Abstract
CCR5 and CXCR4 are the major HIV-1 coreceptors for R5 and X4 HIV-1 strains, respectively, and a threshold number of CD4 and chemokine receptor molecules is required to support virus infection. Therefore, we used a quantitative fluorescence-activated cell sorting assay to determine the number of CD4, CCR5, and CXCR4 antibody-binding sites (ABS) on various T cell lines, T cell subsets, peripheral blood dendritic cells (PBDC), and monocyte-derived macrophages by using four-color fluorescence-activated cell sorting analysis on fresh whole blood. Receptor levels varied dramatically among the various subsets examined and typically varied from 2- to 5-fold between individuals. CCR5 was expressed at much higher levels in CD4+/CD45RO+/CD62L-true memory cells compared with CD4+/CD45RO+/CD62L+ cells. Fresh PBDC had the highest number of CCR5 ABS among the leukocyte subsets examined but had few CXCR4 ABS, affording a strategy for sort-purifying PBDC. In vitro maturation of PBDC resulted in median 3- and 41-fold increases in CCR5 and CXCR4 ABS, respectively. We found that macrophage colony-stimulating factor caused the greatest up-regulation of both CCR5 and CXCR4 on macrophage maturation (from ≈5,000 to ≈50,000 ABS) whereas granulocyte-macrophage colony-stimulating factor caused a marked decrease of CXCR4 (from ≈5,000 ABS to <500) while up-regulating CCR5 expression (from ≈5,000 to ≈20,000 ABS). Absolute ABS for CD4 and the major HIV-1 coreceptors serve as a more quantitative measure of cell surface expression, and we propose that this be used for future studies looking at the modulation of CD4 or chemokine receptor expression by cytokines, HIV-1 infection, or receptor polymorphisms.
HIV-1 entry into cells requires sequential interactions between envelope (Env), CD4, and a coreceptor (1–3). Epidemiological and experimental evidence indicates that CD4 and coreceptor levels affect the efficiency of viral entry and that this may have consequences for the pathogenesis of HIV disease. Individuals homozygous for the Δ32-ccr5 allele have no surface expression of CCR5 and are highly protected against HIV-1 infection, whereas Δ32-ccr5 heterozygotes have lower CCR5 expression levels and progress to AIDS more slowly than individuals without this allele (reviewed in ref. 4). Individuals homozygous for a mutation in the SDF-1 gene also progress more slowly to clinical AIDS (5), perhaps because of increased expression of SDF-1 and modulation of CXCR4 expression. Indeed, in vitro studies have shown that CD4, CCR5, and CXCR4 expression levels impact the efficiency of viral entry (6–8).
Chemokine receptor expression in both peripheral blood lymphocytes and monocyte-derived macrophages (MDM) is sensitive to cytokine-mediated modulation (reviewed in ref. 9). Because the presence of CD4 and either CCR5 and/or CXCR4 on specific leukocytes and MDMs designates these cells as potentially susceptible targets for viral infection, it is important to determine quantitatively the amount of CD4 and the major coreceptors present on various leukocyte and monocyte subpopulations to help clarify the roles these cells may play in the dynamics of viral replication in vivo and to rigorously address the effects of cytokines on coreceptor expression. In this report, we used a quantitative fluorescence-activated cell sorting (QFACS) assay that relies on a series of precalibrated beads that can bind a fixed number of mouse IgG molecules to determine the absolute number of CD4 and coreceptor molecules on the surface of numerous leukocyte subsets, MDMs, and peripheral blood dendritic cells (PBDC). By using this approach, we found great variation in chemokine receptor expression in T cell lines and lymphocyte subsets, in immature versus mature dendritic cells (DC), and in MDM depending on culture conditions. These results provide insight into the types of cells most susceptible to infection by R5 and X4 viruses and an understanding of the discrepancies in the literature regarding CD4 and coreceptor expression in cultured MDM.
MATERIALS AND METHODS
Cell Lines and Infection Studies.
All cell lines were obtained from the American Type Culture Collection or the National Institutes of Health AIDS Reference and Reagent Program (GHOST cells). All cell lines were maintained according to the supplier’s recommendations. Pseudotyped luciferase reporter viruses were used for infection studies as described (10).
Antibodies.
Phycoerythrin-conjugated anti-CD4 (Q4120) was obtained from Sigma. Allophycocyanin-conjugated anti-CD4 (S3.5), anti-CD8 (3B5), anti-HLA-DR (TU36), FITC-conjugated anti-CD11c, and tricolor-conjugated anti-CD3, anti-CD14 (Tuk4), anti-CD16 (3G8), anti-CD19 (SJ35-C1), anti-CD45RA (MEM56), anti-CD45RO (UCHL1), anti-CD56 (NKI-nbl-1), anti-CD62L (DREG-56), anti-CD83 (HB15), and anti-HLA-DR (TU36) were obtained from Caltag (South San Francisco, CA). Cychrome-conjugated anti-CD26, phycoerythrin-conjugated anti-CCR5 (2D7), and anti-CXCR4 (12G5) were obtained from PharMingen. FITC-conjugated CD1a (B-B5) was obtained from BioSource International (Camarillo, CA).
FACS Strategy.
