Abstract
In paroxysmal nocturnal hemoglobinuria (PNH), acquired somatic mutations in the PIG-A gene give rise to clonal populations of red blood cells unable to express proteins linked to the membrane by a glycosylphosphatidylinositol anchor. These proteins include the complement inhibitors CD55 and CD59, and this explains the hypersensitivity to complement of red cells in PNH patients, manifested by intravascular hemolysis. The factors that determine to what extent mutant clones expand have not yet been pinpointed; it has been suggested that existing PNH clones may have a conditional growth advantage depending on some factor (e.g., autoimmune) present in the marrow environment of PNH patients. Using flow cytometric analysis of granulocytes, we now have identified cells that have the PNH phenotype, at an average frequency of 22 per million (range 10–51 per million) in nine normal individuals. These rare cells were collected by flow sorting, and exons 2 and 6 of the PIG-A gene were amplified by nested PCR. We found PIG-A mutations in six cases: four missense, one frameshift, and one nonsense mutation. PNH red blood cells also were identified at a frequency of eight per million. Thus, small clones with PIG-A mutations exist commonly in normal individuals, showing clearly that PIG-A gene mutations are not sufficient for the development of PNH. Because PIG-A encodes an enzyme essential for the expression of a host of surface proteins, the PIG-A gene provides a highly sensitive system for the study of somatic mutations in hematopoietic cells.
Paroxysmal nocturnal hemoglobinuria (PNH) is a chronic hemolytic disorder characterized by intravascular hemolysis, caused by a clonal (1) red blood cell population of variable size that is hypersensitive to complement (2). Cells belonging to this clone have reduced or absent expression on their surface of the complement inhibitors CD55 and CD59, as well as of all other membrane proteins anchored by a glycosylphosphatidylinositol (GPI) moiety (reviewed in ref. 3). Nonerythroid blood and bone marrow cells exhibit the same defect (4). The product of the PIG-A gene, which maps to Xp.21, is responsible for an early step in GPI-anchor biosynthesis and is disrupted by a somatic mutation in virtually all PNH patients (5–8), resulting in the deficiency of all GPI-linked surface molecules.
Although the PNH phenotype is thus rationalized at the molecular level, we cannot yet explain the basis for clonal expansion, which results in PNH cells contributing substantially to hematopoiesis. In fact, evidence from an in vitro model of human hematopoiesis (9) as well as murine models of PNH (10–12) indicate that the PNH clone has no intrinsic growth advantage, in keeping with the clinical observation that the PNH red cell population can spontaneously disappear in some patients (13). Therefore, it has been postulated that there must be a second disease mechanism promoting the relative expansion of the PNH clone (14).
The clinical association between PNH and acquired aplastic anemia (AAA) (15) and the observation that, as in AAA, PNH patients have decreased hematopoietic progenitors (16) may be taken to suggest a common pathogenetic process (17). There is strong evidence that AAA is an autoimmune disease (18), and, like for AAA, bone marrow failure in PNH can be treated successfully with immunosuppression (19); thus, autoimmunity is likely to play a role in PNH as well. Specifically, it has been hypothesized that an autoimmune attack on normal stem cells targets a GPI-linked molecule and therefore preferentially spares the PNH stem cell, which thus has a growth or a survival advantage (or both) in this abnormal environment (14, 17, 20).
A direct implication of this hypothesis is that somatic mutations of the PIG-A gene, if they occur in normal individuals, would not give rise to a sizable clonal population. We show here that rare PNH cells are present in individuals with normal hematopoiesis, strongly reinforcing the notion that, although a PIG-A gene mutation may be necessary for the development of PNH, it is not sufficient.§
MATERIALS AND METHODS
Donors.
After giving informed consent, healthy male volunteers (ages 30–65) donated 50 ml of whole blood. Donor 8, a 64-year-old man with non-Hodgkin’s lymphoma in complete remission 5 years after receiving radiotherapy and cyclophosphamide/doxorubicin/vincristine/prednisone chemotherapy, was diagnosed with hereditary hemochromatosis and kindly donated his therapeutic phlebotomy samples. Donor 9 is a 36-year-old man who is hemizygous for G6PD Mediterranean. All other donors had no clinical problem.
Granulocyte Separation.
Whole blood was anticoagulated with either heparin or acid-citrate-dextrose and was mixed 1:1 with 6% hetastarch/0.9% saline for RBC sedimentation, and the supernatant was centrifuged over Ficoll. After this step, the cells were kept in ice/refrigeration for the remainder of the procedure to prevent activation. The RBCs then were lysed by hypotonic shock, and the remaining cells were resuspended in PBS with 0.1% BSA.
Flow Cytometry Analysis.
Murine anti-human CD55 (Clone BRIC 110) was obtained from Serotec (Oxford), and, for some experiments, we used IA10, an anti-CD55 antibody kindly provided by Victor Nussenzweig (New York University). Murine anti-human CD59 (Clone MEM 43) and IgG2a isotype antibodies were from Research Diagnostics (Flanders, NJ); R-phycoerythrin-conjugated F(ab′)2 fragment rabbit anti-mouse Ig (RAM-PE) and murine anti-human glycophorin A–FITC were from Dako; murine anti-human CD11b conjugated to FITC was from Immunotec (Marseille, France); and goat anti-mouse FITC and IgG1 isotype antibodies were from Becton Dickinson.
