Abstract
We report an intracellular peptide delivery system capable of targeting specific cellular compartments. In the model system we constructed a chimeric protein consisting of the nontoxic B subunit of Escherichia coli heat-labile enterotoxin (EtxB) fused to a 27-mer peptide derived from the DNA polymerase of herpes simplex virus 1. Viral DNA synthesis takes places in the nucleus and requires the interaction with an accessory factor, UL42, encoded by the virus. The peptide, designated Pol, is able to dissociate this interaction. The chimeric protein, EtxB-Pol, retained the functional properties of both EtxB and peptide components and was shown to inhibit viral DNA polymerase activity in vitro via disruption of the polymerase-UL42 complex. When added to virally infected cells, EtxB-Pol had no effect on adenovirus replication but specifically interfered with herpes simplex virus 1 replication. Further studies showed that the antiviral peptide localized in the nucleus, whereas the EtxB component remained associated with vesicular compartments. The results indicate that the chimeric protein entered through endosomal acidic compartments and that the Pol peptide was cleaved from the chimeric protein before being translocated into the nucleus. The system we describe is suitable for delivery of peptides that specifically disrupt protein–protein interactions and may be developed to target specific cellular compartments.
Most cellular and viral processes depend on the coordinated formation of protein–protein interactions (1). The exquisite specificity of such cognate interactions suggests that peptidic compounds that mimic subunit interfaces might function to disrupt protein–protein interactions in a highly specific manner. This prospect first was implicated by studies on the ribonucleotide reductase (RR) from herpes simplex virus 1 (HSV-1) in which a nonapeptide corresponding to the C terminus of one of the enzyme subunits was able to specifically inhibit enzyme activity by disruption of the interaction between the subunits (2, 3). However, addition of the nonapeptide to HSV-1-infected Vero cells failed to inhibit RR or viral replication, presumably because the peptide was unable to gain entry to the cell unaided (2).
Certain bacterial toxins, such as cholera toxin, diphtheria toxin, and Pseudomonas exotoxin A, have an inherent capacity to enter eukaryotic cells (4). Cholera toxin and related diarrhoeagenic toxins from Escherichia coli consist of a single A subunit, with ADP-ribosyltransferase activity, and five B subunits, which bind to a ubiquitous cell-surface glycosphingolipid, GM1 ganglioside (5). B subunit-receptor interaction triggers toxin internalization and results in the A subunit gaining access to the cytosol (6). We recently reported the use of the nontoxic B subunit of E. coli heat-labile enterotoxin (EtxB) as a carrier for the receptor-mediated delivery of a nonapeptide from the small subunit of HSV-1 RR. We showed that an EtxB-nonapeptide fusion protein was able to inhibit specifically viral growth in HSV-1-infected cells by disrupting the cytoplasmic RR (7). Addition of the free peptide or EtxB alone had no effect, implying that the fusion protein mediates entry of the attached peptide to an intracellular compartment, which leads to its subsequent delivery to the cytosol.
To test whether the B subunit could be used to deliver peptides into different cellular compartments we chose to target a viral protein–protein interaction that occurs in the nucleus and is crucial for replication of HSV-1 DNA. This interaction is that between the catalytic subunit (POL) of the DNA polymerase and the UL42 accessory protein, which acts to increase both the rate of incorporation of deoxyribonucleoside triphosphates and the processivity of the enzyme (8). A peptide corresponding to the 27 C-terminal amino acids of POL, which is the region responsible for binding to UL42, specifically blocks the interaction between the two proteins (9).
In this paper we show that a chimeric protein consisting of EtxB extended at its C terminus with such an inhibitory peptide can deliver the peptide into the nucleus to exert selective anti-HSV activity. We also report experiments to elucidate the trafficking pathway that enables the peptide to be transported from the outside of the cell, through endosomal compartments, and into the nucleus.
MATERIALS AND METHODS
Proteins and Peptides.
HSV-1 UL42 and POL proteins were expressed in insect cells from recombinant baculovirus and purified as described (8, 9). EtxB and EtxB-Pol were expressed in a nontoxinogenic vibrio, Vibrio sp. 60, and purified as reported (10, 11). Synthesis of the 27-mer Pol peptide has been described (9).
Antibodies and Reagents.
The polyclonal antiserum 113 was raised against the C-terminal 15 residues of HSV-1 POL (9). The mAb 118–8 anti-EtxB was kindly provided by E. Lundgren and H. Persson (University of Umea, Sweden), whereas the polyclonal antibody anti-EtxB was obtained from M. Pizza (12). mAb OSW2 against human V-ATPase (13) and mAb specific for human transferrin receptor were a gift of S. B. Sato (University of Kyoto, Japan) and C. R. Hopkins (University College London), respectively. The polyclonal antibody against cathepsin D was purchased from DAKO; mAbs against β-tubulin or γ-adaptin were obtained from Sigma. Texas red-conjugated goat anti-rabbit IgG and fluorescein-conjugated goat anti-mouse IgG were purchased from Jackson ImmunoResearch Laboratories and Calbiochem, respectively.