We used phycoerythrin (PE)- and allophycocyanin (APC)-conjugated mAbs for quantification because they do not self-quench at high density (11, 12). Tricolor (Tri) and FITC were the two other fluorochromes used in our four-color FACS analysis. For peripheral blood mononuclear cells (PBMCs), the following panels were used for each donor: (i) CD62L-FITC, CCR5-PE, CD45RO-Tri, CD4-APC; (ii) CD62L-FITC, CXCR4-PE, CD45RA-Tri, CD4-APC; (iii) CD19-FITC, CCR5-PE, CD56-Tri, CD4-APC; (iv) CD19-FITC, CXCR4-PE, CD56-Tri, CD8-APC; (v) CD4-FITC, CCR5-PE, CD26-Tri, HLA-DR-APC; and (vi) CD8-FITC, CXCR4-PE, CD26-Tri, HLA-DR-APC. The strategy for identifying PBDC is illustrated in Fig. 3. For quantification of CD4 and coreceptors on PBDC, the following panels were used on each donor: (i) CD11c-FITC, CCR5-PE, CD3/16/56/19/14-Tri, HLA-DR-APC; (ii) CD11c-FITC, CXCR4-PE, CD3/16/56/19/14-Tri, HLA-DR-APC; and (iii) CD11c-FITC, CD4-PE, CD3/16/56/19/14-Tri, HLA-DR-APC.
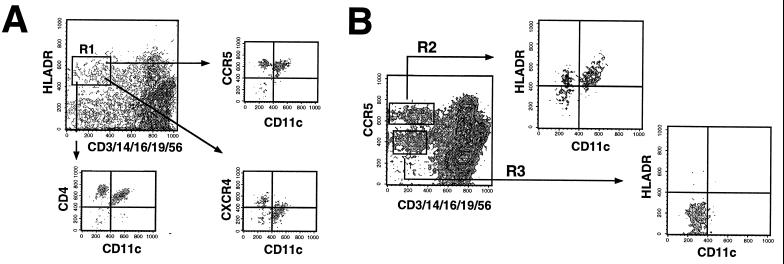
Figure 3.
FACS gating strategy for identifying fresh PBDC. Fresh whole blood was processed as in Fig. 2. (A) Tri-color-conjugated CD3, CD14, CD16, CD19, and CD56 was used to exclude lineage-positive cells. Lineage-negative cells that were HLA-DR+ (R1 gate) were identified as candidate PBDC. R1-gated cells were then analyzed for CD11c vs. CCR5, CXCR4, or CD4 expression to obtain values for ABS. CD1a was alternatively used to analyze R1-gated cells and gave similar profiles to that obtained with CD11c expression (data not shown). (B) A proposed strategy for identifying PBDC. An analysis of lineage markers vs. CCR5 expression reveals two distinct populations of lineage-negative cells that are either CCR5-high (R2 gate) or CCR5-low (R3 gate). R2-gated cells are almost exclusively HLA-DR+, i.e., candidate DCs, which can be either CD11c+(51.2 ± 10.5%) or CD11c− (48.8 ± 10.5%) whereas R3 gated cells are exclusively HLA-DR− and CD11c−.
QFACS.
QFACS was performed by converting the mean channel fluorescence into antibody-binding sites (ABS) by using a standardized microbeads kit (Sigma). This is a mixture of five microbead populations of uniform size, coated with goat anti-mouse antibodies, that have differing abilities to bind mouse antibodies (one population does not bind mouse IgG and is used as a control). Q4120, PE-conjugated anti-CD4 (7 μl); 2D7, PE-conjugated anti-CCR5 (12.5 μl); and 12G5, PE-conjugated anti-CXCR4 (10 μl), were added at saturating amounts to ≈100,000 beads. Beads were incubated with the same concentration of mAb for 1 hour and processed identically as the samples being quantitated. The binding capacities of the stained microbeads were then regressed against the corresponding geometric mean of each bead population, and the MFI of the antigen analyzed on the sample cells was converted to ABS per cell by comparison with the regression curve generated. The MFI of the isotype control for each experiment was converted to ABS and subtracted from the ABS value obtained with the experimental sample. The parameters of the regression curve permit a determination of the linear deviation and hence provide an estimate of the degree of confidence one should have in the values generated. Regression curves were only acceptable if r2 >0.995 and the deviation from linearity was <5%. Additional details on the relationship between ABS and mean fluorescence intensity values can be obtained from the manufacturer (Sigma).
Statistics.
Student’s t test was used to determine any significant differences between expression levels among the various cell types. The simultaneous analysis of multiple markers on the same donor allowed a paired t test two-tailed distribution analysis to be applied on analysis of the leukocyte subsets. For analysis of expression levels on macrophages, an unpaired t test (two-sample unequal variance) was used because data on time points from six to eight different donors from five independent experiments were combined in the analysis.
RESULTS
Choice of Antibodies Used for Quantitative Studies.
Seven transmembrane domain receptors may exist in multiple conformational states, which can affect exposure of specific antigenic epitopes (16). Because QFACS determines the number of ABS on a cell rather than the physical number of cell surface molecules, we selected mAbs that efficiently block Env–receptor interactions. mAb Q4120 competes with gp120 binding on CD4 (13) whereas the epitopes recognized by 2D7 and 12G5 overlap with both the chemokine and HIV-1 gp120 binding sites on CCR5 and CXCR4, respectively (14, 15). Furthermore, in our recent analysis of the antigenic structure of CCR5, we found that 2D7 reacts with CCR5 more efficiently than any of the 18 mAbs examined (16).
Quantification of CD4, CCR5, and CXCR4 on T Cell Lines.
