Abstract
The catalytic mechanisms employed by RNA are chemically more diverse than initially suspected. Divalent metal ions, nucleobases, ribosyl hydroxyl groups, and even functional groups on metabolic cofactors all contribute in the various strategies employed by RNA enzymes. This catalytic breadth raises intriguing evolutionary questions about how RNA lost its biological role in some cases, but not in others, and what catalytic roles RNA might still be playing in biology.
RNA is well-suited for its role as a purveyor of genetic information. The consistent hydrogen bonding potential of each nucleobase is critical for fidelity during replication, transcription and translation. It is easy to imagine evolutionary pressures that would select for residues with such properties. This is achieved in part because none of the nucleobases have ionizable groups near neutrality
Although optimized for reliable base pairing, RNA is also able to catalyze essential biochemical reactions, including RNA processing and protein synthesis. It does this despite significant biophysical handicaps. RNA has a small repertoire of functional groups and these are embedded in a poorly-constrained ribosyl-phosphate backbone heavy with negative charge. The pKas of the bases would appear to be either too low (3.5 for A, 4.2 for C) or too high (9.8 for G, 10.5 for U) for use in efficient general acid or base catalysis. As a group, RNA enzymes, or ribozymes, are less efficient catalysts relative to chemically supercharged proteins, yet in many cases ribozymes provide enough rate enhancement to have escaped replacement by protein alternatives in the evolution from an RNA World to the protein dominated world of modern biochemistry.
In this review we will examine four examples of RNA catalysis, each of which provides a different chemical strategy used to promote a ribozyme reaction. This includes: the group I class of self-splicing introns which employs two divalent metal ions to promote RNA cleavage and ligation reactions; the Hepatitis Delta Virus ribozyme which catalyzes autolytic RNA cleavage using an essential, pKa perturbed cytidine; the glmS ribozyme which is an autolytic riboswitch that uses the small molecule metabolite glucosamine-6-phosphate (GlcN6P) as a catalytic cofactor; and the ribosomal peptidyl transferase center which doesn’t appear to use any of these strategies, but instead forms peptide bonds with the assistance of a functional group on the tRNA substrate. The diversity of solutions employed by RNA to achieve reaction catalysis is unexpected; raising intriguing questions about the evolution and devolution of RNA catalysis. What led RNA to loose its catalytic role in some biological cases but not others, and what other catalytic functions RNA might still be playing in biology?
Group I intron-Two metal mechanism
RNA splicing by the group I intron involves two symmetrical phosphoryl transfer reactions, both catalyzed by the intron itself (Fig. 1). In the first reaction, an exogenous G attacks the 5′-splice site to release the 5′-exon from the intron. In the second reaction, the 5′-exon attacks the 3′-splice site, at the phosphate 3′ to the last G of the intron (ΩG), to produce ligated exons and liberate the intron. The second reaction is essentially the reverse of the first, differing only in whether exogenous G or ΩG is bound in the active site. An early model for group I intron catalysis was proposed by Steitz and Steitz based upon analogy to protein enzymes that catalyze phosphoryl transfer.1 Their proposal involved two active site divalent metal ions bridging across the scissile phosphate positioned 3.9 Å apart, one activating the O3′ nucleophile and the other activating the O3′ leaving group. Although at the time no evidence for this model was provided, it was greeted enthusiastically and efforts were made to find these catalytic metals within the group I intron and other ribozymes.
Figure 1.

Group I intron splicing promoted by two catalytic metal ions. The second of the two-step splicing reactions is shown. The nucleophile (U-1 O3′), scissile phosphate, leaving group (ΩG O3′) and labile bond are shown in orange. Both catalytic metal ions are shown in green. A similar coloring scheme is used in all four figures where the chemically reactive groups and labile bond are shown in orange and the catalytically important groups are shown in green. A. Active site structure adapted from Ref. 7. B. Reaction mechanism.7
Biochemical analysis provided the initial evidence for metal ion participation in group I intron catalysis. Active site metal ion coordination was probed using metal specificity switch analysis. These experiments change the identity of a putative metal ligand from an oxygen to a sulfur. The electronically "soft" sulfur substitution disrupts Mg2+ coordination, rendering the RNA inactive when the substitution is at a site of metal binding. Activity rescue upon addition of a "soft" metal provides evidence for metal ion coordination. Single and pair wise substitutions were made throughout the group I intron active site. Consistent with the Steitz and Steitz model, the biochemical data suggested that metals coordinate the nucleophile, leaving group and scissile phosphate, but the intron also has a metal coordinating the 2′-OH vicinal to the leaving group (Fig. 1). As a result, an alternative three metal mechanism was proposed, in which none of the metals bridged between the scissile phosphate and the ΩG O3′ leaving group.2,3 This active site architecture differed significantly from that proposed for the two metal model.