Brefeldin A (BFA; used at 10 μM), bafilomycin A1 (used at 200 nM), and nocodazole (used at 10 μM) were obtained from Sigma. X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactopyranoside) was purchased from Boehringer Mannheim.
Cells and Viruses.
Vero, HeLa, COS-1, NIH 3T3, MDCK, and 293 cells all were maintained in DMEM containing 10% FCS (GIBCO) and supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin. T84 cells were maintained in DMEM/Ham’s F-12 medium (1:1 mixture) containing 10% FCS.
HSV-1 strain 17+ (14) was used throughout. Recombinant adenovirus type 5 (Ad5.βgal) contained the lacZ gene with the simian virus 40 nuclear localization signal under control of the long terminal repeat of the Rous sarcoma virus (15).
POL/UL42 Physical and Functional Interaction Assays.
The POL/UL42 physical interaction assay was performed on microtiter plates (9). Stimulation of POL activity by UL42 was measured by incorporation of [3H]dTTP into a poly(dA)⋅ oligo(dT) template (16).
Fluorescence Microscopy.
Cells were seeded onto coverslips in 24-well trays and grown for 24 hr. After treatment with toxin and drugs, cells were fixed and incubated with primary and secondary antibodies as reported (17). Optical sections (1 μm thick) of cells were obtained with a Zeiss Axiovert TV-100 fluorescence microscope, equipped with a charge-coupled device camera, by using the metamorf imaging system software. Some samples were examined in dual channels by using a Nikon RCM 8000 confocal microscope, and images were processed by using epoch 2000 software.
Antiviral Assays for HSV.
For the plaque reduction assay, confluent Vero cells were infected at room temperature with HSV-1. After adsorption for 90 min, various amounts of purified EtxB-Pol, EtxB, or free Pol peptide, diluted in DMEM/2% FCS medium containing 0.2% (vol/vol) pooled human IgG, were added to the medium. Plaques were counted after 2 days of incubation at 37°C.
For the virus yield reduction assay, Vero cells infected with HSV-1 at different multiplicities of infection (mois) and treated with EtxB-Pol, EtxB, or free Pol peptide for 48 hr were washed twice with PBS, subjected to three cycles of freeze/thawing, and resuspended in ice-cold PBS. Virus was titered by a standard plaque assay.
The viability and metabolic activity of Vero cells treated with purified EtxB or EtxB-Pol up to 2.8 mg/ml (200 μM of the monomeric form) in DMEM/10% FCS were assessed by using the tetrazolium salts colorimetric method (Boehringer Mannheim).
Southern Blot Analysis and Titration of HSV-1 DNA.
Vero cells were infected with HSV-1 at a moi of 0.1 plaque-forming unit (pfu) per cell and treated with 100 μM EtxB-Pol or EtxB, diluted in DMEM medium. Total DNA was extracted from each sample 24 hr postinfection and digested with BamHI. DNA fragments were separated on a 0.8% agarose gel and blotted onto a nylon filter (Hybond-N; Amersham). Different amounts (0.5, 5, and 50 ng) of the pUC8-HSV plasmid, which contains the 3.6-kb BamHI Q fragment of HSV-1 genome (18), also were loaded on the gel. Blots were hybridized to a digoxigenin-dUTP-labeled probe, complementary to the BamHI Q fragment, which was generated by amplifying a 598-bp region of pUC8-HSV with primers 5′-AGCAAGAAGCCACGGAAGTC-3′ and 5′-GCCAACATAGCCAGGTCAAG-3′. Traces on autoradiograms were quantified at 570 nm by a densitometer (Shimadzu Dual-Wavelength Chromato Scanner CS 930) and correlated to a reference curve constructed with the different amounts of pUC8-HSV.
Antiviral Assay for Adenovirus.
Monolayers of 293 cells were grown up to 80% confluence and infected with Ad5. βgal at a moi of 0.001 pfu/cell for 90 min. After washing with PBS, cells were incubated for 3 days with various amounts of purified EtxB-Pol or EtxB, diluted in DMEM/2% FCS medium. Monolayers subsequently were fixed and stained with X-Gal as described (19), and viral plaques were counted.
RESULTS
EtxB-Pol Fusion Protein Inhibits in Vitro Activity of HSV-1 DNA Polymerase by Disruption of POL/UL42 Complex.