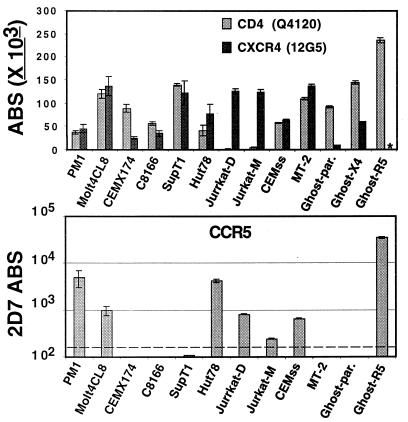
There was marked variation of CD4 and CXCR4 levels among T cell lines commonly used to propagate HIV-1 (Fig. 1). CD4 levels ranged from <1,000 ABS on Jurkat cells to >200,000 ABS on GHOST-CCR5 cells. CXCR4 levels ranged from 20,000 ABS on CEMx174 cells to ≈150,000 ABS on Molt4 clone-8 and MT-2 cells. CCR5 was present on PM1 cells at levels equivalent to that on relevant T lymphocyte subsets (≈7,000 ABS, see Fig. 2). Low levels of CCR5 were also found on Molt4 clone-8, Hut78, Jurkat, and CEMss cells. Of these, we found that Molt4 clone-8, Jurkat-D and CEMss cells were all infectable, albeit inefficiently, by R5 Env (HIV-1 ADA)-pseudotyped luciferase reporter viruses in a 2D7 inhibition-sensitive, CCR5-dependent manner (data not shown). However, the relative infectabilities of these cell lines did not correlate with either their CD4 or coreceptor levels. The QFACS approach allowed data to be computed from multiple experiments without being dependent on the subjective interpretations of slight shifts in MFI. Furthermore, the value for CXCR4 ABS obtained for Jurkat cells was similar to SDF-1 binding sites previously determined by using Scatchard analysis (17).
Figure 1.
CD4, CXCR4, and CCR5 ABS on T cell lines. (Upper) The ABS for CD4 and CXCR4. ∗CXCR4 ABS were not determined for the GHOST-R5 cell line. (Lower) The ABS for CCR5. Jurkat-D and Jurkat-M refers to different sources of Jurkat cells from two independent labs. The dashed line (Lower) indicates the threshold of detection. Experiments were performed at least three independent times, and data are presented as absolute number of ABS ±SEM.
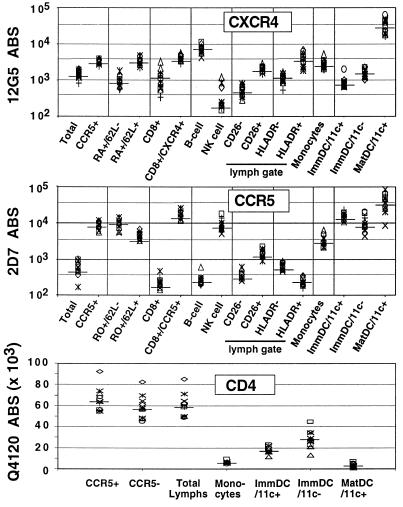
Figure 2.
CXCR4, CCR5, and CD4 ABS on PBMC. Four-color FACS analysis was used to identify the various subpopulations of PBMC indicated. Fresh whole blood was stained with fluorochrome-conjugated mAb immediately after venupucture, and FACS analysis was performed after ammonium chloride-mediated selective red blood cell lysis. B cells are CD19+, NK cells are CD56+, monocytes are the cluster of cells with high forward scatter and low side-scatter profiles (independently confirmed to be CD14+), and ImmDC are immature PBDC (Fig. 3), which can be CD11c+ or CD11c−. MatDC are mature DC from Ficoll-purified PBMC after 24 hours of culture in RPMI medium 1640 with 10% FCS. MatDC are almost exclusively (>95%) CD11c+. ABS for CXCR4 and CCR5 are indicated on a log scale while that for CD4 is indicated on a linear scale. Median values are indicated by a solid bar across each data group. Data is compiled from simultaneous analysis of 12 donors.
CD4, CCR5, and CXCR4 Levels on Fresh Peripheral Blood Leukocytes.
Various cytokines and cell purification protocols can affect coreceptor expression levels. Therefore, we determined the number of CD4, CCR5, and CXCR4 ABS on fresh unstimulated PBMC from healthy donors (wild-type for CCR5) to determine the basal levels of coreceptor expression before any exogenous treatment. Four-color FACS analysis was performed on stained whole blood after selective red blood cell lysis, thus avoiding Ficoll purification, which can acutely affect chemokine receptor expression (10). Table 1 shows the mean percent CCR5/CXCR4-positive cells and the median CCR5/CXCR4 ABS on each leukocyte subset. In general, CCR5 and CXCR4 levels varied from 2- to 5-fold between donors, depending on the subset examined. In the lymphocyte gate and its various subsets, CCR5 could usually be gated into distinctly positive and negative populations. In contrast, the expression of CXCR4 was heterogenously spread through the lymphocyte gate and many of its subsets. Therefore, the ABS values for CCR5 and CXCR4 on both total lymphocytes and on the CXCR4- or CCR5-positive gates are shown (CXCR4+ or CCR5+, see Fig. 2 and Table 1). Note that although CXCR4 was expressed on a higher percentage of total lymphocytes compared with CCR5 (mean 69.6% vs. 14.1%), the expression of CCR5 on the CCR5-positive gate (median 8,345 ABS) was higher than CXCR4 (median 3,387 ABS) on the corresponding CXCR4-positive gate.