Before resuspension, antibody was added to the centrifuged pellet to ensure staining of all cells. The cells were sequentially stained in three separate steps, each for at least 30 min, first with anti-CD55 mixed together with anti-CD59, then with RAM-PE followed by CD11b-FITC. In some initial experiments, cells were fixed in 1% parafomaldyhyde before fluorescence-activated cell sorter analysis. This step resulted in a decreased proportion of PCR reactions yielding a visible product, and it was therefore discontinued.
Magnetic Bead Enrichment for PNH Granulocytes.
To increase the number of PNH cells available for analysis, in three experiments (donors 3, 7, and 8), before staining for fluorescence-activated cell sorting, a part of the sample was depleted of GPI-positive cells by incubating with a murine (IgM) anti-CD16 antibody (Becton Dickinson) and separating with rat anti-mouse IgM magnetic Dynabeads (Dynal, Oslo). This procedure increased the frequency of CD55(−) CD59(−) cells, on average, by a factor of 10, proving that the CD55(−) CD59(−) cells in normal donors are also CD16(−), supporting the notion that they are deficient in all GPI-linked molecules.
Ham Test.
A type AB donor whose serum had been shown to be effective in the Ham test was selected. Serum was immediately separated from whole blood and was stored in liquid nitrogen. For a negative control, thawed aliquots were heat-inactivated at 56°C for 30 min before use. Serum (0.5 ml) was acidified with 0.05 ml of 200 mM HCl, with MgCl2 added to a final concentration of 4.1 mM. Washed red blood cells (0.05 ml) suspended in saline were incubated with the serum for 1 hour at 37°C, after which they were washed and stained sequentially with anti-CD55 and anti-CD59, RAM-PE, and glycophorin A–FITC.
Flow Cytometric Sorting.
Cells were analyzed and sorted on a Becton Dickinson FACStarPlus from cold polystyrene tubes. Some experiments involving only analysis were performed on a Becton Dickinson FACScan. Granulocytes were identified by using forward/side scatter gating as well as FL1 (green) fluorescence, indicating expression of the non-GPI-linked granulocyte marker CD11b. Granulocytes with low FL2 fluorescence (based on isotype controls) were defined as PNH cells and were sorted into refrigerated polypropylene tubes. GPI(+) cells also were collected as a control. To detect rare PNH cells, at least 106 events were collected for analysis.
DNA Isolation.
On average, the cytometer collected 120 PNH granulocytes from each experiment. These were divided into Eppendorf tubes in aliquots of 10–50 cells per tube and were centrifuged at 16,000 × g at 4°C. After gentle aspiration of the supernatant, 12 μl of 50 mM NaOH was added. The sample then was heated at 100°C for 5 min and was neutralized with 8 μl of 150 mM Tris (pH 7.8).
PIG-A Gene Amplification.
Exons 2 and 6 were amplified in nested PCR reactions with 100 μl of “PCR supermix” (GIBCO), under heavy mineral oil, using a Perkin–Elmer thrermocycler with the following conditions: initial denaturation at 94°C for 10 min, and 35 cycles with 55°C for 1 min, 72°C for 3 min, and 94°C for 1 min. The DNA extraction mixture (10 μl), containing on average 8 (range 4–25) sorted PNH granulocytes, served as the template for the first reaction, and sorted GPI(+) cells served as a positive control; reagents without cells served as a negative control. In the first reaction, the following intronic primers were added at a concentration of 1 μM each to amplify exons 2 and 6 together: exon 2 forward, 5′AATATCTAGATCTGGATTTGTA; exon 2 reverse, 5′GGGAATTCTTTGCTATAATTTATAG; exon 6 forward: 5′TAAGGATATCATTACTATGAC; and exon 6 reverse: 5′AAAATATTGAATGATATAGAGGTAGCATAA.
For the second PCR reaction, the following intronic primers were added at a concentration of 0.5 μM in separate reactions for exons 2 and 6: exon 2 forward: 5′TGGAATGTGTTTTTGTTTCTGAGCTG; exon 2 reverse: 5′CAAGTATTCAACAGCTTTCTATAG; exon 6 forward: 5′TTTATAAAATGTTCCTGCAGGATC; and exon 6 reverse: 5′CTTTCTAGAACTATTACAAATGTT. A portion (5 μl) of the first reaction served as the template for the second reaction.
Single-Strand Conformation Polymorphism (SSCP) and Heteroduplex Analysis.
PCR products were analyzed on an agarose gel; those reactions yielding a visible product were reamplified in the presence of 1 μCi 32P α-dCTP (Amersham Pharmacia) and were digested and analyzed by SSCP and heteroduplex analysis (HA) as described (6–8). Digested PCR products from normal cells served as controls.