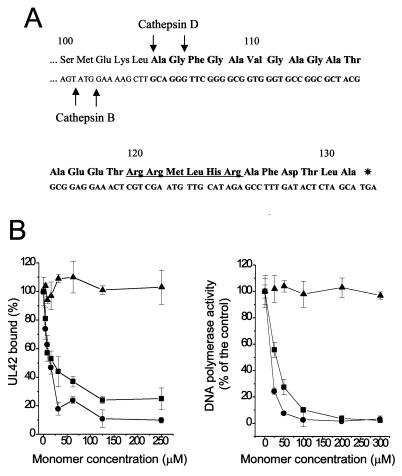
We previously reported (11) the construction and purification of a fusion protein, EtxB-Pol, consisting of the 27 C-terminal amino acids of POL linked to the C terminus of EtxB (Fig. 1A). The chimera was properly assembled into pentamers and bound GM1 gangliosides as wild-type EtxB (11). We now tested whether EtxB-Pol could interfere with the POL/UL42 physical and functional interaction. The IC50 of fusion protein was approximately 15 μM (Fig. 1B Left; concentrations for both EtxB and EtxB-Pol are given for the monomeric forms to have a better comparison with the free Pol peptide). This value is comparable to that (25 μM) for the free 27-mer. EtxB alone was unable to inhibit POL/UL42 interaction, indicating that the inhibitory activity of the fusion protein derives from the attached peptide. Similar IC50 values were obtained by adding inhibitors to the preformed POL/UL42 complex (data not shown), suggesting that both free 27-mer and EtxB-Pol fusion also could dissociate the complex. In addition, EtxB-Pol chimera and free Pol peptide inhibited the stimulation of HSV-1 POL activity by UL42 with IC50 values of approximately 10 μM and 25 μM, respectively (Fig. 1B Right). Again, EtxB showed no inhibition. We conclude that fusion of the 27-aa Pol peptide to the C terminus of EtxB generates a chimera that retains the essential properties of both EtxB and the Pol peptide.
Figure 1.
Sequence and in vitro activity of the EtxB-Pol chimera. (A) Sequence of the C-terminal 32 residues of the fusion protein is reported, with the amino acids numbered from the first residue of mature EtxB. Boldface letters indicate the region coding the attached Pol peptide. Putative endoprotease cleavage sites are indicated by an arrow; a putative nuclear localization signal is also indicated (underlined sequence). (B) Effect of EtxB (▴), EtxB-Pol (●), and Pol peptide (■) on the POL/UL42 physical interaction as measured by the ELISA interaction assay (Left) and on incorporation of [3H]dTTP into a poly(dA)⋅oligo(dT) template by POL in the presence of UL42 (Right). The graphs show the average of data from three independent experiments together with the SD.
EtxB-Mediated Delivery of Pol Peptide into the Cell Nucleus.
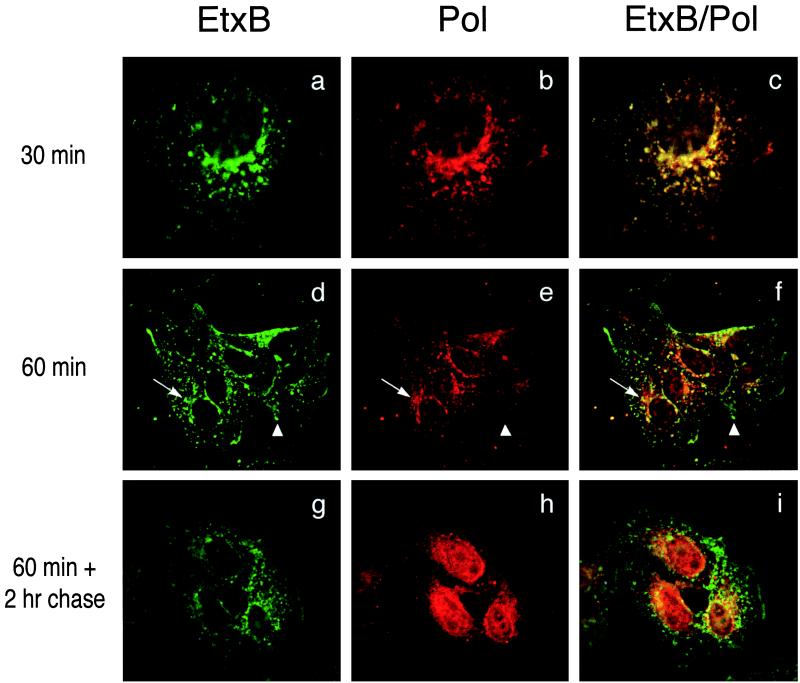
To study the uptake and intracellular trafficking of the chimera, Vero cells were treated for different times with 10 μM EtxB-Pol and then stained both with an anti-EtxB mAb and an anti-Pol peptide antiserum. Analysis by deconvolution fluorescence microscopy showed that the EtxB-Pol chimera bound to cell surface receptors and was internalized in a manner resembling that of wild-type EtxB. After 30 min of incubation, both domains of EtxB-Pol fusion colocalized in punctated intracellular compartments that appeared either scattered throughout the cytoplasm or in the perinuclear area (Fig. 2 a–c). After 1 hr the two moieties of EtxB-Pol were beginning to dissociate, such that in addition to EtxB+/Pol+ compartments in the perinuclear region, numerous EtxB+/Pol− vesicles were visible in the surrounding cytoplasm and some Pol signal, with no EtxB, was detectable in the charioplasm (Fig. 2 d–f). In cells incubated with EtxB-Pol for 1 hr, washed, and chased for a further 2 hr the latter pattern was emphasized (see Fig. 2 g–i): with some EtxB+/Pol+ staining in the perinuclear region, the majority of the EtxB+/Pol− signal scattered in the cytosol and a very strong nuclear-located Pol signal was observed, suggesting that the Pol peptide had concentrated in the nucleus. Longer chase times revealed that the Pol peptide was detectable in the nucleus of all cells after 24 hr posttreatment (p.t.) and still was present in a significant fraction of nuclei after 48 hr p.t. (data not shown). As a control, the fate of free Pol peptide also was followed and, as expected, failed to bind the cell surface or to be internalized into cell.
