Abstract
The SR superfamily of splicing factors and regulators is characterized by arginine/serine (RS)-rich domains, which are extensively modified by phosphorylation in cells. In vitro binding studies revealed that RS domain–mediated protein interactions can be differentially affected by phosphorylation. Taking advantage of the single nonessential SR protein–specific kinase Sky1p in Saccharomyces cerevisiae, we investigated RS domain interactions in vivo using the two-hybrid assay. Strikingly, all RS domain–mediated interactions were abolished by SKY1 deletion and were rescuable by yeast or mammalian SR protein–specific kinases, indicating that phosphorylation has a far greater impact on RS domain interactions in vivo than in vitro. To understand this dramatic effect, we examined the localization of SR proteins and found that SC35 was shifted to the cytoplasm in sky1Δ yeast, although this phenomenon was not obvious with ASF/SF2, indicating that nuclear import of SR proteins may be differentially regulated by phosphorylation. Using a transcriptional repression assay, we further showed that most LexA-SR fusion proteins depend on Sky1p to efficiently recognize the LexA binding site in a reporter, suggesting that molecular targeting of RS domain–containing proteins within the nucleus was also affected. Together, these results reveal multiple phosphorylation-dependent steps for SR proteins to interact with one another efficiently and specifically, which may ultimately determine the splicing activity and specificity of these factors in mammalian cells.
Keywords: SR proteins, SRPK1, SRPK2, Clk, Sty, RS domains
Pre-mRNA splicing takes place in an RNA machine known as the spliceosome, which consists of small nuclear ribonucleoprotein particles (snRNPs)1 and non-snRNP protein factors. The RNA components in the spliceosome form the catalytic core through a series of dynamic RNA–RNA interactions which are likely to be mediated and/or stabilized by non-snRNP protein factors (for recent reviews see Nilsen, 1998; Staley and Guthrie, 1998). Among the best characterized non-snRNP splicing factors are SR proteins which contain one or two RNA recognition motifs and a signature RS domain containing multiple serine and arginine repeats (for reviews see Fu, 1995; Manley and Tacke, 1996). The RNA recognition motifs bind to RNA sequences in a coordinated fashion to determine splicing specificity (Chandler et al., 1997) and commit pre-mRNA substrates to the splicing pathway (Fu, 1993), whereas the RS domains mediate specific protein– protein interactions in a number of spliceosomal assembly steps (Wu and Maniatis, 1993; Kohtz et al., 1994; Roscigno and Garcia-Blanco, 1995; Tronchère et al., 1997). In fact, the RS domain is present in a large number of proteins, many of which are proven splicing factors and/or regulators (for review see Fu, 1995).
SR proteins are especially interesting because their functions in splice site selection and spliceosome assembly appear to be regulated by phosphorylation. In vitro, phosphorylation of RS domains can enhance the interactions between RS domain–containing proteins (Xiao and Manley, 1997; Wang et al., 1998) and prevent their nonspecific binding to RNA (Tacke et al., 1997), which may explain the requirement of phosphorylated SR proteins to initiate spliceosome assembly at early steps (Mermoud et al., 1994; Cao et al., 1997). On the other hand, dephosphorylation appears to be required for the resolution of the spliceosome to produce spliced products (Tazi et al., 1993; Mermoud et al., 1994; Cao et al., 1997). Thus, the phosphorylation–dephosphorylation cycle of RS domains appears to accompany, and likely regulate, the splicing cycle.
The impact of phosphorylation on SR protein function in vivo is further manifested by the localization and intracellular trafficking of splicing factors in mammalian cells. Most splicing factors are unevenly distributed in the nucleus, giving rise to a speckled appearance in all somatic animal cells examined to date (for review see Spector, 1993). Although nuclear speckles contain polyadenylated RNA (Xing et al., 1993) and are associated with some actively transcribing genes (Xing et al., 1995), most splicing reactions appear to take place cotranscriptionally in the nucleoplasm and, therefore, splicing factors need to be recruited to sites of transcription (Misteli et al., 1997, 1998). Importantly, this intranuclear movement of splicing factors appears to be mediated by RS domain phosphorylation (Gui et al., 1994a; Colwill et al., 1996a; Cáceres et al., 1997; Misteli et al., 1997, 1998; Duncan et al., 1998). Because SR proteins are known to affect splice site selection in a concentration-dependent manner, such phosphorylation-dependent localization of splicing factors among different nuclear pools may be a mechanism to regulate alternative splicing. Indeed, it was demonstrated recently that alternative splicing in vivo can be modulated by phosphorylation, presumably through modifying SR proteins (Duncan et al., 1997; Kanopka et al., 1998). These observations illustrate the importance of understanding the function and regulation of SR proteins by specific kinases in cells.