Table 1.
Comparison of percent CCR5/CXCR4-positive cells with number of 2D7 (CCR5) and 12G5 (CXCR4) ABS
| Cell | Mean CCR5 positive, % (range) | Mean CXCR4 positive, % (range) | Median 2D7 ABS (range) | Median 12G5 ABS (range) |
|---|---|---|---|---|
| Total lymph gated | 14.1 (5.7–24.1) | 69.6 (51.7–80.1) | 593 (167–1,006) | 1,572 (834–1,961) |
| Gated (CCR5+ or CXCR4+ cells only) | 8,345* (5,606–11,933) | 3,387* (2,498–3,759) | ||
| Monocytes | 47.0 (24.2–90.2) | 70.9 (55.0–95.5) | 2,875 (2,117–6,526) | 2,491 (2,015–5,113) |
| CD4+/CD45RO+/CD62L− (true memory) | 74.9 (50.8–92.7) | 36.8 (26.0–51.5) | 9,576* (5,416–14,626) | 505 (348–655) |
| CD4+/CD45RO+/CD62L+ | 48.5 (29.0–82.9) | 63.9 (50.1–73.2) | 4,741* (3,246–6,802) | 1,013 (653–1,377) |
| CD4+/CD45RA+/CD62L− | 0‡ | 40.8 (16.6–70.8) | 0† | 902 (554–1,762) |
| CD4+/CD45RA+/CD62L+ (true naive) | 2.1 (0.3–9.3) | 91.3 (82.9–96.6) | 0† | 3,386 (2,233–4,644) |
| CD8+ (total lymph) | 26.0 (15.4–39.9) | 54.9 (24.3–84.1) | 216 (141–474) | 1,030 (344–3,272) |
| Gated (CD8+/CCR5+ or CD8+/CXCR4+) | 16,082* (11,907–25,783) | 4,033* (2,915–5,680) | ||
| NK Cells (CD56+) | 22.2 (7.9–49.3) | 21.0 (2.6–58.9) | 7,223* (5,065–18,037) | 229 (152–1,189) |
| B cells (CD19+) | 4.2 (1.1–7.8) | 91.0 (80.9–98.2) | 245 (189–605) | 7,402 (4,227–10,941) |
| CD26− | 29.3 (18.4–47.3) | 54.7 (36.7–67.6) | 325 (259–607) | 546 (289–791) |
| CD26+ | 64.1 (45.0–76.1) | 94.9 (92.3–96.6) | 1,400 (888–2,203) | 2,130 (1,508–2,784) |
| HLADR− | 42.2 (33.4–53.5) | 77.9 (62.2–87.3) | 571 (469–908) | 1,035 (545–1,668) |
| HLADR+ | 19.2 (6.5–25.4) | 88.6 (79.1–96.3) | 228 (159–349) | 4,089 (1,915–6,734) |
| Immature DC/CD11c+ | 96.1 (93.2–98.7) | 17.1 (7.92–47.2) | 15,620 (10,756–23,200) | 800 (681–1,962) |
| Immature DC/CD11c− | 81.6 (69.7–92.0) | 47.3 (36.7–60.3) | 8,794 (4,390–20,511) | 1,511 (1,031–2,259) |
| Mature DC | 96.3 (87.6–100) | 96.9 (93.1–100) | 44,647 (8,584–84,737) | 35,175 (16,598–62,375) |
Percent positive gate is defined as the histogram gate giving <2% positive cells using an isotype control. Leukocyte subsets that can be distinctly gated into coreceptor-positive or -negative populations are indicated (∗), and the corresponding ABS values were calculated based on the gated positive population only. Values are given with background already substracted.
Always less than threshold of detection.
No appreciable CD4+ cells in CD45RA+/CD62L−gate.
We confirmed that CCR5 and CXCR4 were expressed predominantly on CD4+/CD45RO+ and CD4+/CD45RA+ cells, respectively (refs. 18 and 19; Fig. 2 and Table 1). Analysis of these memory and naive T cells revealed that CCR5 was more highly expressed on CD45RO+/CD62L− (true quiescent memory cells, median 9,576 ABS) compared with CD45RO+/CD62L+ cells (median 4,741 ABS), whose nature is presently unclear. Conversely, CXCR4 was expressed at significantly higher levels on CD45RA+/CD62L+ cells (true naive cells, median 3,386 ABS) compared with CD45RA+/CD62L− (median 901 ABS) cells that may be memory revertants. CCR5 was not expressed on CD45RA+ (naive) cells, and CXCR4 was expressed at significantly lower amounts on total CD45R0+ memory cells compared with CD45RA+ naive cells (P < 0.001).
Significant amounts of CXCR4 were expressed on CD19+ B cells (≈7,500 ABS) with almost undetectable levels present on CD56+ NK cells (<250 ABS). The converse was true for CCR5 on B cells (≈250 ABS) and NK cells (≈7,000 ABS). CD26 and HLA-DR, considered as markers for acute activation (20), were used to determine whether coreceptor expression differed depending on the activation status of the cell. Although CD26 is not a specific marker for T cell activation, our results show that both CCR5 and CXCR4 were expressed at significantly higher levels in CD26+ vs. CD26− lymphocytes (P < 0.0001 for both). CCR5 and CXCR4 also were differentially expressed on HLA-DR+ and HLA-DR− cells, with CCR5 being lower and CXCR4 higher on HLA-DR+ cells (P < 0.0001 for both cases). However, because HLA-DR and CD26 expression may coincide with CD45 isoform expression (memory vs. naive), in addition to being markers of different activation states (i.e., expression of HLA-DR and CD26 are almost mutually exclusive), the relevance of these subset differences in CCR5 and CXCR4 expression awaits further refined analysis.