X-ray crystal structures of the group I intron became available in the past few years. Structures of three different introns in three different reaction states were reported in 2004.4–6 These provided three-dimensional views of the group I intron, but the RNA in each of these structures lacked at least one of the active site functional groups known to coordinate a catalytic metal ion. The structure of a catalytically active complex, including the complete intron and both exons and all metal ligands, was reported a year later (Fig. 1).7 The overall structure of the intron was unchanged from the previous structures, but significant differences were observed within the active site. The structure revealed two well-ordered and extensively coordinated Mg2+ ions positioned 3.9 Å apart, both making inner sphere coordination to the scissile phosphate. Metal M1 is coordinated to the terminal O3′ of the 5′-exon and the pro-RP oxygen of the scissile phosphate. Metal M2 also makes inner sphere coordination to the pro-RP oxygen of the scissile phosphate, as well as the O3′and O2′ of ΩG. Both metals have a total of five inner sphere coordinations including interactions with phosphate oxygens within the intron. The two metals are coordinated by all of the biochemically predicted ligands, including two ligands identified within the intron.
This two-metal ion structure suggests that the group I intron active site is mechanistically equivalent to a large number of protein-based phosphoryl transferases, including all known DNA and RNA polymerases. RNA enzymes and protein enzymes could not be evolutionarily related, so the equivalence of group I intron and polymerase active sites must result from convergent evolution. The intron uses the phosphate backbone in a manner equivalent to aspartate site chains in proteins for active site metal ion coordination, including one phosphate within the intron that uses both of its non-bridging oxygens to directly coordinate both active site metals. That macromolecular evolution arrived independently at the same solution in RNA and proteins implies an intrinsic catalytic capacity of the two-metal-ion catalytic architecture for phosphoryl transfer.
What remains unknown is whether other RNA enzymes, such as the group II intron, RNase P or the spliceosome, use the same catalytic strategy. Two RNase P structures were reported recently, but both complexes are of apoenzymes without the tRNA substrate.8,9 Biochemical experiments strongly implicate metal ions in catalysis,10 but conformational changes or metal ion rearrangements might accompany substrate binding. No high resolution structural views of the group II intron or splicesome exist, however biochemical data implicate divalent metal ions in catalysis in both systems. 11,12
HDV ribozyme- Nucleobase catalysis
Divalent metals are not the only solution RNA has found to catalyze its reactions. The symmetry of the two metal mechanism led to the premature conclusion that all ribozymes are metalloenzymes. Not every protein enzyme uses metals for phosphoryl transfer, and it is now clear that this is also true for ribozymes. For example, ribonuclease A employs two catalytic histidines to activate the nucleophile and stabilize the leaving group during the RNA cleavage reaction it promotes. Histidine, with its pKa of 6.8, is ideally suited for these roles at physiological pH. Because RNA lacks any nucleobase functional groups with a pKa near neutrality, nucleobase participation appeared to be an unlikely contributor to ribozyme chemistry. But it was then observed that several catalytic RNAs, including the HDV ribozyme, retained reasonable activity in the presence of high concentrations of monovalent ions, or other polyvalent ions that could not make inner sphere coordination to RNA ligands.13 This suggested alternative mechanisms must be in play.