Figure 2.
Pol peptide is internalized by EtxB-Pol fusion protein and localizes into the cell nucleus. Vero cells were treated with EtxB-Pol for 30 min (a–c) or 1 hr (d–f) and fixed immediately. Alternatively, cells were treated with the fusion protein for 1 hr, washed, and fixed 2 hr after treatment (g–i). All samples then were probed with mAb 118–8, specific for EtxB (EtxB, Left), and with polyclonal antiserum 113, specific for the Pol peptide (Pol, Center) and secondary anti-mouse fluorescein-conjugated or anti-rabbit Texas red-conjugated antibodies, respectively. Images were collected by using a ×100 objective with a deconvolution fluorescence microscope and superimposed (EtxB/Pol, Right). Arrows and arrowheads point at representative EtxB+/Pol+ and EtxB+/Pol− compartments, respectively.
Similar nuclear staining with antibodies to Pol also was detected in other cell types, including 293, COS-1, HeLa, NIH 3T3, MDCK, and polarized T84 epithelial cells, when they were treated with EtxB-Pol (data not shown).
We therefore conclude that the EtxB allows for entry into cells of an attached peptide, which subsequently localizes into the nucleus.
Intracellular Trafficking of EtxB-pol Involves Different Stages of the Endocytic Pathway.
To identify the compartments involved in the intracellular trafficking of EtxB-Pol, we investigated its colocalization with a number of well characterized markers of different membrane compartments: transferrin receptor (TFR) for early-recycling endosomes, vacuolar-type H+-ATPase (V-ATPase) for acidic late endosomes and lysosomes, cathepsin D for lysosomes, and γ-adaptin for the trans-Golgi network (TGN).
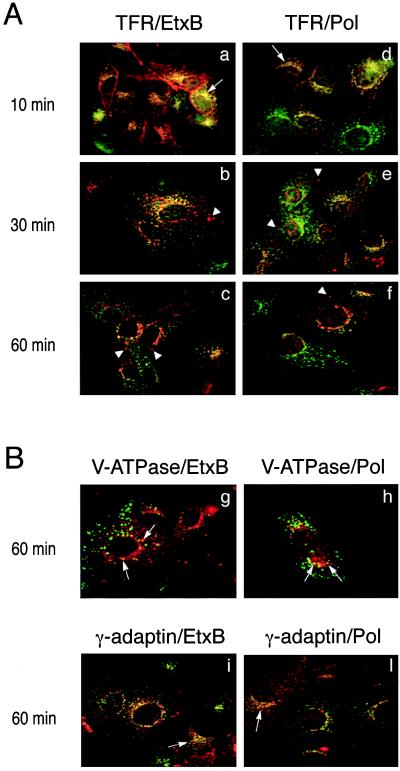
Double-fluorescence labeling with antibodies to either EtxB or Pol peptide and to the TFR showed that after 10 min of incubation, both domains of EtxB-Pol were almost exclusively present in the early-recycling compartment (Fig. 3 a and d). After 30 min and, particularly, after 60 min of endocytosis, an increasing amount of EtxB and Pol peptide was sorted from the TFR+ compartments (Fig. 3 b and e, and c and f).
Figure 3.
Colocalization of the two EtxB-Pol domains with markers of intracellular compartments. (A) Vero cells were treated with EtxB-Pol for 10, 30, or 60 min and probed either with an antibody specific for TFR and an antibody against EtxB (TFR/EtxB, a–c) or with an antibody specific for TFR and the antiserum 113 against the Pol peptide (TFR/Pol, d–f). Images were analyzed as described in the legend to Fig. 2. Arrows point at an example of TFR+/EtxB+ or TFR+/Pol+ compartments; arrowheads indicate TFR−/EtxB+ or TFR−/Pol+ vesicles. (B) Vero cells were treated with EtxB-Pol for 60 min and probed either with the antibody OSW2 specific for V-ATPase and an antibody specific for EtxB (V-ATPase/EtxB, g), with the antibody OSW2 and the antiserum 113 (V-ATPase/Pol, h), with an antibody specific for γ-adaptin and an antibody against EtxB (γ-adaptin/EtxB, i), or with an antibody specific for γ-adaptin and the antiserum 113 (γ-adaptin/Pol, l). Colocalization of EtxB and Pol moieties of EtxB-Pol with V-ATPase or with γ-adaptin are indicated by arrows.