Mammalian cells express several SR protein–specific kinases, of which the best characterized are the SRPK and Clk/Sty families of kinases (Gui et al., 1994a; Colwill et al., 1996ab; Duncan et al., 1998; Kuroyanagi et al., 1998; Wang et al., 1998). These two kinase families are structurally distinct, exhibit different substrate specificities, and are differentially expressed (Gui et al., 1994b; Colwill et al., 1996b; Wang et al., 1998). The SRPK and Clk/Sty kinases are also differentially localized in cells: SRPKs are present in both the cytoplasm and the nucleus (Wang et al., 1998), whereas Clk/Sty kinases appear to localize exclusively in the nucleus (Colwill et al., 1996a; Duncan et al., 1998). The existence of multiple SR protein kinases with distinct substrate specificity and localization in mammalian cells suggests that RS domain–mediated interactions may be subject to coordinate regulation by phosphorylation in a temporal- and spatial-specific manner. Therefore, it is important to study phosphorylation regulation of pre-mRNA splicing at both the biochemical and cellular levels.
In this study, we established a novel model system using Saccharomyces cerevisiae to investigate the importance of phosphorylation in RS domain–mediated protein–protein interactions in vivo. Unlike mammalian cells that express many SR protein kinases, budding yeast express only a single conserved SR protein–specific kinase, Sky1p, which was cloned and characterized recently in our lab (Siebel et al., 1999). Structural and functional studies demonstrate that Sky1p is a SRPK family member that has the same high activity and specificity for mammalian SR proteins as SRPK1 and SRPK2. Moreover, increasing evidence suggests that budding yeast express proteins that are structurally and functionally related to mammalian RS domain– containing proteins. Therefore, yeast provide a powerful genetic system to study SR protein function in vivo, which will provide clues to the function and regulation of SR proteins in mammalian cells. Here, we took advantage of the fact that SKY1 is not essential for vegetative growth in yeast. Therefore, we were able to express mammalian RS domain–containing proteins and study their interactions in cells with and without SRPK-mediated phosphorylation, an approach that was not possible in mammalian cells. Our results demonstrate that RS domain–mediated protein– protein interactions in vivo are entirely dependent on the activity of Sky1p, which can be substituted by SRPK1 or Clk/Sty from mammalian cells. Further examination of RS domain interactions in this system reveal that, in addition to the modulation of protein affinities, SRPKs appear to play an important role in mediating nuclear import of SR proteins as well as in finding their targets within the nucleus.
Materials and Methods
In Vitro Binding
GST pull-down assays were performed as described (Wang et al., 1998). Bacterially expressed GST-ASF/SF2 (100 μg) was phosphorylated in vitro using baculovirus-expressed GST-SRPK1 (10 U, see Gui et al., 1994b) for 6 h at 30°C with or without 1 mM ATP. Proteins were desalted on G-50 columns (Pharmacia) and rebound to glutathione beads before binding to TNT (Promega) translated U1-70K or U2AF35. Empty beads or GST-loaded beads were used as controls.