The HDV ribozyme is a small autonucleolytic RNA that shares a common chemistry with the other nucleolytic ribozymes. The 2′-OH next to the scissile phosphate acts as nucleophile to displace the O5′ resulting in two RNA products, one with a 2′-3′ cyclic phosphate terminus and one with a 5′-OH terminus (Fig. 2). This is the same chemical reaction catalyzed by ribonuclease A. The first HDV crystal structure, of the product form of the ribozyme, showed that the 5′-OH was within hydrogen bonding distance to the N3 of C75, an invariant residue indispensable for efficient cleavage.14 The inactive C75U mutation could be rescued with imidazole and the pKa of the reaction appeared to track with the pKa of nucleobases substituted at the key residue.15,16 This suggested that C75 might function as a general acid to protonate the O5′ leaving group. However, subsequent crystal structures of the substrate form of the ribozyme showed that the C75 N3 is closer to the nucleophile, instead implicating C75 as a general base to deprotonate the 2′-OH.17 This controversy was biochemically resolved in an elegant experiment showing that an enzyme with an activated leaving group no longer required this C for function.18 These data strongly favor the original model; C75 acts as a general acid to protonate the leaving group and a divalent metal ion activates the nucleophile (Fig. 2). Although C75 is not as efficient as histidine in this role due to its acidic pKa, the microenvironment of the enzyme active site appears to perturb the pKa toward neutrality.
Figure 2.
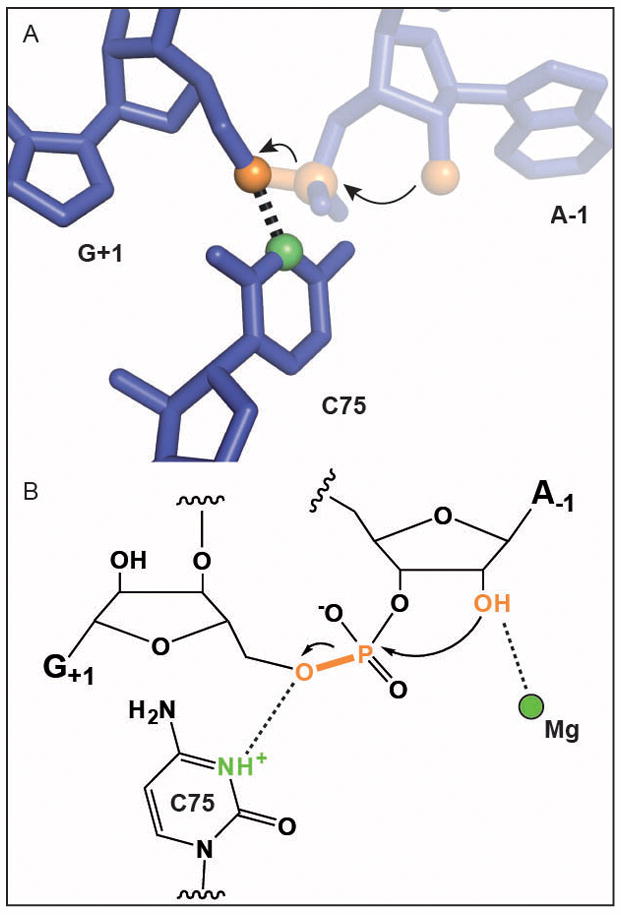
Autolytic cleavage of RNA by the HDV ribozyme. Color scheme as in Fig. 1. A. Active site structure adapted from Ref. 14. Residue A-1 was modeled into the coordinates for ease of comparison to the chemical scheme. B. Reaction mechanism.16,18
The precedent of general acid catalysis by the HDV ribozyme argues that other catalytic RNAs could employ such a strategy. Nucleobase catalysis has now been implicated in all of the autolytic ribozymes, but the diversity of the bases involved has come as a surprise. A critical A has been implicated in the VS ribozyme.19 The crystal structure of the full-length hammerhead ribozyme reveals two critical Gs in the active site.20 The N1 of G12 is near the O2′ nucleophile and the 2′-OH of G8 is close to the O5′ leaving group. The participation of G in RNA catalysis is unexpected as it has a alkaline pKa and the deprotonated form is anionic. It is relatively easy to envision how the highly anionic environment of an RNA could perturb a cationic pKa higher, but its harder to understand how the electrostatics of RNA could perturb an anionic pKa lower. Yet, Gs are also implicated in catalysis by the hairpin, VS and glmS (see below) ribozymes. 21,22 Proposed models for the role these Gs play in the reaction include simple substrate alignment by hydrogen bonding or proton transfer through tautomerization of the base. 23,24
Ribosomal Peptide Bond Formation by Substrate Assistance
The peptidyl transferase center of the ribosome is also a ribozyme.25 Initial mechanistic models for this reaction employed nucleobase involvement in catalysis (specifically A2451), however mutational analysis of this and other active site residues found that they contributed relatively little to the rate of peptide bond formation.26–28 The effects were larger for the hydrolysis reaction that occurs at termination, but even in that reaction the largest rate decreases were still about 100-fold. Further, all crystallographic efforts to identify active site metal ions have also been unsuccessful.29 While there is an obligate Mg2+ requirement for peptidyl transferase activity, the contribution does not appear to result from direct chemical participation. Relative to the rate in solution, the peptidyl transferase center of the ribosome provides a rate enhancement of approximately 107 fold, a contribution that has been largely attributed to entropy.30 Beyond substrate alignment, how else might the ribosome promote this biologically essential reaction?