Cloning and Sequencing. PCR products showing shifted bands were reamplified, were digested with the appropriate restriction enzymes, were purified (GenecleanII kit, Bio 101), and were ligated with T4 ligase into an M13 vector that had been digested with the same enzymes and had been dephosphorylated with calf alkaline phosphatase. Ligation reactions were transformed into Escherichia coli strain DH5αF′IQ, (GIBCO). Pools of plaques were inoculated into fresh PCR reaction mix and were analyzed by SSCP/HA. Plaques with the same shifted bands originally seen on SSCP/HA were selected for dideoxynucleotide sequence analysis by using the T7 Sequenase v2 kit (Amersham Pharmacia). For donor 9, automated sequencing (Applied Biosytems ABI 377) was performed. In the aim to rule out Taq polymerase and E. coli DNA polymerase errors, we have listed only mutations identified in at least two independent M13 clones: In a detailed analysis, it has been demonstrated that such errors typically give rise only to single abnormal M13 clones (21).
RESULTS
Development of Methodology.
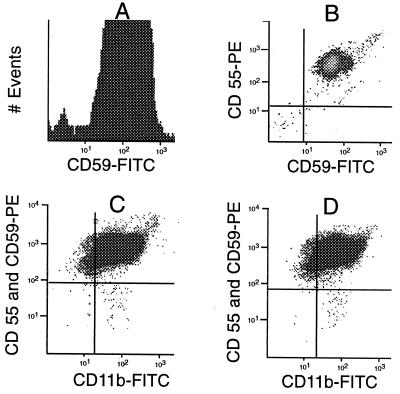
In pilot studies, granulocytes from normal individuals were stained with either of two antibodies recognizing GPI-linked proteins. We found that 0.17% of cells were negative with anti-CD59 (Fig. 1A), and 1.36% of cells were negative with anti-CD55. If these negative cells represented the “tail” of a distribution curve reflecting continuous variation in expression of each of these antigens, we would not expect them to correlate closely with each other. On the other hand, if cells are CD59(−) because of a PIG-A mutation, they would be expected to be CD55(−) also. By two-dimensional analysis (Fig. 1B), we observed that 0.04% of granulocytes are, in fact, both CD59(−) and CD55(−). When we increased the intensity of the fluorescence signal in normal cells by substituting RAM-PE for goat anti-mouse FITC, we obtained a more conservative estimate of 0.01% (100 per million) CD59(−) CD55(−) cells. To eliminate events that might originate from cell debris or other contaminating particles rather than from intact granulocytes, we needed a positive stain, and we chose CD11b, a non-GPI linked granulocyte antigen. By positive staining with anti-CD11b, we eliminated, on average, 62% of the CD59(−) CD55(−) events (range 16–97%). For instance, in donor 8, we now could demonstrate by triple staining a discrete population of CD55(−) CD59(−) CD11b(+) cells (Fig. 1C) having a frequency of 7.5 per million. This tiny population of cells, visible in the lower right quadrant of the display, has identical staining characteristics as the large PNH clone from a patient with florid PNH (Fig. 2A).
Figure 1.
Development of the Methodology. (A) Histogram of normal granulocytes (donor 1) stained with anti-CD59 and goat anti-mouse FITC. A portion (0.17%) of the cells are CD59(−) (fluorescence <101). (B) Two-dimensional scatter analysis of normal granulocytes (donor 1) stained with anti-CD59 and anti-CD55. Here, 0.04% (400 per million)—seen in the lower left quadrant—express neither antigen. (C) Scatter analysis of granulocytes from donor 8, stained with anti-CD55 and anti-CD59, followed by RAM-PE and then CD11b-FITC. A distinct population of PNH cells is seen in the lower right quadrant at a frequency of 7.5 per million and appears similar to the large PNH clone from patient MSK11 (compare with Fig. 2A). (D) Granulocytes from donor 8 stained in parallel to sample in C. The pattern is virtually identical, and the PNH population is present at a frequency of 9.6 per million, confirming the reproducibility of the assay.
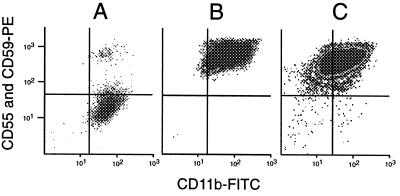
Figure 2.
(A) Granulocytes from PNH patient MSK11 stained with anti-CD55, anti-CD59, and CD11b-FITC, demonstrating a large PNH clone representing 80% of the cells. (B) Post-sort analysis of collected normal GPI(+) granulocytes from donor 8, demonstrating no residual PNH cells (compare with Fig. 1C). This confirms the ability of the cytometer to accurately sort a rare population. (C) A simulated experiment with PNH cells from patient MSK11 added to sorted GPI(+) cells from donor 1 at a frequency of 25 per million. The PNH population is clearly seen in the lower right quadrant, again confirming the ability of the instrument to identify this rare population. Events, most likely representing debris, also are seen in the lower left quadrant, demonstrating the usefulness of anti-CD11b for excluding these events.
Isolation of Rare PNH Cells by Flow Sorting.
To test the ability of the flow cytometer to collect rare PNH cells, we performed post-sort analysis on the normal cells from donor 8 (upper right quadrant of Fig. 1C). Before sorting, the frequency of PNH cells was 7.5 per million; by contrast, after sorting, we found none [of 1.3 million cells reanalyzed (Fig. 2B)]. Thus, the sorting apparatus was able to separate physically the rare PNH cells.