CD4 levels were uniform among lymphocyte subsets that were CD4+. Of interest, CD4 ABS were higher on CCR5+ than CCR5− lymphocytes from all 12 donors (P < 0.001). The mean lymphocyte CD4 ABS value (65,339 ± 6,049) was equivalent to that reported in the literature with the same whole blood lysis protocol (21).
Fresh PBDC Can Be Identified by High CCR5 Expression.
There is evidence that mucosal DC are one of the initial targets of HIV-1 infection and that they help transmit the virus to the T lymphocyte pool (reviewed in ref. 22). Although there have been numerous studies on chemokine receptor expression in cultured monocyte-derived DC or explanted, skin-derived DC (23–26), it is also important to determine the levels of CD4 and coreceptor expression on fresh unstimulated PBDC, generally known to be immature DC (27, 28) that may serve as the initial targets of infection when they target the mucosa, or the responsible agent for transmitting virus when they retarget the lymph node. The strategy for phenotypically defining PBDC was modified from published protocols (ref. 28; Fig. 3). Because PBDC compose <1% of PBMC and fresh whole blood was used, ≈200,000 events were collected from each donor to obtain sufficient candidate PBDC for analysis. CD11c was used to gate on lineage-negative/HLA-DR-positive candidate DC, with results similar to that reported in the literature (28). These candidate PBDC (lineage-negative/HLA-DR-positive) had ≈10-fold more CCR5 ABS than CXCR4 ABS, with slight differences between the CD11c− and CD11c+ groups (Fig. 2, Table 1). The increasing levels of CD4 from monocytes to 11c+ PBDC to 11c− PBDC is in agreement with published reports (28). When the same gating strategy was employed on Ficoll-purified PBMC maintained overnight in media with 10% FCS, these lin−/HLADR+ PBDC became exclusively CD11c+ and up-regulated CD83 (from <0.5% CD83+ cells to >20% CD83+ cells), an established marker for DC maturation (29, 30). When compared with the CD83−lin−/HLADR+ immature PBDC, there was a dramatic up-regulation of CXCR4 compared with CCR5 (average 40- vs. 2.9-fold increase; P < 0.0001) in these mature DC.
Because CCR5 was so highly expressed on PBDC, we asked whether CCR5 in combination with lineage markers could be used to correctly identify PBDCs. Fig. 3B shows a contour plot indicating that lineage-negative cells could be distinctly gated in either CCR5high or CCR5low/neg populations. The lineage-negative CCR5high population was predominantly HLA-DR+ (R2 gate) whereas the lineage-negative CCR5low/neg population was exclusively HLA-DR− (R3). We propose that PBDC can be defined as lineage−/CCR5high cells, and sorted cells with this phenotype can be investigated to see whether they recapitulate DC functions in vitro.
Modulation of CD4, CCR5 and CXCR4 Levels on MDM.
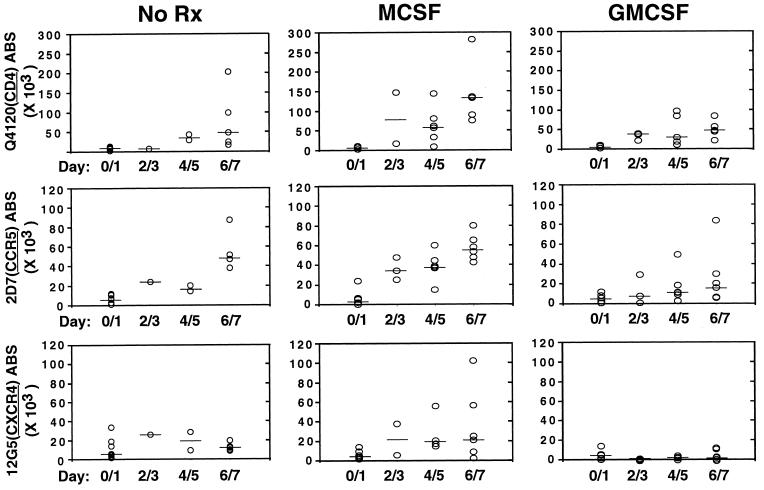
MDM are usually generated by culturing monocytes in the presence of human sera or with the addition of macrophage colony-stimulating factor (M-CSF) or granulocyte-macrophage colony-stimulating factor (GM-CSF). There have been a multitude of studies examining the effects of these cytokines on the replication of HIV and on the expression of CCR5 and CXCR4 (refs. 9, 31, and 32, and refs. therein). However, donor variability and greatly increased autofluorescence of MDM complicates interpretation of any studies regarding the variation in coreceptor levels as MDM mature. However, the QFACS assay is relatively immune to the problem of increasing autofluoresence because the background at every stage always is converted to an ABS value that is subtracted from the value obtained with the cognate antibody. MDM were matured for 7 days in the presence of human serum alone or with the addition of M-CSF or GM-CSF. ABS were determined over time for CD4, CCR5, and CXCR4. Although there was considerable donor variability (Fig. 4), M-CSF increased expression of CD4, CCR5, and CXCR4 on average when compared with GM-CSF (P < 0.0001, Fig. 4). Second, CXCR4 expression was virtually eliminated in the presence of GM-CSF (median <200 ABS) in contrast to M-CSF after 6–7 days of differentiation. Third, cytokine treatment usually increased the variability of expression levels when compared with the untreated (human sera alone) controls.