The peptidyl transferase reaction involves two substrate tRNAs, an aminoacyl-tRNA that binds to the ribosomal A site and a peptidyl tRNA that binds to the P site. The nascent peptide in the P site is linked via an ester to the 3′-O of the terminal nucleoside of the tRNA, A76 (Fig. 3).31 The reaction involves aminolysis of the P-site ester by the A-site α-amino group. The ribosome aligns the two substrates such that the amine approaches the sc face of the P-site ester, resulting in a chiral transition state with an S stereochemistry. 29
Figure 3.

Peptide bond formation promoted by the ribosome. The α-amine, carbonyl carbon, O3′ leaving group and scissile bond are shown in orange. The critical 2′-OH is in green. A. Active site structure adapted from Ref. 29. B. Proposed reaction mechanism.29,34
The A76 2′-OH vicinal to the P-site ester is essential for the reaction.32 Deletion or alteration of this functional group results in a complete loss of peptidyl transferase activity (>106 fold loss of activity). Without this hydroxyl group, the rate of spontaneous ester hydrolysis is faster than the rate of peptide bond formation. This contribution is significantly greater than that made by any of the rRNA nucleobases within the active site, though the 2′-OH of A2451 has also been shown to contribute to the reaction.33
Hydroxyl groups are unlikely catalytic players in a reaction that appears to involve proton transfer. With a pKa of approximately 12, the 2′-OH is less likely than the nucleotide bases to be involved in proton transfer even if the pKa were significantly perturbed. Furthermore, there is nothing about the ribosome active site that would lead to such a perturbation. All unexplained density near the reaction center appears to be water, including one molecule positioned within the oxyanion hole.29 Mechanistic models must therefore explain the role of the 2′-OH without invoking a significant degree of protonation or deprotonation.
The ribosome positions the critical A76 2′-OH between the α-amino nucleophile and the 3′-O leaving group (Fig. 3). As the amine must lose a proton and the O3′ must gain one, mechanisms have been proposed that involve concerted proton transfer from the α-amine to the A76 O3′ via the A76 2′-OH. 29,34–37
These mechanisms explain the need for the 2′-OH without having it assume a significant charge, consistent with its elevated pKa. Several mechanisms have been proposed that invoke this role for the 2′-OH, each differing in the extent of peptide bond formation and the degree of leaving group dissociation in the transition state. Although the mechanisms share common themes, they make significantly different predictions regarding the nature of the transition state and the contribution the ribosome could make to catalysis. The answer will require enzymatic analysis to define the degree of bonding and distribution of charge in the transition state. Only then will it be clear exactly what transition state the ribosome stabilizes during peptide bond formation and the precise role of the 2′-OH of A76.