Consistent with this, at 60 min a significant fraction of EtxB and Pol peptide colocalized with V-ATPase, a marker for late endosomal and lysosomal vesicles (Fig. 3 g and h). Similarly, the EtxB signal was found to be increasingly colocalized with the lysosomal marker, cathepsin D, as sorting from the recycling compartments progressed (data not shown). Such observations indicate that at least some of the chimera traffics into the late stages of the endocytic pathway. Other experiments using antibodies to γ-adaptin demonstrated that both the EtxB and Pol domains also can reach the TGN in the perinuclear region (Fig. 3 i and l). In contrast, staining of the nuclear envelope, diagnostic of retrograde transport to the endoplasmic reticulum, was not evident at any time.
Bafilomycin A1 Blocks Intranuclear Localization of Pol Peptide.
The above results indicate that after endocytosis into the early-recycling compartment, the EtxB-Pol chimera can follow two distinct pathways, the first of which continues toward lysosomes whereas the second diverges to the TGN. To investigate the role of these two compartments in processing and/or the translocation of the Pol peptide, we studied EtxB-Pol trafficking in the presence of drugs known to specifically affect different intracellular compartments.
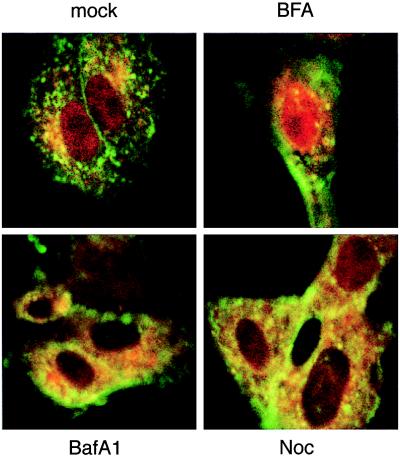
Vero cells were treated with BFA, which disrupts the Golgi apparatus and inhibits classical vesicle-mediated secretion (20), for 1 hr before addition of EtxB-Pol for a further 1-hr period. Cells then were washed and incubated with BFA for another 2 hr before being analyzed by confocal fluorescence microscopy. The action of BFA on Golgi disruption was almost immediate, as evidenced by the diffuse cytoplasmic staining of γ-adaptin, a Golgi marker (ref. 21 and data not shown). In these conditions, the Pol peptide still was able to accumulate into the nucleus (Fig. 4, BFA), suggesting that nuclear localization of Pol peptide is not dependent on sorting of EtxB-Pol to the Golgi.
Figure 4.
Effect of different drugs on intranuclear localization of Pol peptide. Vero cells were treated for 1 hr with BFA, bafilomycin A1 (BafA1), or nocodazole (Noc) or not treated (mock). EtxB-Pol was added for 1 hr, and then monolayers were washed and incubated for a further 2 hr in the presence of the inhibitors. Double immunofluorescence was carried out with mAb 118–8 (anti-EtxB, green fluorescence) and rabbit polyclonal serum 113 (anti-Pol peptide, red fluorescence). Images of the same field for both stains were taken with a confocal microscope at a ×100 magnification and superimposed.
To target the acidic endosomal and lysosomal vesicular compartments we used bafilomycin A1 (BafA1), a specific inhibitor of the V-ATPase, which is responsible for acidification of organelles of the endocytic pathway (22) and which colocalizes with the EtxB-Pol domains (Fig. 3). Pretreatment of cells with this pH-neutralizing drug, before addition of the chimeric toxin, resulted in the complete block of Pol peptide entry into the nucleus (Fig. 4, BafA1).
The effect of the microtubule-depolymerizing drug nocodazole also was studied because it has been reported that the microtubule network is involved in the vesicular transport from late endosomes to lysosomes (23). Pretreatment of cells with nocodazole for 1 hr before addition of EtxB-Pol resulted both in the complete depolymerization of the microtubule network, as evidenced by diffuse β-tubulin staining (data not shown), and in less Pol peptide targeting to the nucleus than that observed in untreated cells (Fig. 4, Noc).
Taken together, these results support the importance of endosomal/lysosomal organelles for the EtxB-Pol processing and/or its translocation.
EtxB-Pol Chimera Specifically Inhibits HSV-1 Replication in Vero Cells.