Next, to assess the purity of the sorted PNH cells, we simulated a sorting experiment by using granulocytes from a male patient, MSK11, in whom we previously had identified a mutation in exon 2 (259delC), which eliminates an HaeIII restriction site (7). Granulocytes from this patient and normal donor 1 were stained in parallel, and the GPI(+) population from donor 1 was purified by sorting. Stained cells from patient MSK11 (Fig. 2A) were added to this GPI (+) population at a frequency of 25 per million, and the PNH population is clearly seen in the lower right quadrant of Fig. 2C. This population was recovered by sorting with a yield of 50%, and the collected cells were separated into four aliquots of five cells each, from which DNA was extracted. Exon 2 was amplified and digested with HaeIII. Analysis of the digests on an agarose gel revealed the normal (digested) bands in all four aliquots and the mutant (undigested) band in two of the four aliquots. Thus, although there was some contamination by normal cells, we proved that it was possible to retrieve PIG-A gene mutations from a very small number of PNH cells sorted from a vast excess of normal cells.
Reproducibility of the Assay.
A blood sample from donor 8 was split before granulocyte separation, and two aliquots were stained and analyzed in parallel (Fig. 1 C and D). The patterns obtained were virtually identical, and the frequency of PNH cells in the two samples was 7.5 and 9.6 per million, respectively. From this and similar experiments, we estimate that the reproducibility of our method is of the order of ±15%.
PNH Blood Cells in Healthy Donors.
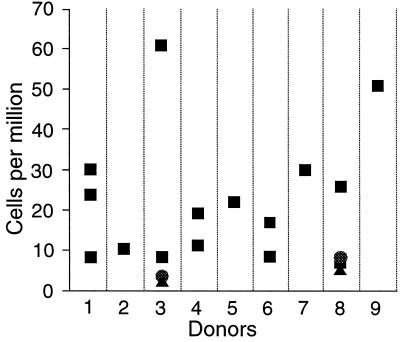
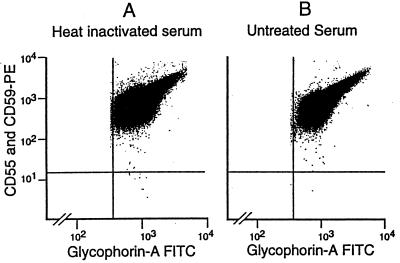
By the techniques described, we found PNH cells in 19 of 19 samples from nine normal donors. The frequency of PNH cells ranged from 10 to 51 per million, with a mean of 22 per million (Fig. 3). Because the aim of our work included identification of PIG-A mutations, and because red cells cannot be used for this purpose, all of this work was carried out on granulocytes. However, PNH red cells can be demonstrated as well. For instance, red cells from donor 8 were washed and incubated as for a Ham test with either untreated serum or heat-inactivated serum, followed by sequential staining with anti-CD55 and anti-CD59, RAM-PE, and glycophorin A–FITC. In the sample incubated with heat-inactivated serum, there was a distinct population of CD55(−) CD59(−) cells, at a frequency of eight per million (Fig. 4A), which is similar to the proportion of PNH granulocytes in this donor. In the sample incubated with untreated serum, this population almost completely disappears, indicating that it has been lysed by complement in the Ham test (Fig. 4B).
Figure 3.
Frequency of PNH granulocytes in nine volunteer donors. Each data point represents the results from a separate blood sample. Three symbols (■, ●, ▴) are used to indicate overlapping data points. PNH cells were identified in all 19 analyses from the donors. The average frequency of PNH cells is 22 per million. The samples from donors 1, 3, 6, and 8 were drawn over a time interval of 6–8 months.
Figure 4.
PNH red blood cells in donor 8. Red cells are positively identified by light scatter characteristics and by gating to include glycophorin A(+) events. (A) Flow analysis of red blood cells incubated in a mock Ham test with heat-inactivated serum. PNH cells are clearly seen in the lower right hand quadrant at a frequency of eight per million. (B) A parallel analysis of red blood cells from donor 8 after incubation in the Ham test with untreated serum. The population in the lower right quadrant is reduced by a factor of 6, confirming that these PNH cells exhibit complement sensitivity.
PIG-A Mutation Analysis.
Reaction conditions for nested PCR amplification of exon 2 and exon 6 were optimized by using DNA from normal granulocytes titrated down to <10 cells per reaction. Overall, 62% of the reactions carried out by using as template DNA from an average of 8 CD59(−) CD55(−) CD11b(+) sorted cells yielded a visible product on an agarose gel. Products amplified from these cells were compared with those amplified from normal cells by SSCP and HA. Shifted bands were observed in 19 of 63 PCR products from exon 2 and in 3 of 27 PCR products from exon 6. The PCR products with shifted bands that appeared more abundant by visual inspection were selected for cloning and sequencing.
Base pair changes predicting single amino acid substitutions were identified in PNH cells from donors 3, 5, 6, and 9 (Table 1). In donor 6, we identified a missense mutation, 55C→T, in all 15 M13 sequences cloned from one PCR product. The same mutation also was seen in four M13 sequences cloned from one PCR product from donor 5. This mutation has been described as an inherited polymorphic mutation that by itself does not interfere with the expression of GPI-linked antigens (22). In our donors, this substitution could not have been inherited because it was found only in PNH cells and not in the respective control cells from these donors.