Figure 4.
Cytokine effects on CD4, CXCR4, and CCR5 expression on MDM. Absolute numbers of CD4, CC5, and CXCR4 ABS were determined on MDM differentiated in the presence of human serum alone (NoRx) plus either M-CSF (50 units/ml) or GM-CSF (10 ng/ml). Data from six to eight donors in five independent experiments were used for this analysis. Median values for each data group are indicated by the solid bars. Although there was considerable donor variability, M-CSF coordinately increased CD4, CCR5, and CXCR4 ABS significantly compared with GM-CSF (P < 0.0001). GM-CSF significantly reduced CXCR4 expression compared with M-CSF (P < 0.01). The differences in CD4 and coreceptor expression were not as significant when comparing NoRx with either M-CSF (P = 0.05) or GM-CSF (P = 0.06).
DISCUSSION
In vitro studies indicate that R5 and X4 viral strains can have differential requirements for CD4 and coreceptor levels (6, 7). Platt et al. (8) determined that, where CD4 levels were “limiting” (≈104), levels of CCR5 below a threshold of 1–2 × 104 molecules significantly affected the efficiency of infection. However, when CD4 levels were high (≈4.5 × 105), minimal levels of CCR5 (2 × 103) supported maximal viral infectivity. In addition, primary X4-viruses appear more dependent on CD4 levels than lab-adapted strains (7, 8). Considering that CD4 antigen density on PBMC is <105 molecules per cell (ref. 21; Fig. 2), it is likely that endogenous CCR5 levels will impact the efficiency of viral entry. Although a number of studies have correlated coreceptor expression levels with cytokine stimulation and with HIV status or disease progression (reviewed in ref. 9), we sought a more sophisticated method for measuring coreceptor levels because data presented as MFI or percent positive cells are always relative to controls that are specific for any given experiment. In addition, variations in emission filters used on different flow cytometers combined with differences in quantum efficiency and Stokes shifts of the various fluorophores used can result in markedly different fluorescence values. Therefore, we used a QFACS assay that is independent of these limitations to determine the absolute number of CD4 and coreceptor molecules on both cell lines and relevant primary cell types to shed light on their susceptibility to HIV-1 infection.
We confirmed that CCR5 and CXCR4 were differentially expressed on memory vs. naive T cells (18, 19), but we also discovered significant differences in coreceptor expression when CD62L was used to define precisely the true naive (RA+/62L+) and memory (RO+/62L−) subsets (see Table 1) (33–35). Specifically, whereas CCR5 was expressed only on CD45RO+ memory cells, the order of mean CXCR4 ABS was RA+/62L+ > RA+/62L− ≈ RO+/62L+ > RO+/62L− (Table 1). Memory T cells are the main responders to B-chemokines (36), and the high expression of CCR5 on true quiescent memory cells (RO+/62L−) is consistent with their role in being highly responsive to chemokine gradients generated at sites of immune and inflammatory reactions (37). Although there are reports indicating that CD45RA+ naive T cells are less susceptible to virus infection in vitro (33, 38), it remains to be determined whether these subpopulations in vivo differ in their susceptibility to infection by either R5 or X4-viruses. We also found that, for most peripheral blood lymphocytes subsets, CCR5 and CXCR4 levels varied by 2- to 5-fold between donors.
Immature DC in blood (PBDC) that migrate to body mucosa have been posited to be the initial targets of HIV-1 infection (39), preferentially via R5 viruses (24, 40). Although there have been reports describing the expression of chemokine receptors on in vitro cultured DC or explanted Langerhans cells (23–26), the expression of chemokine receptors on fresh uncultured PBDC has received scant attention. Our findings that PBDC have the highest number of CCR5 ABS among all leukocyte subsets examined is consistent with the putative role of DC as being the sentinel targets of HIV infection and may contribute to the preferential transmission of R5 viruses. However, the vanishingly small number of PBDC has made it difficult to determine whether these cells are infected with HIV in vivo. In addition, whether this level of CCR5 is maintained on DC interdigitation in the mucosa is unknown. However, on in vitro maturation in media with 10% FCS, the number of CXCR4 ABS increased by ≈40-fold with a 2- to 3-fold increase in CCR5 ABS. This pattern of CXCR4 up-regulation on DC maturation is similar to that seen with in vitro cultured MDDC (data not shown). However, the moderate increase in CCR5 seen in our mature PBDC is contrary to what has been reported in the literature (26). This discrepancy may be due to the culture systems employed. Indeed, we also saw down-regulation of CCR5 with maturation of our MDDC (30,603 ± 1,224 to 18,023 ± 720 ABS). But our fresh PBDC were matured in the presence of all of the leukocyte subsets present in peripheral blood, as opposed to cytokine-cultured MDDC and explanted skin-derived Langerhan cells. These mature PBDC may represent a novel subset of DC that warrants further characterization. The small but significant differences in CCR5 expression between the CD11c+ and CD11c− subgroups of PBDC (P = 0.008) also lends credence to the hypothesis that CD11c (+) or (−) DC constitute functionally different subsets of DC (28). We also suggest that the high expression of CCR5 on fresh PBDC allows an alternative way for sort-purifying PBDC for analysis of uncultured blood-derived DC. The use of one (CCR5) versus two (HLA-DR and CD11c) positive markers may have advantages of preserving more of the native DC phenotype for in vitro studies.