To test whether EtxB-Pol inhibited HSV-1 replication in Vero cells, purified EtxB-Pol, EtxB, or Pol peptide was applied to infected monolayers and both viral plaque formation and infectious virus yield were compared with untreated controls. Only treatment with EtxB-Pol inhibited plaque formation, with an IC50 of approximately 11 μM (Fig. 5A). A similar inhibitory effect of EtxB-Pol also was observed on HSV-2 replication (data not shown). The yield of infectious virus harvested from Vero cell monolayers treated with 100 μM of EtxB-Pol fusion protein was reduced by about 2 logs up to a moi of 0.1 pfu/cell (Fig. 5B). Both wild-type EtxB and free Pol peptide showed no antiviral effect up to a concentration of 500 μM. Vero cell viability and metabolic activity were unaffected by treatment with concentrations of EtxB-Pol and EtxB up to 200 μM (data not shown).
Figure 5.
The EtxB-Pol fusion protein inhibits HSV-1 replication and DNA synthesis in Vero cells. (A) Inhibition of plaque formation. Confluent Vero cells were infected with HSV-1 at a moi of 0.001 and treated with EtxB-Pol (●), EtxB (▴), or the Pol peptide (■) for 48 hr. The plaque-forming efficiency was calculated as a percentage of plaques compared with an untreated control. The graph shows the mean values and SDs from three independent experiments. (B) Inhibition of virus yield. Vero cells were infected with HSV-1 at different mois and treated with various amounts of EtxB-Pol for 48 hr. Virus progeny was harvested from the infected cells and titered by using a standard plaque assay. (C) Inhibition of viral DNA synthesis. Total DNA was extracted from Vero cells infected with HSV-1 and treated with EtxB-Pol or EtxB or cells that were infected and untreated (HSV-1) or not infected (mock). Samples were digested with BamHI, blotted, and hybridized to a probe complementary to the HSV-1 BamHI Q fragment. Plotted values represent the number of viral genomes relative to autoradiographic bands, quantified by densitometry and correlated with a reference curve as described in the text.
A quantitative Southern blot analysis was performed to characterize further the effect of EtxB-Pol on HSV-1 DNA replication. The amount of viral DNA synthesized in HSV-1-infected Vero cells treated with 100 μM of EtxB-Pol was reduced by approximately 70% compared with that synthesized in untreated cells or in cells treated with EtxB (Fig. 5C). This demonstrates that EtxB-Pol can inhibit HSV-1 replication by impairing viral DNA synthesis.
To obtain additional evidence for the specificity of the anti-HSV effect, we tested whether EtxB-Pol could also inhibit replication of adenovirus, a double-stranded DNA virus that, like herpesviruses, replicates in the cell nucleus (24). Monolayers of 293 cells were infected either with HSV-1 or with a recombinant adenovirus (Ad5.βgal) (15) and treated with increasing concentrations of EtxB-Pol or EtxB. The number of HSV-1 or Ad-5 plaques in toxin-treated samples was counted after 48 or 72 hr, respectively, and compared with that of an untreated control. At a concentration of 100 μM of EtxB-Pol there was no effect on adenovirus replication, whereas there was a 68% decrease in HSV-1 replication (data not shown), confirming that the antiviral activity of EtxB-pol is specific to HSV-1.
DISCUSSION
Peptides and peptide mimetics capable of disrupting protein–protein interactions have such exquisite specificity that they merit consideration as lead compounds for the development of novel therapeutic agents (25). However, the use of peptides has been thwarted by problems related to their cell permeation, intracellular localization, and stability. In this paper we demonstrate that EtxB, a nontoxic receptor-binding component of E. coli enterotoxin, can be used to mediate the intracellular delivery of a peptide that subsequently gets access to the nucleus. For this purpose we selected a peptide corresponding to the C-terminal 27 aa of HSV-1 DNA polymerase (POL), which normally is involved in cognate interaction with UL42. Although the free peptide or EtxB alone had no effect on virally infected cells, the EtxB-Pol fusion protein selectively inhibited HSV-1 replication. The antiviral activity of the chimera could be explained only by the dissociation of the nuclear complex between POL and UL42, because both EtxB-Pol and the 27-mer inhibited this interaction in vitro. The specificity of the inhibitory effect of EtxB-Pol was confirmed by lack of interference with adenovirus growth and lack of cytotoxicity.
Experiments were performed to elucidate the intracellular pathway and fate of the fusion protein. After endocytosis, EtxB-Pol traffics through different locales of the endocytic pathway, initially as an entire fusion protein and later as two separated components. Dissociation of the EtxB-Pol components seems to take place in acidic organelles. This suggestion is supported both by the block in nuclear localization of the Pol peptide caused by bafilomycin A1, which specifically inhibits the V-ATPase responsible for acidification of late endosomes and lysosomes, and by colocalization experiments performed with markers specific for acidic vesicles. Cleavage of the Pol peptide from the fusion protein therefore is likely to be dependent on late endosomal or lysosomal endoproteases. Consistent with this hypothesis are the presence of a number of putative cathepsin cleavage sites in the C terminus of EtxB and the junction with the Pol peptide (see Fig. 1A) and our preliminary biochemical evidences supporting specific proteolysis (not shown).