Table 1.
PIG-A gene mutations identified in PNH cells from volunteer donors
| Donor | Exon* | Mutation† | Amino acid replacement | PCR reactions‡ | M13 clones§ |
|---|---|---|---|---|---|
| 3 | 6 | 1381 C → T | ARG → TRP | 1 | 2 |
| 4 | 2 | 196 Ins AT | FRAMESHIFT¶ | 2 | 7 |
| 5‖ | 2 | 614 T → A | VAL → ASP | 2 | 6 |
| 5 | 2 | 55 C → T | ARG → TRP | 1 | 4 |
| 6 | 2 | 55 C → T | ARG → TRP | 1 | 15 |
| 8 | 2 | 229 C → T | ARG → STOP | 3 | 12 |
| 9 | 2 | 167 T → C | LEU → PRO | 1 | 4 |
*Only exons 2 and 6 were tested.
Numbers indicate position in the cDNA sequence starting with the first coding nucleotide (5).
Number of PCR reactions in which the same mutation was found.
Number of sequenced M13 clones in which the mutation listed was found.
Predicted truncation of protein after amino acid 68.
In this donor, we also observed a T → C substitution in position 30 of intron 2 in five M13 clones from one PCR reaction and A → G substitution in position 31 of intron 2 in six M13 clones from another PCR reaction.
In donor 5, we also found the mutation 614T→A, resulting in a VAL→ASP substitution in exon 2, in a total of five independent M13 clones from one PCR product and one additional M13 clone from a separate PCR product. In donor 3, a mutation in exon 6, 1381C→T, causing an ARG→TRP substitution was identified in two independent M13 clones from one PCR product. In this initial sample, the frequency of PNH cells was four per million. In a sample taken 51 days later, the 1381C→T mutation was not found, but the frequency of PNH cells was still three per million. In donor 9, a mutation in exon 2, 167T→C, resulting in a LEU→PRO substitution, was identified in four separate M13 clones from one PCR product.
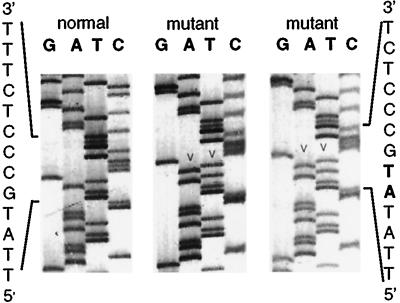
In donor 4, a 2-bp insertion/duplication at position 196, identified in seven independent M13 clones, resulted in a frameshift in exon 2 (Fig. 5). An oligonucleotide primer (5′GGGTGACAATTATAACCACAT3′) was used to set up a customized amplification-created restriction site assay, such that mutant sequences are cut by NdeI and normal sequences are uncut. PNH cells then were isolated from samples taken from this donor at 164 and 192 days after the initial sample, and exon 2 was amplified and reanalyzed by amplification-created restriction site assay in the presence of 32P on an acrylamide gel. At 164 days, 1 of 13 PCR products demonstrated a faint band indicating digestion by NdeI. This mutation was not detected by amplification-created restriction site assay in sorted GPI(+) cells, indicating that the mutation was confined to the cell population identified as PNH-like by flow cytometry. At 192 days, none of seven PCR products from PNH cells demonstrated any digestion by NdeI. We conclude that the stem cell clone bearing this mutation had stopped producing mature granulocytes by the time the third sample was obtained.
Figure 5.
Insertion 196AT in PNH granulocytes from donor 4. Sequence analysis of the noncoding strand is shown for one normal and two representative mutant M13 clones. The insertion/duplication at position 196 (arrow heads, bold type) results in a frameshift and premature chain termination.
In donor 8, a nonsense mutation in exon 2 (229C→T) was identified in a total of eight independent M13 clones isolated from two separate PCR products amplified from sorted PNH cells collected from an initial blood sample. In this sample, the frequency of PNH cells was 26 per million. This mutation eliminates a TaqI restriction site and is thus readily identified, yet it was not seen, as expected, in any of 20 PCR products arising from sorted GPI(+) or unseparated cells. The same mutation was identified again in all M13 clones from 1 of 12 PCR products, each one using as template DNA from 10 PNH granulocytes isolated from a blood sample collected 65 days after the initial sample. In this sample, the frequency of PNH cells was seven per million. In a third blood sample obtained 174 days after the initial sample, this mutation was not found in any of 12 PCR products, despite the continued presence of a distinct PNH population by flow cytometry at a frequency of nine per million. We infer that the clone with the 229C→T mutation is gradually disappearing.
DISCUSSION
We have identified rare PNH granulocytes from normal individuals and PIG-A mutations in them. In so doing, we have confirmed an important prediction of our model of PNH: Namely, that PIG-A mutations by themselves are not sufficient for the development of the disease. Previous work by others (refs. 23 and 24; P. Hillmen, personal communication) has led to the demonstration of GPI(−) lymphocytes in patients who did not have PNH, and who—as part of anti-lymphoma treatment—were administered CAMPATH, an antibody directed against the GPI-linked protein CD52. It is not known whether hematopoietic cells, in addition to lymphocytes, were involved in those patients. In our study, we have demonstrated the occurrence in normal people of PNH granulocytes and red cells, which must arise from the same sort of precursors from which such cells are derived in patients with PNH.