Independent of these mechanistic details, the observation that a functional group on the tRNA substrate provides a major catalytic contribution invokes an additional role for tRNAs in translation. Even before the discovery of tRNA, Crick proposed the adaptor hypothesis, which predicted a molecule capable of two functions: interaction with an mRNA and delivery of the correct amino acid. tRNA plays both these roles, but the adaptor model suggests a passive role for tRNA during translation. tRNA is aminoacylated by synthases and used by the ribosome for decoding, but the tRNAs are the object, rather than the subject of action in these roles. In a recent review Woese called for a reevaluation of tRNA during translation,38 and there are now indications that tRNA does play a significant proactive role, including a spring-like distortion of the tRNA during accommodation into the A site where it provides the key functional group for peptide bond formation to proceed.32,39
GlmS ribozyme- Use of a catalytic cofactor
And when it appeared that all permutations for RNA catalysis had been exhausted, a ribozyme was identified that uses still another mechanistic strategy (Fig. 4). The glmS riboswitch is located in the 5′-untranslated region of the gene encoding glucosamine-6-phosphate (GlcN6P) synthetase.40 In the presence of GlcN6P, it cleaves its own mRNA, which down-regulates the production of the synthetase. GlcN6P is essential for efficient cleavage activity, though other primary amines with a vicinal hydroxyl group can also activate the ribozyme in vitro, including serinol and Tris. 41 The primary amine is essential for the reaction; substitution of the amine with a hydroxyl results in complete loss of activity. Glucose-6 phosphate (Glc6P) is still able to bind to the ribozyme, but functions as a competitive inhibitor.
Figure 4.
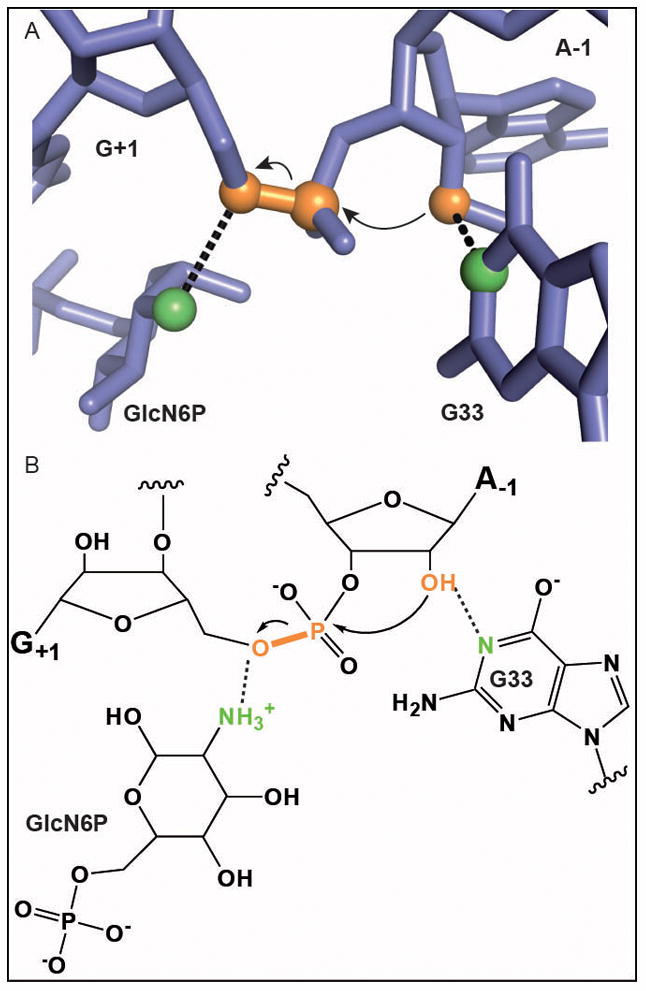
Autolytic cleavage of RNA by the glmS ribozyme in the presence of GlcN6P. The color scheme is as in Fig. 1 with the critical amine of GlcN6P and the N1 of G33 shown in green. A. Active site structure adapted from Ref. 45. B. Proposed reaction mechanism.45 The identity of G33 is important to the reaction, but its role remains to be fully defined.
Two possible models for the role of GlcN6P in glmS catalysis can be envisioned. In traditional riboswitches, metabolite binding induces a conformational change in the RNA that affects the transcription or translation of the gene.42 In this model for riboswitch function, binding of the GlcN6P would stabilize the active ribozyme conformation and lead to catalysis. In the alternative model, GlcN6P binding does not affect the conformation of the RNA, but instead the critical amine on the metabolite participates directly in chemistry.
Biochemical and structural data both indicate that the contribution made by GlcN6P is not conformational, but is instead chemical.40,43–45 Initial inline probing experiments showed that the conformational flexibility of the RNA is unchanged upon GlcN6P binding.40 Hydroxyl radical footprinting experiments showed that the RNA had an equivalent solvent accessible surface in both the bound and free states.43 Consistent with these observations, independent crystal structures revealed that the RNA adopts the same conformation in the metabolite bound, inhibitor bound and free states.44,45 Thus, glmS is not a traditional riboswitch, changing its conformation upon metabolite binding, instead it uses the chemical capability of the metabolite in catalysis. The pKa of the GlcN6P amine is ~8.3, a value reasonably well-suited for a role in proton transfer. This demonstrates that RNA, like protein enzymes, can employ the chemical potential of small molecules to promote catalytic activity.