Two-Hybrid Interactions
Two-hybrid assays were performed as described using the LexA system (Gyuris et al., 1993; Wu and Maniatis, 1993). Strain EGY48 was deleted of the SKY1 gene by recombination with a kanamycin resistance expression unit flanked by SKY1 genomic sequences (Wach et al., 1994). The deletion encompassed 316 bp 5′ of the initiation codon to 541 bp 5′ of the termination codon, and was confirmed by PCR. Bait and prey plasmids were gifts from J. Wu (Wu and Maniatis, 1993) or were constructed in pEG202 and pJG4-5 with modified polylinkers (Wang et al., 1997). Expressed proteins were verified by Western blotting with a monoclonal anti-LexA antibody (Clontech) using the enhanced chemiluminescent detection system (Pierce) in conjunction with HRP-labeled goat anti–mouse antibodies. Kinase expression plasmids were constructed in p415GAL1 (Mumberg et al., 1994) for selecting Leu+ phenotype (Wu and Maniatis, 1993). Deletions of the RS domains in U2AF65 (residues 1–65) and U2AF35 (residues 191– 240) were made by PCR. β-Galactosidase activity is expressed as Miller units: OD420 × 1,000/OD600 × time (min) × culture volume assayed (ml), determined at 30°C.
Immunolocalization
Freshly streaked yeast clones bearing prey plasmids (HA tagged) were inoculated in 5 ml YPGal/Raff and grown for 3 h at 30°C. Yeast were fixed with 0.7 ml 37% formaldehyde and processed for immunofluorescence as described (McConnell et al., 1990), using the 12CA5 antibody at 1:100.
GFP-ASF/SF2 Expression
Humanized GFP (pGreenLantern-1; Life Technologies) was fused in frame to the NH2 terminus of ASF/SF2 wild-type cDNA or the KS substitution mutant (Cáceres and Krainer, 1993). The fusion protein was expressed from pcDNA3 (Invitrogen) in HeLa cells for 48 h before visualization of live cells. Whole cell extracts (200 μg) were treated with 500 U CIP (Boehringer Mannheim; Misteli et al., 1998) and subjected to Western analysis using an anti-GFP antibody (Chemicon).
Transcriptional Repression
Both the parent EGY48 and corresponding sky1Δ strains were grown on 5-FOA plates to eliminate the pSH18-24 two-hybrid reporter (lacZ) plasmid. The resulting strains were cotransformed with pEG202-based bait plasmids and pJK101, a transcriptional repression reporter (Golemis and Brent, 1992), then assayed for β-galactosidase activity. Maximal transcription was determined using a modified bait plasmid lacking LexA.
Results
Differential Modulation of RS Domain Interactions In Vitro
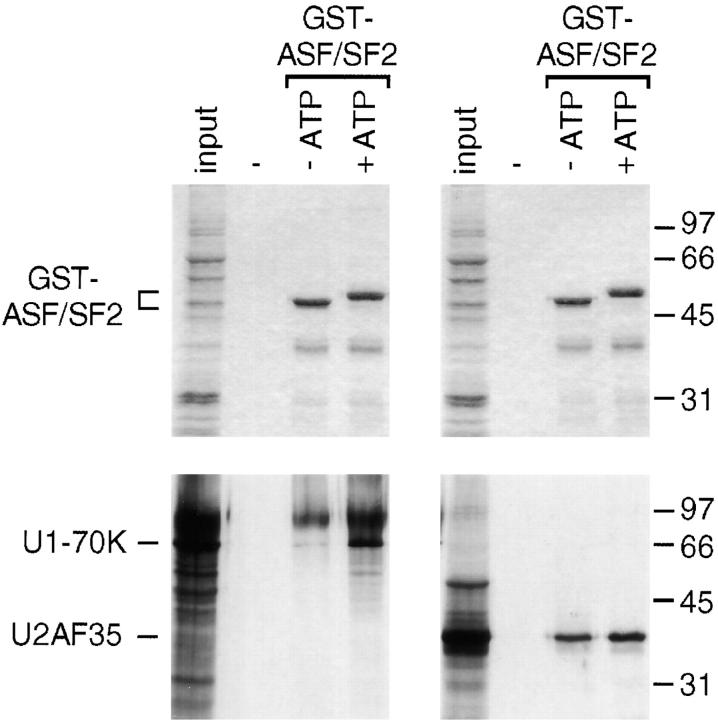
Phosphorylation of the SR protein ASF/SF2 by SRPK1, SRPK2, or Clk/Sty has been shown previously to enhance its interaction in vitro with the U1 snRNP-specific 70-kD protein (U1-70K) in a GST pull-down assay (Xiao and Manley, 1997; Wang et al., 1998; Fig. 1). This enhancement may result from the neutralization of charges, allowing RS domains to interact efficiently (Xiao and Manley, 1997). To test the generality of this principle, we examined the effect of SRPK1-mediated phosphorylation on ASF/SF2 binding to another RS domain–containing splicing factor, U2AF35, an interaction proposed for pairing the 5′ and 3′ splice sites (Wu and Maniatis, 1993). Unexpectedly, binding of unphosphorylated ASF/SF2 to U2AF35 was readily detectable, and this interaction was largely unaffected by SRPK1-mediated phosphorylation (Fig. 1). These results indicate that phosphorylation differentially modulates the affinity between RS domain–containing splicing factors, depending on the pairs examined. A similar observation was also made independently by Xiao and Manley (1998).