Methodology for Identifying Rare PIG-A Mutant Cells.
It is well known that artifactual single base changes (usually A→G or T→C) turn up in PCR products (25), but we have shown specifically with respect to PIG-A that nesting the PCR reactions does not increase the rate of artifact and that artifacts are extremely unlikely to be seen in more than one M13 clone (21). We have therefore included in our results only mutations identified in at least two independent M13 clones. In three cases (donors 4, 5, and 8), we have gone further and in fact have identified the same mutation in more than one separate PCR product (Taq polymerase errors would not be likely to be identical in multiple PCR reactions). In addition, a dinucleotide insertion, such as that seen in donor 4, could hardly be produced by a polymerase error at all, whereas it is precisely the type of mutation found commonly in patients with PNH (7–8).
Although we found PIG-A mutations in most donors, the proportion of mutations detected in this study represents a minimum estimate of all those present for several reasons: (i) We have chosen to amplify only exons 2 and 6 because that is where most mutations in PNH patients are found (7); however, some of our donors may have had mutations in other exons or in noncoding regions; (ii) any deletions or rearrangements involving regions to which the primers are annealed would be missed; and (iii) the sensitivity of SSCP/HA is ≈80% (26). Thus, PIG-A mutations may be even more common that we have demonstrated.
Significance of PIG-A Mutations.
Of the PIG-A mutations that we have identified (see Table 1), at least two (a nonsense mutation and a frameshift mutation in exon 2) must be inactivating PIG-A function by truncation of the protein; these mutations are typically representative of those most frequently encountered in PNH (7–8). The missense mutations in donors 3, 5, and 9 are also likely to interfere with PIG-A function, as they entail fairly drastic amino acid replacements. By contrast, we know that the ARG→TRP substitution in amino acid position 19 does not by itself result in the PNH phenotype (7, 8, 22). However, it is interesting that the corresponding mutation, a known inherited polymorphism in the PIG-A gene, was observed in as many as five patients with PNH (refs. 7 and 8; data not shown) and now in two normal donors as a somatically acquired mutation. It seems reasonable to infer that this site is a mutational “hot spot.” It is also conceivable that this mutation could cause the PNH phenotype in conjunction with some other mutation that we have not yet identified.
The Lifespan of PIG-A Mutant Clones.
Although rare PNH cells were found in all samples, we have documented in donors 3,4, and 8 that individual PIG-A mutant clones are transient. The mutation from donor 3 could no longer be found after 51 days. In donor 4, the 196Ins AT mutation was still found 164 days after but not 192 days after the initial sample. In donor 8, the 229C→T mutation was found again at 65 days but not at 174 days after the initial sample. Thus, the PIG-A mutant clone can continue to produce mature granulocytes for at least up to 164 days. Because these clones are likely to have existed for some time before their detection, this represents a minimum estimate of their lifespan. The persistence of a small PNH population revealed by flow cytometry in these donors could be explained if new mutant clones were to arise by somatic mutation as old clones die out. Longer-term follow-up of these donors will be needed to determine whether there is a sort of steady state in the turnover of these clones. In those donors in whom we were able to obtain longitudinal data (donors 1, 3, 4, 6, and 8 in Fig. 3), the frequency of PNH cells generally varies within a fairly narrow range.
PNH Clones in Normal Donors and the Pathophysiology of PNH.
The findings we have reported confirm our hypothesis that, in PNH patients, there must exist a second mechanism, beyond PIG-A mutations, promoting the expansion of PNH clones. The transient nature of individual PIG-A mutant clones in our donors indicates that these clones have no selective advantage in a normal marrow environment. Thus, it does not seem that the PIG-A mutation by itself results in impaired apoptosis (27), consistent with the recent suggestion that impaired apoptosis may be instead a feature of bone marrow failure (28, 29).
Our findings point, by contrast, to an abnormal marrow environment as being required for expansion of the PNH clone in patients with the disease (14, 20, 17). Of course, we cannot formally rule out the possibility that, in patients with PNH, unlike in our donors, the PNH clone expands because of the coexistence of a somatic mutation in another gene or because of some special property of the cell in which the PIG-A mutation occurs. Such explanations have been invoked to explain how normal individuals without lymphoma or leukemia can harbor t(14;18) translocations (30) or bcr-abl translocations (31). However, PNH behaves very differently from a malignancy. Leukemic transformation, though documented (32), is very rare (13), and, conversely, spontaneous long-term remissions are more frequent (13). In addition, allogeneic bone marrow transplantation following a nonmyeloablative conditioning regimen can result in complete remission, unlike for other clonal disorders (33). These observations strongly suggest that the second pathogenetic mechanism is not intrinsic to the PNH clone. Reports of multiple PNH clones in the same patient are also consistent with a mechanism extrinsic to the PNH clone (21, 34–36). Given the association between PNH and AAA, this mechanism is, in all likelihood, an autoimmune reaction that selectively spares the PNH stem cell (14, 20, 18, 17). The GPI(−) phenotype could protect the PNH clone if, for example, normal GPI-anchor assembly were to be important for the presentation of the autoantigen or the expression of an adhesion or costimulatory molecule.