How does GlcN6P serve as a catalytic cofactor? The authors of the initial structure of the Glc6P inhibitor bound ribozyme suggested that the amine might serve as a general base to deprotonate the O2′ nucleophile.44 Although the C2 hydroxyl is not well-positioned for this role, the authors proposed that it could act through a chain of two water molecules present in the active site. The GlcN6P bound structure superimposes on the Glc6P structure, but no water molecules for proton shuttling to the O2′ were observed.45 In both structures the amine (or its hydroxyl substitute) is within hydrogen bond distance of the O5′ leaving group (Fig. 4). This suggests a role for the amine as a general acid to activate the leaving group. A definitive answer to this question awaits the kind of analysis that was performed on the HDV ribozyme.18 The closest functional group to the O2′ nucleophile is not on GlcN6P, but is the N1 of G33 (Fig. 4). Consistent with a role in ribozyme function, mutation of G33 disrupts activity.45 However the precise contribution of this functional group to glmS ribozyme catalysis is also unknown. Neverthelss, the primary catalytic contribution is expected to come through the metabolic cofactor, as activation of the leaving group is the chemically difficult step in phosphoryl transfer reactions.
Implications
The mechanisms employed by RNA to promote chemistry are substantially more diverse than once thought. They include metals, nucleobases, ribosyl groups and even small molecule cofactors. Such observations raise questions regarding the evolution of biocatalysis and the diversity of reactions that RNA might catalyze.
The observation that the group I intron has a polymerase-like active site built of RNA leads to the question: What selective pressure led to the evolution of the current protein based system for replication and transcription? All nucleic acid polymerases are built from protein and all appear to use the same mechanism, namely two divalent metal ions that bridge across the α-phosphate of the incoming nucleotide triphosphate. The reaction catalyzed by the protein based polymerase and the RNA based intron are virtually equivalent. The nucleophile is the same in both cases, namely a 3′-OH (of the primer strand for polymerase, of the 5′-exon for splicing). The leaving group is different (G for the intron, pyrophosphate for the polymerase) but the organization of the metals is identical. For an RNA World to be viable, there must have been some kind of RNA-based polymerase to catalyze RNA replication. The group I structure demonstrates that RNA can fold and bind metals to create a polymerase-like active site, but it appears that biology has abandoned this catalyst. The reason for this transformation is likely to involve issues of fidelity, processivity and reaction rates, all of which are expected to be greater in protein based systems.
Why hasn’t a similar selective pressure led protein to usurp RNA as the catalyst of peptide bond formation? In spite of the fact that all nucleotide polymerases are made of protein, all amino acids are still polymerized within an RNA active site. The role of rRNA in this reaction appears to be largely organizational, binding to the CCA ends of each tRNA and positioning the reactive groups for chemistry. The key functional group is a 2′-OH located at the end of the tRNA, the substrate of the reaction. Under sufficient evolutionary pressure, protein should have taken over the role of aminoacyl polymerase just as it did for the nucleotidyl polymerase. If the issues are fidelity, processivity and reaction rates, why didn’t these same parameters lead to a protein based system for peptide bond formation? This hasn’t happened, or at least hasn’t hadn’t yet. The amino terminus of protein L27 is intriguingly close to the peptidyl transferase center as observed in the recent 70S crystal structure from Thermus thermophilus.46 L27 is not conserved and so is not essential for protein synthesis in all organisms, but this terminal extension is essential in E. coli.47 L27 may represent an adaptation, possibly an evolutionary intermediate between an RNA and a protein based active site for protein synthesis.