The mechanism of Pol peptide translocation from the lumen of endocytic organelles into the cytosol remains to be explained. Studies on the mechanism of internalization of diphtheria toxin have shown that a pH-dependent perturbation in its α-helical-rich translocation domain occurs, with protonation of acidic residues in a helical hairpin permitting insertion into the lipid bilayer (26). The last 36 aa of POL have been shown to adopt a hairpin-like structure with two α-helical regions interrupted by a nonhelical region that contains two glutamate residues (27). Thus, the requirement for EtxB-Pol entry into acidic organelles might also stem from the need to enter a pH environment that causes protonation of the two glutamate residues and, thus, permits insertion and translocation across the membrane. Furthermore, the 27-mer Pol peptide contains an N-terminal stretch of 12 hydrophobic and nonpolar amino acids with an intrinsic propensity to penetrate lipid bilayers.
The nuclear localization of Pol peptide and its stationing in this compartment for a longer period than in any other subcellular location indicate that the peptide may contain specific information for nuclear targeting. Indeed, the Pol peptide contains the sequence RRMLHR (underlined in Fig. 1A), which resembles a nuclear localization signal (NLS) (28, 29). In contrast to the Pol peptide, our previous work on the EtxB-mediated delivery of a C-terminal nonapeptide of HSV-1 RR had provided evidence that the peptide functioned by inhibiting RR, which is located in the cytosol (7). Although the nonapeptide is much shorter than the Pol peptide, they possess a number of similar features, namely an uninterrupted stretch of hydrophobic and nonpolar amino acids and some acidic residues. However, the nonapeptide lacks an NLS, which may account for its localization in the cytosol.
Among the biological systems proposed for the delivery of peptides into living cells for either experimental or therapeutic use, which include viruses and proteins of eukaryotic, bacterial, or viral origin (30–33), only the tegument protein VP22 (viral protein 22) of HSV-1 has been shown to be capable of the nuclear import of heterologous peptide molecules (33). However, this is a viral product whose intracellular function and biosafety profile have not yet been defined completely. On the other hand, other bacterial toxins have been proposed for peptide translocation in the cytosol, such as diphtheria toxin (31) and Pseudomonas exotoxin A (32). However, their receptors are not as ubiquitous as those for the B subunits of cholera toxin and E. coli enterotoxin (34), and they need genetic detoxification and attain only low intracellular concentrations of the translocated material (31).
The ability of EtxB to enter the endocytotic pathway, in which a proportion of the material traffics to the Golgi, suggests that EtxB can find widespread application as a delivery system. EtxB-based chimeras might be engineered by introduction of specific signals into attached peptides to either enhance or prevent cleavage, potentiate or inhibit insertion across endocytic membranes, and ensure targeting to other intracellular compartments, such as Golgi apparatus, endoplasmic reticulum, or mitochondria. Our demonstration that the Pol peptide, when delivered by EtxB, retains its pharmacological characteristics heralds the prospect that B subunit-peptide conjugates could be used as a screen for bioactive peptides that inhibit intracellular enzymes and macromolecular complexes relevant to the pathogenesis of microbial infections and malignant diseases. One could also envisage, as an in vivo application of this system, the intracellular delivery of immunodominant epitopes as a conceptually new type of vaccines that exploit the intrinsic, immune-adjuvant activity of EtxB and its mucosal tropism (35, 36).
Acknowledgments
We thank Dr. E. Cancellotti for his assistance with the EtxB and EtxB-Pol purification and Dr. G. McVey for purification of HSV-1 POL and UL42 proteins. This work was funded by the Istituto Superiore di Sanità of Italy (to G.P.) and the Medical Research Council of Great Britain (to T.R.H. and H.S.M.).