Figure 1.
SRPK1-mediated phosphorylation differentially modulates the affinity of RS domain interactions in vitro. In a GST pull-down assay, mock-phosphorylated (−ATP) or SRPK1-treated GST-ASF/SF2 (+ATP) were bound to 35S-labeled, in vitro translated U1-70K (left four lanes) or U2AF35 (right four lanes) followed by analysis on SDS-PAGE. Coomassie-stained bands are shown in the top panel to illustrate the mobility shift due to phosphorylation, and the lower panel shows an autoradiograph of the same gel. A band above the U1-70K could be a modified form of U1-70K because it is only present in U1-70K RNA-programmed in vitro translation reactions.
Detection of Phosphorylation-dependent Interactions In Vivo
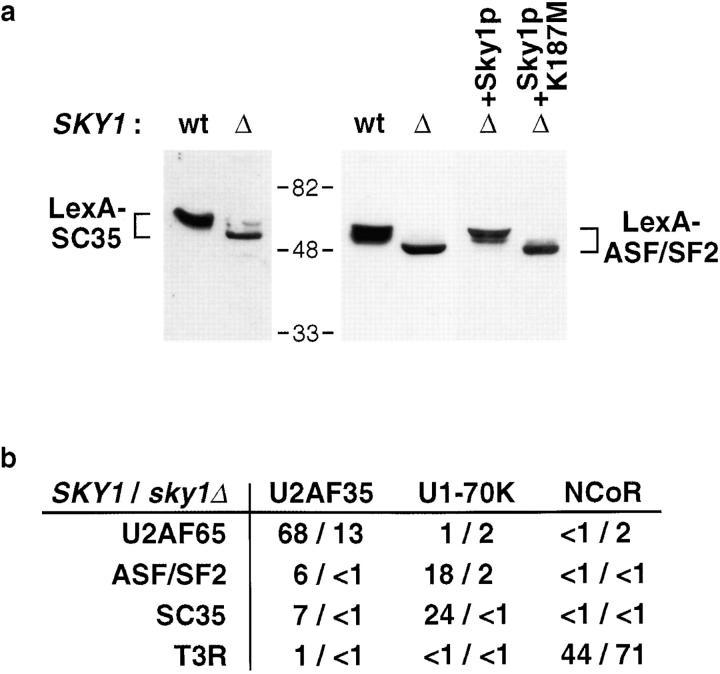
To examine the role of phosphorylation in protein–protein interactions under physiological conditions, we took advantage of the single conserved SR protein–specific kinase in S. cerevisiae to investigate RS domain interactions in the presence or absence of phosphorylation. Mammalian SR proteins expressed in both wild-type and SKY1 deletion (sky1Δ) yeast strains were soluble, but differed in their phosphorylation states, as determined by mobility shift on SDS-PAGE as well as Western blotting with mAb 104, a monoclonal antibody specific for the phosphoepitope present in all SR proteins (data not shown; Siebel et al., 1999). As shown in Fig. 2 a, LexA-ASF/SF2 and LexA-SC35 expressed in wild-type yeast migrated more slowly than those from sky1Δ yeast (left four lanes), indicative of a change in phosphorylation states (see Xiao and Manley, 1997). Reexpression of Sky1p, but not an ATP binding site mutant (Sky1p K187M), restored the phosphorylated form of ASF/SF2 (right two lanes). Therefore, Sky1p appears to be the only endogenous kinase capable of efficiently phosphorylating mammalian SR proteins expressed in yeast. This system offers a unique opportunity to test the phosphorylation dependence of RS domain interactions in vivo using the two-hybrid strategy.