Studies of the glmS ribozyme revealed that RNA can use small molecules as catalytic cofactors. With an expanded chemical repertoire made possible by small organic compounds, what other reactions did (or still does) RNA catalyze in biological systems? To date, riboswitches responsive to 10 different small molecules have been identified.42 The metabolites include coenyzme B12 (B12), flavin mononucleotide (FMN), thiamine pyrophosphate (TPP), S-adenosyl methionine (SAM), guanine, adenine, glycine, lysine and GlcN6P. The intriguing nature of this list is that many of the metabolites that are recognized by riboswitches are also among the most ubiquitous cofactors used by protein enzymes, particularly B12, FMN, TPP and SAM. Protein enzymes use these cofactors and substrates for radical chemistry, oxidation-reduction reaction and carbon-carbon bond formation. To date, chemistry promoted by naturally occurring RNAs has been confined to phosphoryl transfer and transesterification. Naturally occurring RNAs bind these small molecules. The use of their chemical moieties in chemistry could significantly expand the catalytic repertoire of RNA. Ribozymes with radical, redox and methyltransferase activities were needed for metabolism in the RNA World. Which, if any of them, have persisted into modern biology remains to be determined.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shan S, Yoshida A, Sun S, Piccirilli JA, Herschlag D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc Natl Acad Sci U S A. 1999;96:12299–304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shan SO, Herschlag D. Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry. 1999;38:10958–75. doi: 10.1021/bi990388e. [DOI] [PubMed] [Google Scholar]
- 4.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 5.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–62. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol. 2005;12:82–9. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 7.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 8.Kazantsev AV, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–7. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–7. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 10.Warnecke J, Held R, Busch S, Hartmann R. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J Mol Biol. 1999;290:433–45. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 11.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–4. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon PM, Fong R, Piccirilli JA. A second divalent metal ion in the group II intron reaction center. Chem Biol. 2007;14:607–12. doi: 10.1016/j.chembiol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–95. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 14.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–74. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 15.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–6. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 16.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–7. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 17.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–5. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 18.Das S, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem Bio. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 19.Lafontaine DA, Wilson TJ, Norman DG, Lilley DM. The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol. 2001;312:663–74. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- 20.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–20. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–4. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 22.Wilson TJ, McLeod AC, Lilley DM. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzmin Y, Da Costa C, Fedor M. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J Mol Biol. 2004;340:233–51. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 24.Pinard R, et al. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–42. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen P, Hansen J, Ban N, Moore P, Steitz T. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J, et al. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyl transferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci U S A. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polacek N, Gaynor M, Yassin A, Mankin AS. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature. 2001;411:498–501. doi: 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- 28.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–99. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 29.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–48. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Sievers A, Beringer M, Rodnina MV, Wolfenden R. The ribosome as an entropy trap. Proc Natl Acad Sci U S A. 2004;101:7897–901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang KS, Weinger JS, Butler EB, Strobel SA. Regiospecificity of the peptidyl tRNA ester within the ribosomal P site. J Am Chem Soc. 2006;128:3108–9. doi: 10.1021/ja0554099. [DOI] [PubMed] [Google Scholar]
- 32.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–6. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 33.Erlacher MD, et al. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J Am Chem Soc. 2006;128:4453–9. doi: 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 34.Dorner S, Polacek N, Schulmeister U, Panuschka C, Barta A. Molecular aspects of the ribosomal peptidyl transferase. Biochem Soc Trans. 2002;30:1131–1137. doi: 10.1042/bst0301131. [DOI] [PubMed] [Google Scholar]
- 35.Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci U S A. 2005;102:12395–400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Changalov M, et al. 2′/3′-O-peptidyl adenosine as a general base catalyst of its own external peptidyl transfer: Implications for the ribosome catalytic mechanism. ChemBioChem. 2005;6:992–996. doi: 10.1002/cbic.200400349. [DOI] [PubMed] [Google Scholar]
- 37.Rodnina MV, Beringer M, Wintermeyer W. Mechanism of peptide bond formation on the ribosome. Q Rev Biophys. 2006;39:203–25. doi: 10.1017/S003358350600429X. [DOI] [PubMed] [Google Scholar]
- 38.Woese CR. Translation: in retrospect and prospect. RNA. 2001;7:1055–67. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–80. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–6. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy TJ, et al. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–6. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–63. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 43.Hampel KJ, Tinsley MM. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–71. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- 44.Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–6. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 45.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 47.Maguire B, Beniaminov A, Ramu H, Mankin A, Zimmermann R. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol Cell. 2005;20:427–35. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]