The Rate of Somatic Mutations in Hematopoietic Stem Cells.
In this study, we have found PNH granulocytes at an average frequency of 22 per million in nine donors and PNH red blood cells at a frequency of 8 per million in one donor. This frequency is of the same order of magnitude as the estimated mutation frequency in normal individuals in Hprt (2–10 per million), glycophorin A (10 per million), and HLA (30 per million) (reviewed in ref. 37). We believe that, quite apart from its relevance to the pathogenesis of PNH, the analysis of the PIG-A gene may serve for the detection and quantitation of somatic mutations in hematopoietic cells in general. From this point of view, there are at least four advantages: (i) The PIG-A gene is X-linked; therefore, one mutation is sufficient to abolish or drastically reduce the amount of the gene product; (ii) because the PIG-A gene product is part of an enzyme, it acts catalytically; therefore, the effect of mutations in the gene are amplified; (iii) because many proteins are able to acquire the GPI anchor, further amplification of the effect of the mutation consists in affecting not just one but numerous proteins; and (iv) because GPI-anchored proteins are membrane proteins, they can be conveniently detected, and mutant cells can be sorted, as we have shown in this work. This method may complement other existing assays for occult somatic mutations, and it may help to assess mutational load in hematopoietic stem cells in relation to either an inherited or environmentally induced increase in mutation rate.
Acknowledgments
With this paper we pay tribute to the memory of our young co-author, “Kriang,” who sadly died in an airplane accident just before our manuscript was submitted. We thank Patrick Anderson, Tom Delohery, Jason Angell, and Jianlin Xu for their technical assistance, Valerie Charles for help with the manuscript, the blood donors who contributed samples, and the late Dr. John Lindenbaum for his help and encouragement. We also thank Drs. Monica Bessler, Anastasios Karadimitris, and Rosario Notaro for their assistance and advice. This work was supported by the Aplastic Anemia Foundation of America New Researcher Award, the Norman and Rosita Winston and the Charles A. Dana Foundations, and National Institutes of Health Grants 5K-12 CA01712-04 and 5RO1 HL56778-03.
ABBREVIATIONS
- PNH
paroxysmal nocturnal hemoglobinuria
- AAA
acquired aplastic antemia
- GPI
glycosylphosphatidylinositol
- SSCP
single-strand conformation polymorphism
- HA
heteroduplex analysis
- RAM-PE
F(ab′)2 fragment of rabbit antimouse immunoglobulin conjugated to R-phycoerythrin
Note Added in Proof
Since submitting this paper, we have analyzed granulocytes from one additional volunteer, a 61-year-old man being phlebotomized for hemochromatosis, and we found PNH cells at a frequency of 9 per million. In three independent PCR reactions (each amplified from approximately five sorted PNH cells) we identified a C → A mutation at position 294 in exon 2, which introduces a stop in codon 98. This was confirmed in a second sample taken 8 weeks later. Of interest, this same mutation has been previously reported in a patient with PNH (38). Thus, the very same PIG-A mutation that causes PNH in one person does not cause PNH in another person.
Footnotes
Preliminary results on part of this work were presented at the 40th Meeting of the American Society of Hematology, Dec. 5–8, 1998, Miami Beach, FL, Abstract 1943.
References
- 1.Oni S-B, Osunkoya B-O, Luzzatto L. Blood. 1970;36:145–152. [PubMed] [Google Scholar]
- 2.Ham J-T, Dingle J-H. J Clin Invest. 1939;80:7–12. doi: 10.1172/JCI101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosse W-F, Ware R-E. Blood. 1995;86:3277–3286. [PubMed] [Google Scholar]
- 4.Terstappen L-W, Nguyen M, Huang S, Lazarus H, Medof M-E. Br J Hematol. 1993;84:504–514. doi: 10.1111/j.1365-2141.1993.tb03108.x. [DOI] [PubMed] [Google Scholar]
- 5.Miyata T, Yamada N, Iida Y, Nishimura J, Takeda J, Kitani T, Kinoshita T. N Engl J Med. 1994;330:249–255. doi: 10.1056/NEJM199401273300404. [DOI] [PubMed] [Google Scholar]
- 6.Bessler M, Mason P-J, Hillmen P, Miyata T, Yamada N, Takeda J, Luzzatto L, Kinoshita T. EMBO J. 1994;13:110–117. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nafa K, Mason P-J, Hillmen P, Luzzatto L, Bessler M. Blood. 1995;86:4650–4655. [PubMed] [Google Scholar]
