supported by National Institutes of Health Grant CA54358.
ABBREVIATIONS
- BWS
Beckwith–Wiedemann syndrome
- LOI
loss of imprinting
- IGF
insulin-like growth factor
- RT-PCR
reverse transcription–PCR
- EST
expressed sequence tag
- SNP
single-nucleotide polymorphism
References
- 1.Feinberg A P. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 95–107. [Google Scholar]
- 2.Ping A J, Reeve A E, Law D J, Young M R, Boehnke M, Feinberg A P. Am J Hum Genet. 1989;44:720–723. [PMC free article] [PubMed] [Google Scholar]
- 3.Koufos A, Grundy P, Morgan K, Aleck K A, Hadro T, Lampkin B C, Kalbakji A, Cavenee W K. Am J Hum Genet. 1989;44:711–719. [PMC free article] [PubMed] [Google Scholar]
- 4.Hoovers J M N, Kalikin L M, Johnson L A, Alders M, Redeker B, Law D J, Bliek J, Steenman M, Benedict M, Wiegant J, et al. Proc Natl Acad Sci USA. 1995;92:12456–12460. doi: 10.1073/pnas.92.26.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mannens M, Hoovers J M N, Redeker E, Verjaal M, Feinberg A P, Little P, Boavida M, Coad N, Steenman M, Bliek J, et al. Eur J Hum Genet. 1994;2:3–23. doi: 10.1159/000472337. [DOI] [PubMed] [Google Scholar]
- 6.Henry I, Bonaiti-Pellie C, Chehensse V, Beldjord C, Schwartz C, Utermann G, Junien C. Nature (London) 1991;351:609–610. doi: 10.1038/351665a0. [DOI] [PubMed] [Google Scholar]
- 7.Lee M P, Hu R-J, Johnson L A, Feinberg A P. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 8.Weksberg R, Shen D R, Fei Y L, Song Q L, Squire J. Nat Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 9.Steenman M J C, Rainier S, Dobry C J, Grundy P, Horon I L, Feinberg A P. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 10.Joyce J A, Lam W K, Catchpoole D J, Jenks P, Reik W, Maher E R, Schofield P N. Hum Mol Genet. 1997;6:1543–1548. doi: 10.1093/hmg/6.9.1543. [DOI] [PubMed] [Google Scholar]
- 11.Hatada H, Ohashi Y, Fukushima Y, Kaneko M, Inoue Y, Komoto A, Okada S, Ohishi A, Nabetani H, Morisaki M, et al. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-171. [DOI] [PubMed] [Google Scholar]
- 12.Lee M P, DeBaun M, Randhawa G S, Reichard B A, Feinberg A P. Am J Hum Genet. 1997;61:304–309. doi: 10.1086/514858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Keefe D, Dao D, Zhao L, Sanderson R, Warburton D, Weiss L, Anyane-Yeboa K, Tycko B. Am J Hum Genet. 1997;61:295–303. doi: 10.1086/514854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka S, Thompson J S, Edwards M C, Barletta J M, Grundy P, Kalikin L M, Harper J W, Elledge S J, Feinberg A P. Proc Natl Acad Sci USA. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M P, Feinberg A P. Cancer Res. 1998;58:1052–1056. [PubMed] [Google Scholar]
- 16.Lee M P, Reeves C, Schmitt A, Su K, Connors T D, Hu R-J, Brandenburg S, Lee M J, Miller G, Feinberg A P. Cancer Res. 1998;58:4155–4159. [PubMed] [Google Scholar]
- 17.Rainier S, Johnson L A, Dobry C J, Ping A J, Grundy P E, Feinberg A P. Nature (London) 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 18.Qian N, Frank D, O’Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B. Hum Mol Genet. 1997;6:2021–2029. doi: 10.1093/hmg/6.12.2021. [DOI] [PubMed] [Google Scholar]
- 19.Dao D, Frank D, Qian N, O’Keefe D, Vosatka R J, Walsh C P, Tycko B. Hum Mol Genet. 1998;7:597–608. doi: 10.1093/hmg/7.4.597. [DOI] [PubMed] [Google Scholar]
- 20.Cooper P R, Smilinich N J, Day C D, Nowak N J, Reid L H, Pearsall R S, Reece M, Prawitt D, Landers J, Housman D E, et al. Genomics. 1998;49:38–51. doi: 10.1006/geno.1998.5221. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg A P, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 22.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuya, K., Meguro, M., Lee, M. P., Katoh, M., Schulz, T. C., Kugoh, H., Yoshida, M. A., Niikawa, N., Feinberg, A. P. & Oshimura, M. (1998) submitted to Hum. Mol. Genet. [DOI] [PubMed]
- 24.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Nature (London) 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 26.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 27.Webber A L, Ingram R S, Levorse J M, Tilghman S M. Nature (London) 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- 28.Thorvaldsen J L, Duran K L, Bartolomei M S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa O, Becroft D M, Morison I M, Eccles M R, Skeen J E, Mauger D C, Reeve A E. Nat Genet. 1993;5:408–412. doi: 10.1038/ng1293-408. [DOI] [PubMed] [Google Scholar]
- 30.Leighton P A, Ingram R S, Eggenschwiler J, Efstratladis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 31.Sun F L, Dean W L, Kelsey G, Allen N D, Reik W. Nature (London) 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P, Liegeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 33.Caspary T, Cleary M A, Baker C C, Guan X J, Tilghman S M. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junien C. Curr Opin Genet Dev. 1992;2:431–438. doi: 10.1016/s0959-437x(05)80154-6. [DOI] [PubMed] [Google Scholar]
- 35.Henry I, Peuch A, Riesewijk A, Ahnine L, Mannens M, Beldjord C, Bitoun P, Tournade M F, Landrieu P, Junien C. Eur J Hum Genet. 1993;1:19–29. doi: 10.1159/000472384. [DOI] [PubMed] [Google Scholar]
- 36.Haig D, Graham C. Cell. 1991;64:1045–1046. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee, M. P., Brandenburg, S., Landes, G. M., Adams M., Miller, G. & Feinberg, A. P. (1999) Hum. Mol. Genet., in press. [DOI] [PubMed]