Figure 2.
RS domain interactions in vivo require phosphorylation. (a) Western blotting analysis of LexA-SC35 and LexA-ASF/SF2 expressed in wild-type or sky1Δ S. cerevisiae (left four lanes). Whole cell yeast extracts were probed with anti-LexA antibodies. The right two lanes show extracts from sky1Δ yeast coexpressing ASF/SF2 with Sky1p or an ATP binding site mutant, Sky1p K187M. (b) Two-hybrid analysis of protein–protein interactions in wild-type and sky1Δ yeast. Baits are listed on the left and preys are listed across. Data are expressed as fold over background using the empty prey vector in wild-type versus mutant yeast.
The results of two-hybrid assays are shown in Fig. 2 b. As a control, the interaction between the nuclear receptor transcription factor T3R and its corepressor NCoR, which are not SRPK1 substrates (data not shown), was unaffected by the SKY1 deletion. In contrast, the interactions between RS domain–containing splicing factors, all SRPK substrates, were largely eliminated in sky1Δ yeast. Growth assays of these interactions agreed with the quantitative assays (data not shown). In particular, RS domain–mediated interactions between U1-70K and either ASF/SF2 or SC35 were completely abolished in sky1Δ yeast, consistent with the in vitro binding data. As expected, the interaction between ASF/SF2 or SC35 with U2AF35 was detectable in wild-type yeast, but we never detected any interaction between these three RS domain–containing splicing factors in parallel experiments in sky1Δ yeast. Surprisingly, the two-hybrid interaction between U2AF65 and U2AF35 was also severely affected by the SKY1 deletion, despite the fact that their interaction takes place outside their RS domains (Zhang et al., 1992; see below). These results clearly indicate that phosphorylation has a far greater impact on RS domain interactions in vivo than in vitro, reflecting additional phosphorylation-dependent events for the interaction between RS domain proteins in cells.
Functional Rescue by Yeast and Mammalian SR Protein Kinases
To further demonstrate that Sky1p-mediated phosphorylation is critical for RS domain–containing proteins to interact with one another, we conducted kinase rescue experiments. As shown in Table I, Sky1p, but not its ATP binding site mutant, was able to restore the two-hybrid interactions examined, proving that Sky1p kinase activity is required. Because mammalian cells express more than one kinase family for SR proteins, the yeast model system provides an opportunity to evaluate functional similarities between different mammalian SR protein–specific kinases. Therefore, we extended the rescue experiments to SRPK1 and Clk/Sty from mammalian cells. As shown in Table I, both SRPK1 and Clk/Sty, but not corresponding ATP binding site mutants, were able to restore the interactions of U1-70K with both ASF/SF2 and SC35. In contrast, an unrelated kinase, the catalytic subunit of protein kinase A, did not rescue these interactions. Therefore, kinases from both the SRPK and Clk/Sty families appear to be functionally equivalent in this assay. The restoration of these two-hybrid interactions by different SR protein kinases strongly argues against the possibility that the lack of RS domain–mediated interactions in sky1Δ yeast is due to a general defect resulting from the SKY1 deletion. Furthermore, the ability of Clk/Sty to restore the two-hybrid interactions in sky1Δ yeast implies that there may be no endogenous kinase with the same activity and specificity as Clk/ Sty in S. cerevisiae, whereas mammalian cells have evolved multiple SR protein kinases to regulate RS domain–containing splicing factors.
Table I.
SR Protein–specific Kinase Rescue of Two-Hybrid Interactions
| Strain | Kinase expressed | SC35:U1-70K | ASF/SF2:U1-70K | |||
|---|---|---|---|---|---|---|
| SKY1 | None | 100 | 100 | |||
| sky1Δ | None | 0 | 0 | |||
| sky1Δ | Sky1p | 43 | 35 | |||
| sky1Δ | Sky1p K187M | 0 | 0 | |||
| sky1Δ | SRPK1 | 104 | 100 | |||
| sky1Δ | SRPK1 K109M | 0 | 0 | |||
| sky1Δ | Clk/Sty | 86 | 98 | |||
| sky1Δ | Clk/Sty K 190R | 1 | 2 | |||
| sky1Δ | PKA | 1 | 1 |
β-Galactosidase activities were determined in SKY1 or in sky1Δ yeast coexpressing either ASF/SF2 or SC35 and U1-70K in addition to the indicated kinases or mutated kinase controls. Results are expressed as a percentage of the maximal two-hybrid interaction for each pair.