- 8.Nafa K, Bessler M, Castro-Malaspina H, Jhanwar S, Luzzatto L. Blood Cells Mol Dis. 1998;24:370–384. doi: 10.1006/bcmd.1998.0203. [DOI] [PubMed] [Google Scholar]
- 9.Maciejewski J-P, Sloand E-M, Sato T, Anderson S, Young N S. Blood. 1997;89:1173–1181. [PubMed] [Google Scholar]
- 10.Rosti V, Tremml G, Soares V, Pandolfi P-P, Luzzatto L, Bessler M. J Clin Invest. 1997;100:1028–1036. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, Kinoshita T, Takeda J. Blood. 1996;87:3600–3606. [PubMed] [Google Scholar]
- 12.Dunn D-E, Yu J, Nagarajan S, Devetten M, Weichold F-F, Medof M-E, Young N-S, Liu J-M. Proc Natl Acad Sci USA. 1996;93:7938–7943. doi: 10.1073/pnas.93.15.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillmen P, Lewis S-M, Bessler M, Luzzatto L, Dacie J. N Engl J Med. 1995;33:1253–1258. doi: 10.1056/NEJM199511093331904. [DOI] [PubMed] [Google Scholar]
- 14.Rotoli B, Luzzatto L. Baillieres Clin Haematol. 1989;2:113–138. doi: 10.1016/s0950-3536(89)80010-1. [DOI] [PubMed] [Google Scholar]
- 15.Dacie J-V, Lewis S-M. Ser Haematol. 1972;5:3–23. [PubMed] [Google Scholar]
- 16.Rotoli B, Robledo R, Luzzatto L. Blood. 1982;60:157–159. [PubMed] [Google Scholar]
- 17.Luzzatto L, Bessler M, Rotoli B. Cell. 1997;88:1–4. doi: 10.1016/s0092-8674(00)81850-4. [DOI] [PubMed] [Google Scholar]
- 18.Young N-S, Maciejewski J. N Engl J Med. 1997;336:1365–1372. doi: 10.1056/NEJM199705083361906. [DOI] [PubMed] [Google Scholar]
- 19.Stoppa A-M, Vey N, Sainty D, Arnoulet C, Camerlo J, Cappiello M-A, Gastaut J-A, Maraninchi D. Br J Hematol. 1996;93:42–44. doi: 10.1046/j.1365-2141.1996.453993.x. [DOI] [PubMed] [Google Scholar]
- 20.Young N-S. Blood. 1992;79:1385–1392. [PubMed] [Google Scholar]
- 21.Nafa K, Bessler M, Deeg H-J, Luzzatto L. Blood. 1998;92:3422–3427. [PubMed] [Google Scholar]
- 22.Endo M, Ware R-E, Vreeke T-M, Howard T-A, Parker C-J. Br J Haematol. 1996;93:590–593. doi: 10.1046/j.1365-2141.1996.d01-1701.x. [DOI] [PubMed] [Google Scholar]
- 23.Taylor V-C, Sims M, Brett S, Field M-C. Biochem J. 1997;322:919–925. doi: 10.1042/bj3220919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertenstein B, Wagner B, Bunjes D, Duncker C, Raghavachar A, Arnold R, Heimpel H, Schresenmeier H. Blood. 1995;86:1487–1492. [PubMed] [Google Scholar]
- 25.Kaeohavong P, Thilly W-G. Proc Natl Acad Sci USA. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grompe M. Nat Genet. 1993;5:111–117. doi: 10.1038/ng1093-111. [DOI] [PubMed] [Google Scholar]
- 27.Brodsky R-A, Vala M-S, Barber J-P, Medof M-E, Jones R-J. Proc Natl Acad Sci USA. 1997;94:8756–8760. doi: 10.1073/pnas.94.16.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ware R-E, Nishimura J, Moody M-A, Smith C, Rosse W, Howard T-A. Blood. 1998;92:2541–2550. [PubMed] [Google Scholar]
- 29.Horikawa K, Nakakuma H, Kawaguchi T, Iwamoto N, Nagakura S, Kagimoto T, Takatsuki K. Blood. 1997;90:2716–2722. [PubMed] [Google Scholar]
- 30.Liu Y, Hernandez A-M, Shibata D, Cortopassi G-A. Proc Natl Acad Sci USA. 1994;91:8910–8914. doi: 10.1073/pnas.91.19.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose S, Deininger M, Gora-Tybor J, Goldman J-M, Melo J-V. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 32.Stafford H-A, Nagarajan S, Weinberg J-B, Medof M-E. Br J Haematol. 1995;89:72–78. doi: 10.1111/j.1365-2141.1995.tb08908.x. [DOI] [PubMed] [Google Scholar]
- 33.Antin J-H, Ginsburg D, Smith B-R, Nathan D-G, Orkin S-H, Rappeport J-M. Blood. 1985;66:1247–1250. [PubMed] [Google Scholar]
- 34.Bessler M, Mason P, Hillmen P, Luzzatto L. Lancet. 1994;343:951–953. doi: 10.1016/s0140-6736(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 35.Nishimura J, Inoue N, Wada H, Ueda E, Pramoonjago P, Hirota T, Machii T, Kageyama T, Kanamaru A, Takeda J, et al. Blood. 1997;89:3470–3476. [PubMed] [Google Scholar]
- 36.Ostendorf T, Nischan C, Schubert J, Grussenmeyer T, Scholz C, Zielinska-Skowronek M, Schmidt R-E. Blood. 1995;85:1640–1646. [PubMed] [Google Scholar]
- 37.Albertini R-J, Nicklas J-A, O’Neill P-O, Robinson S-H. Annu Rev Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 38.Savoia A, Ianzano L, Lunardi C, De Sandre G, Carotenuto M, Musto P, Zelante L. Hum Genet. 1996;97:45–48. doi: 10.1007/BF00218831. [DOI] [PubMed] [Google Scholar]