Macrophages are also important targets for HIV-1 infection, although the literature is inconsistent about whether CD4 is down- (41–43) or up-regulated (44–46) on macrophage maturation and whether CXCR4 can be used for viral entry in a macrophage context (41, 43, 46–48). Part of the difficulty in monitoring CD4 and coreceptor expression is due to the inconsistent accounting for the increased autofluoresence on macrophage maturation and the fact that MDMs can be obtained in several ways. We found that M-CSF was the most potent inducer of CD4, CCR5, and CXCR4 expression, whereas GM-CSF was the most effective suppressor of CXCR4 expression. Human sera alone had intermediate effects on CD4 and coreceptor expression. We also determined that absolute CD4 ABS actually increased with macrophage maturation (M-CSF > GM-CSF ≈ no treatment). The significant donor variability present in MDM may also explain some of the inconsistent results in the literature regarding the restrictive tropism of X4 viruses for MDM (47–50). Quantifying the levels of CD4 and CXCR4 under defined culture conditions would be useful when revisiting the issue of X4 viral restriction in MDM.
In conclusion, we have obtained baseline values for absolute CD4, CCR5, and CXCR4 ABS on various subsets of peripheral blood lymphocytes, PBDC, and differentially conditioned MDM. Most peripheral blood lymphocytes and DC subsets examined had CD4 levels close to the threshold (≈104) number of molecules where CCR5 levels become important for determining the efficiency of viral entry (8). However, examination of peripheral blood may not reflect changes that occur within secondary lymphoid tissues, where the bulk of active viral replication takes place. Therefore, the same QFACS assay should be used to examine leukocyte populations from lymph nodes and tonsillar tissues. In addition, reexamining coreceptor levels in HIV disease states using this technique, especially in light of the various promoter polymorphisms in ccr5 known to be correlated with disease progression (51, 52), would increase our understanding of the role of coreceptor regulation in HIV-1 pathogenesis.
Acknowledgments
We thank Joe Rucker and Ben Doranz for helpful comments. B.L. was supported by a Measey Foundation Fellowship for Clinicians (Wistar Institute) and by National Institutes of Health Grant K08 HL03923-01. L.J.M. was supported in part by National Institutes of Health Grants AI43206 and AI 44304. D.W. was supported by a grant from the W. W. Smith Charitable Trust. R.W.D. was supported by National Institutes of Health Grant R01 40880.
ABBREVIATIONS
- ABS
antibody-binding sites
- DC
dendritic cells
- PBDC
peripheral blood DC
- MDM
monocyte-derived macrophages
- FACS
fluorescence-activated cell sorting
- QFACS
quantitative FACS
- PE
phycoerythrin
- APC
allophycocyanin
- PBMC
peripheral blood mononuclear cells
- M-CSF
macrophage colony-stimulating factor
- GM-CSF
granulocyte-macrophage CSF
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bieniasz P D, Cullen B R. Front Biosci. 1998;3:44–58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 2.Doms R W, Peiper S C. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 3.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 4.McNicholl J M, Smith D K, Qari S H, Hodge T. Emerg Infect Dis. 1997;3:261–271. doi: 10.3201/eid0303.970302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, et al. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 6.Kabat D, Kozak S L, Wehrly K, Chesebro B. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platt E J, Wehrly K, Kuhnman S E, Chesbro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, B. & Montaner, L. J. (1999) J. Leukoc. Biol., in press. [DOI] [PubMed]
- 10.Lee B, Doranz B J, Rana S, Yi Y, Mellado M, Frade J M R, Martinez-A C, O’Brien S J, Dean M, Collman R G, Doms R W. J Virol. 1998;72:7450–7458. doi: 10.1128/jvi.72.9.7450-7458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka C, Lehnett B E, Lennett N M, Jones G M, Sklar L A, Steinkamp J A. Cytometry. 1996;25:271–279. doi: 10.1002/(SICI)1097-0320(19961101)25:3<271::AID-CYTO8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Haugland R P. Handbook of Fluorescent Probes and Research Chemicals. Eugene, OR: Molecular Probes Inc.; 1996. [Google Scholar]
- 13.Healy D, Dianda L, Moore J P, McDougal J S, Moore M J, Estess P, Buck D, Kwong P D, Beverly P C, Sattentau Q J. J Exp Med. 1990;172:1233–1242. doi: 10.1084/jem.172.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doranz B J, Orsini M J, Turner J D, Hoffman T D, Berson J F, Lu Z-H, Hoxie J A, Peiper S C, Brass L F, Doms R W. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, et al. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B, Sharron M, Blainpain C, Doranz B J, Vakili J, Setoh P, Tsang M, Parmentier M, Chang C N, Doms R W. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 17.Hesselgesser J, Liang M, Hoxie J, Greenberg M, Brass L F, Orsini M J, Taub D, Horuk R. J Immunol. 1998;160:877–883. [PubMed] [Google Scholar]