Nuclear Localization of SC35 Affected by SKY1 Deletion
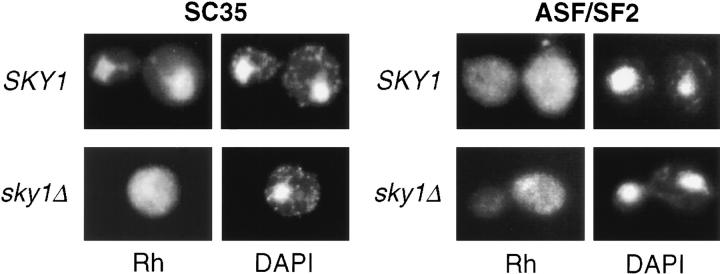
The absolute dependence of RS domain–mediated interactions in vivo on phosphorylation is in sharp contrast to the affinity changes modulated by phosphorylation in vitro. This difference raises the possibility that other aspects of SR protein function may be affected by phosphorylation in vivo and, therefore, offers an opportunity to investigate additional phosphorylation-dependent events. For instance, hypophosphorylated SR proteins may be sequestered in the nucleus due to nonspecific binding to other proteins or nucleic acids, as suggested by in vitro RNA binding studies (Tacke et al., 1997; Xiao and Manley, 1997). Therefore, we examined the localization of expressed SR proteins in yeast by immunohistochemistry. As shown in Fig. 3, SC35 expressed from the prey vector was largely localized in the nucleus in wild-type yeast, but dispersed uniformly in both the cytoplasm and the nucleus in sky1Δ yeast. In other experiments using a prey vector lacking an exogenous nuclear localization signal, SC35 was uniformly distributed in wild-type cells, but excluded from the nucleus in sky1Δ yeast, leaving a “hole” in the immunofluorescence image (data not shown). These unanticipated findings suggest that Sky1p-mediated phosphorylation may regulate nuclear import of SC35 expressed in yeast. However, the localization of ASF/SF2, U1-70K, and U2AF35 seemed mostly unaffected by the SKY1 deletion as they were generally uniformly distributed in both the nucleus and cytoplasm of wild-type and sky1Δ yeast (Fig. 3 and data not shown). These observations suggest for the first time that SRPK-mediated phosphorylation plays an important role in nuclear import of SR proteins, although not all SR proteins are affected in the same way.
Figure 3.
Localization of SR proteins in yeast is differentially affected by Sky1p-mediated phosphorylation. HA-tagged SC35 or ASF/SF2 was expressed from the prey vector in wild-type or sky1Δ yeast. The expressed proteins were detected with a monoclonal antibody (12CA5) followed by rhodamine-labeled goat anti–mouse antibodies (left panels); yeast DNA was stained with DAPI (right panels).