ABBREVIATIONS
- EtxB
Escherichia coli heat-labile enterotoxin B subunit
- HSV-1
herpes simplex virus 1
- RR
ribonucleotide reductase
- moi
multiplicity of infection
- TFR
transferrin receptor
- BFA
brefeldin A
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Eisenstein E, Schachman H K. In: Protein Function: A Practical Approach. Creighton T E, editor. Oxford: IRL; 1989. pp. 135–175. [Google Scholar]
- 2.Dutia B M, Frame M C, Subak-Sharpe J H, Clark W N, Marsden H S. Nature (London) 1986;321:439–441. doi: 10.1038/321439a0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen E A, Gaudreau P, Brazeau P, Langelier Y. Nature (London) 1986;321:441–443. doi: 10.1038/321441a0. [DOI] [PubMed] [Google Scholar]
- 4.Montecucco C, Papini E. Trends Microbiol. 1995;3:165–167. doi: 10.1016/s0966-842x(00)88911-8. [DOI] [PubMed] [Google Scholar]
- 5.Spangler B D. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastiaens P I H, Majoul I V, Verveer P J, Söling H D, Jovin T M. EMBO J. 1996;15:4246–4253. [PMC free article] [PubMed] [Google Scholar]
- 7.Marcello A, Loregian A, Cross A, Marsden H S, Hirst T R, Palù G. Proc Natl Acad Sci USA. 1994;91:8994–8998. doi: 10.1073/pnas.91.19.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlieb J, Marcy A I, Coen D M, Challberg M D. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsden H S, Murphy M, McVey G L, MacEachran K A, Owsianka A M, Stow N D. J Gen Virol. 1994;75:3127–3135. doi: 10.1099/0022-1317-75-11-3127. [DOI] [PubMed] [Google Scholar]
- 10.Amin T, Hirst T R. Protein Expression Purif. 1994;5:198–204. doi: 10.1006/prep.1994.1031. [DOI] [PubMed] [Google Scholar]
- 11.Loregian A, Hirst T R, Marsden H S, Palù G. Protein Expression Purif. 1996;8:381–389. doi: 10.1006/prep.1996.0114. [DOI] [PubMed] [Google Scholar]
- 12.Pizza M, Fontana M R, Giuliani M M, Domenighini M, Magagnoli C, Gianelli V, Nucci D, Hol W, Manetti R, Rappuoli R. J Exp Med. 1994;180:2147–2153. doi: 10.1084/jem.180.6.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato S B, Toyama S. J Cell Biol. 1994;127:39–53. doi: 10.1083/jcb.127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown S M, Ritchie D A, Subak-Sharpe J H. J Gen Virol. 1973;18:329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 15.Stratford-Perricaudet L D, Makeh I, Perricaudet M, Briand P. J Clin Invest. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenney D J, Hurlburt W W, Bifano M, Stevens J T, Micheletti P A, Hamatake R K, Cordingley M G. J Virol. 1993;67:1959–1966. doi: 10.1128/jvi.67.4.1959-1966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papini E, Gottardi E, Sapino B, De Bernard M, Massari P, Telford J, Rappuoli R, Sato S B, Montecucco C. J Med Microbiol. 1996;45:84–89. doi: 10.1099/00222615-45-2-84. [DOI] [PubMed] [Google Scholar]
- 18.Palù G, Summers W P, Valisena S, Tognon M. J Med Virol. 1988;24:251–262. doi: 10.1002/jmv.1890240303. [DOI] [PubMed] [Google Scholar]
- 19.Lemarchand P, Jaffe H A, Danel C, Cid M C, Kleinman H K, Stratford-Perricaudet L D, Perricaudet M, Pavirani A, Leicocq J-P, Crystal R G. Proc Natl Acad Sci USA. 1992;89:6482–6486. doi: 10.1073/pnas.89.14.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M, Kries T. Cell. 1992;69:129–138. doi: 10.1016/0092-8674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- 22.Bowman E J, Siebers A, Altendorf K H. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Brabander M, Nuydens R, Geerts H, Hopkins C R. Cell Motil Cytoskeleton. 1988;9:30–47. doi: 10.1002/cm.970090105. [DOI] [PubMed] [Google Scholar]
- 24.Shenk T. In: Fields Virology. Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 2111–2148. [Google Scholar]
- 25.Marsden H S. Semin Virol. 1992;3:67–75. [Google Scholar]
- 26.Kaul P, Silverman J, Shen W H, Blanke S R, Huynh P D, Finkelstein A, Collier R J. Protein Sci. 1996;5:687–692. doi: 10.1002/pro.5560050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Digard P, Williams K P, Henley P, Brooks I S, Dahl C E, Coen D M. Proc Natl Acad Sci USA. 1995;92:1456–1460. doi: 10.1073/pnas.92.5.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons R H, Ferguson B Q, Rosenberg M. Mol Cell Biol. 1987;7:2451–2456. doi: 10.1128/mcb.7.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff B, Dickson R B, Hanover J A. Trends Pharmacol Sci. 1987;8:119–121. [Google Scholar]
- 30.Seth P. Biochem Biophys Res Commun. 1994;203:582–587. doi: 10.1006/bbrc.1994.2222. [DOI] [PubMed] [Google Scholar]
- 31.Stenmark H, Moskaug J O, Madshus I H, Sandvig K, Olnes S. J Cell Biol. 1991;113:1025–1032. doi: 10.1083/jcb.113.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donnelly J J, Ulmer J B, Hawe L A, Friedman A, Shi X P, Leander K R, Shiver J W, Oliff A I, Martinez D, Montgomery D, Liu M A. Proc Natl Acad Sci USA. 1993;90:3530–3534. doi: 10.1073/pnas.90.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott G, O’Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 34.Eidels L, Proia R L, Hart D A. Microbiol Rev. 1983;47:596–620. doi: 10.1128/mr.47.4.596-620.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nashar T O, Webb H M, Eaglestone S, Williams N A, Hirst T R. Proc Natl Acad Sci USA. 1996;93:226–230. doi: 10.1073/pnas.93.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Akker F, Pizza M, Rappuoli R, Hol W G. Protein Sci. 1997;6:2650–2654. doi: 10.1002/pro.5560061220. [DOI] [PMC free article] [PubMed] [Google Scholar]