- 18.Bleul C, Wu L, Hoxie J, Springer T, Mackay C. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishimoto T. Leucocyte Typing VI: White Cell Differentiation Antigens: Proceedings of the 6th International Workshop and Conference. New York: Garland; 1996. [Google Scholar]
- 21.Lenkei R, Andersson B. J Immunol Methods. 1995;183:267–277. doi: 10.1016/0022-1759(95)00064-h. [DOI] [PubMed] [Google Scholar]
- 22.Steinman R M, Granelli-Piperno A, Tenner-Racz K, Racz P, Frankel S, Delgado E, Ignatius R, Pope M. In: Dendritic Cells: Biology and Clinical Applications. Lotze M T, Thomson A W, editors. San Diego: Academic; 1999. pp. 422–434. [Google Scholar]
- 23.Delgado E, Finkel V, Baggiolini M, Clark-Lewis I, Mackay C R, Steinman R M, Granelli-Piperno A. Immunobiology. 1998;198:490–500. doi: 10.1016/s0171-2985(98)80073-9. [DOI] [PubMed] [Google Scholar]
- 24.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman R M. J Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubbert A, Combadiere C, Ostrowski M, Arthos J, Dybul M, Machado E, Cohn M A, Hoxie J A, Murphy P M, Fauci A S, Weissman D. J Immunol. 1998;160:3933–3941. [PubMed] [Google Scholar]
- 26.Zaitseva M, Blauvelt A, Lee S, Lapham C K, Klaus-Kovton V, Mostowski H, Manischewitz J, Golding H. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 27.Weissman D, Li Y, Ananworanich J, Zhou L J, Adelsberger J, Tedder T F, Baseler M, Fauci A S. Proc Natl Acad Sci USA. 1995;92:826–830. doi: 10.1073/pnas.92.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Doherty U, Peng M, Gezelter S, Swiggard W J, Betjes M, Bhardwaj N, Steinman R M. Immunology. 1994;82:487. [PMC free article] [PubMed] [Google Scholar]
- 29.Weissman D, Li Y, Orenstein J M, Fauci A S. J Immunol. 1995;155:4111–4117. [PubMed] [Google Scholar]
- 30.Zhou L J, Tedder T F. Proc Natl Acad Sci USA. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Roderiquez G, Oravecz T, Norcross M. J Virol. 1998;72:7642–7647. doi: 10.1128/jvi.72.9.7642-7647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen O J, Kinter A, Fauci A S. Immunol Rev. 1997;159:31–48. doi: 10.1111/j.1600-065x.1997.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 33.Roederer M, Raju P, Mitra D, Herzenberg L, Herzenberg L. J Clin Invest. 1997;99:1555–1564. doi: 10.1172/JCI119318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okumura M, Fujii Y, Takeuchi K, Inada K, Nakahara K, Matsuda H. Eur J Immunol. 1993;23:1057–1063. doi: 10.1002/eji.1830230512. [DOI] [PubMed] [Google Scholar]
- 35.Picker L, Treer J, Ferguson D, Collins P, Buck D, Terstappen L. J Immunol. 1993;150:1105–1121. [PubMed] [Google Scholar]
- 36.Schall T J, Bacon K, Camp R D R, Caspari J W, Goeddel D V. J Exp Med. 1993;177:1821–1826. doi: 10.1084/jem.177.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loetscher P, Seitz M, Baggiolini M, Moser B. J Exp Med. 1996;184:569–577. doi: 10.1084/jem.184.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spina C, Prince H, Richman D. J Clin Invest. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinman R M, Granelli-Piperno A, Tenner-Racz K, Racz P, Frankel S, Delgado E, Ignatius R, Pope M. In: Dendritic Cells: Biology and Clinical Applications. Lotze M T, Thomson A W, editors. San Diego: Academic; 1999. pp. 422–434. [Google Scholar]
- 40.Reece J C, Handley A J, Anstee E J, morrison W A, Crowe S M, Cameron P U. J Exp Med. 1998;187:1623–1631. doi: 10.1084/jem.187.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiMarzio P, Tse J, Landau N R. AIDS Res Hum Retroviruses. 1998;14:129–138. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- 42.Lewin S R, Sonza S, Irving L B, McDonald C F, Mills J, Crowe S M. AIDS Res Hum Retroviruses. 1996;12:877–883. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- 43.Tuttle D L, Harrison J K, Anders C, Sleasman J W, Goodenow M M. J Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collman R, Nathanson N. Semin Virol. 1992;3:185–202. [Google Scholar]
- 45.Kazazi F, Mathijs J M, Foley P, Cunningham A L. J Gen Virol. 1989;70:2661–2672. doi: 10.1099/0022-1317-70-10-2661. [DOI] [PubMed] [Google Scholar]
- 46.Naif H M, Li S, Alali M, Sloane A, Wu L, Kelley M, Lynch G, Lloyd A, Cunningham A L. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmons, G., McKnight, A., Takeuchi, Y., Hoshino, H. & Clapham, P. R. (1995) Virology209. [DOI] [PubMed]
- 50.Schmidtmayerova H, Bolmont C, Baghgiguian S, Hirsch I, Chermann J C. Virology. 1992;190:124–133. doi: 10.1016/0042-6822(92)91198-4. [DOI] [PubMed] [Google Scholar]
- 51.McDermott D H, Zimmerman P A, Guignard F, Kleeburger C A, Leitman S F, Murphy P M. Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 52.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N, Lee B, Doms R W, Margolick J, Buchbinder S, et al. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]