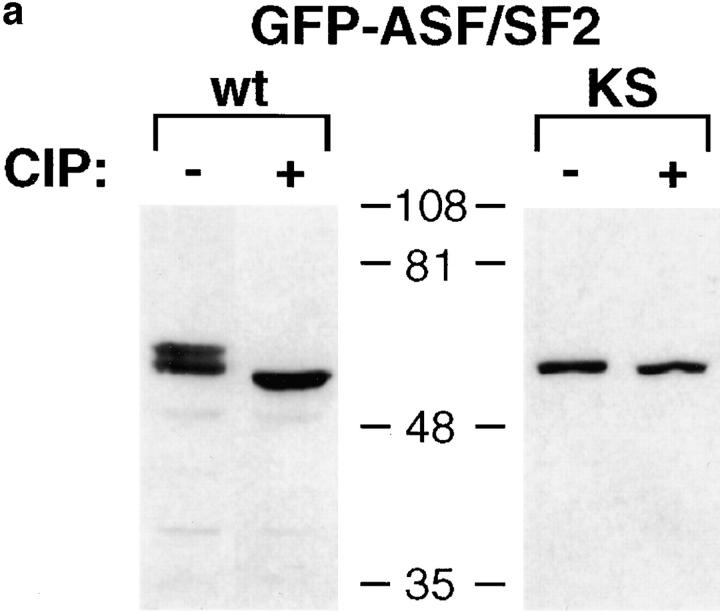
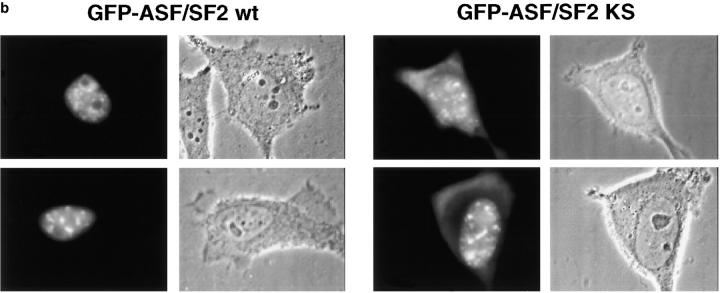
To obtain further evidence that SRPK-mediated phosphorylation plays a role in nuclear localization of RS domain proteins in mammalian cells, we took advantage of the KS substitution mutant of ASF/SF2 in which arginines in the RS domain are replaced by lysines (Cáceres and Krainer, 1993). We have shown previously that ASF/SF2-KS is not a substrate for the SRPK family of kinases, but can be phosphorylated by Clk/Sty (Colwill et al., 1996b). Consistent with the observation that there appears to be no Clk/Sty-like kinase activity in yeast, ASF/SF2-KS did not interact with U1-70K in the two-hybrid assay even in wild-type yeast (data not shown). We expressed wild-type ASF/SF2 and ASF/SF2-KS as GFP fusion proteins in HeLa cells, and expressed proteins were examined for phosphorylation-dependent mobility shifts on SDS-PAGE (Fig. 4 a). Treatment with alkaline phosphatase caused an increase in the mobility of wild-type ASF/SF2, but had no effect on that of the KS mutant, suggesting that GFP-ASF/ SF2-KS expressed in mammalian cells is not phosphorylated. The localization of these proteins in HeLa cells is shown in Fig. 4 b. Wild-type GFP-ASF/SF2 localized in the nucleus in a speckled pattern characteristic of endogenous splicing factors. In contrast, GFP-ASF/SF2-KS showed significant accumulation in the cytoplasm. ASF/ SF2-KS was also localized in the nucleus in enlarged speckles, which is consistent with a role of SRPKs in dissociating splicing factors from nuclear speckles, as suggested earlier (Gui et al., 1994a). These observations reinforce the idea that SRPK-mediated phosphorylation plays an important role in both nuclear import and intranuclear localization of SR proteins.
Figure 4.
Evidence for a role of SRPK-mediated phosphorylation in ASF/SF2 localization in mammalian cells. (a) Whole cell extracts of GFP-ASF/SF2 wild-type or GFP-ASF/SF2-KS expressed in HeLa cells were either mock (−) or phosphatase (+) treated, then probed with an antibody to GFP. (b) Wild-type GFP-ASF/SF2 (left) or mutant GFP-ASF/SF2 KS (right) fusion proteins were expressed and visualized in live HeLa cells.
Evidence for Phosphorylation-dependent Molecular Targeting in the Nucleus
Although inefficient nuclear localization may be a contributing factor for impaired protein–protein interactions involving SC35, it cannot explain the absolute phosphorylation dependence of all the RS domain–mediated interactions we tested in sky1Δ yeast. Furthermore, the two-hybrid interaction between U2AF65 and U2AF35 in sky1Δ yeast was also substantially reduced (Fig. 2 b) despite the fact that this interaction is not RS domain–mediated (Zhang et al., 1992). When the RS domains of U2AF65 and U2AF35 were deleted, their two-hybrid interaction became highly efficient in both wild-type and sky1Δ yeast, even surpassing the interaction between full-length proteins in wild-type yeast (Table II). These observations suggest that the phosphorylation state of their RS domains may influence the U2AF65-35 interaction in eukaryotic cells, even though this interaction can take place in bacteria (Rudner et al., 1998). In an attempt to correlate inefficient two-hybrid interactions with a defect in heterodimer formation between U2AF65 and U2AF35, we conducted gel filtration and coimmunoprecipitation experiments and detected similar amounts of U2AF65 and 35 in large protein complexes from both wild-type and sky1Δ yeast (data not shown). Because these complexes may contain additional proteins, we cannot determine whether phosphorylation directly affects the efficiency of heterodimer formation. Alternatively, Sky1p may indirectly affect the outcome of the two-hybrid interaction assay by regulating molecular targeting of its substrates to each other and/or to the reporter gene in the nucleus.
Table II.
RS Domain Phosphorylation Influences In Vivo Interactions Mediated by Non-RS Domains
| SKY1/sky1Δ | U2AF35 | U2AF35ΔRS | ||
|---|---|---|---|---|
| U2AF65 | 124/3 | 308/93 | ||
| U2AF65ΔRS | 340/8 | 347/460 |
Two-hybrid analysis of U2AF65-35 interactions in wild-type and sky1Δ yeast. Baits are listed on the left and preys are listed across. Quantitative assays are expressed as fold over background using the empty prey vector in wild-type versus sky1Δ strains.
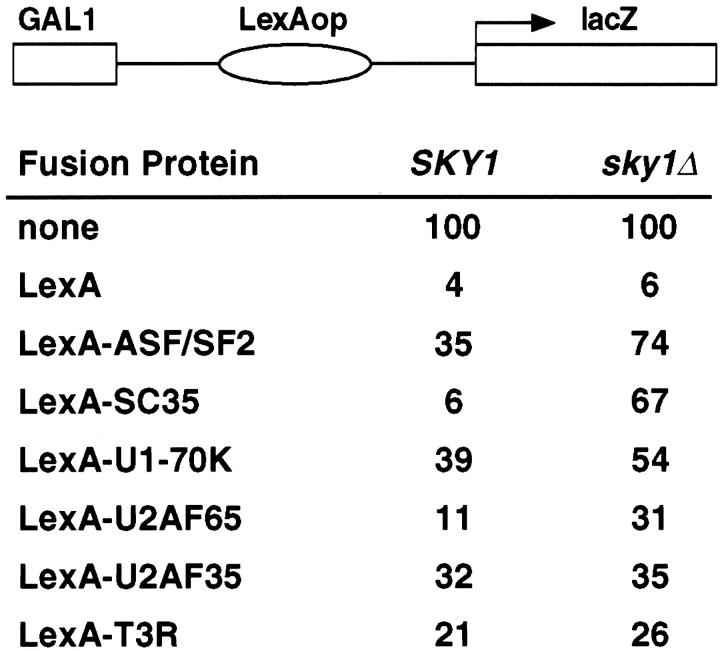
To demonstrate that SRPKs play a direct role in assisting RS domain–containing proteins to locate their specific protein or RNA targets, we adapted the following transcriptional repression assay (Golemis and Brent, 1992), which allowed a quantitative measure of phosphorylation-dependent molecular targeting of RS domain proteins in vivo. As diagrammed in Fig. 5, LexA operators were inserted between a GAL1 upstream activating sequence (UAS) and the transcriptional start site for lacZ such that LexA or LexA fusion proteins bind to the operators and repress lacZ expression. In both wild-type and sky1Δ yeast, lacZ expression was constitutive in the absence of LexA, and repressed in its presence. The ability of a control LexA-T3R fusion protein to repress lacZ expression was not affected by the SKY1 deletion. In contrast, the targeting of LexA-SR fusion proteins was dramatically, although not equally, affected by the SKY1 deletion in this assay (Fig. 5). In particular, the LexA-SC35 fusion protein fully repressed lacZ expression in wild-type yeast, but showed little repression in sky1Δ yeast, probably reflecting the significant impact of Sky1p-mediated phosphorylation on the fusion protein at both nuclear localization and intranuclear targeting steps. A similar SKY1-dependent effect was also seen, although less pronounced, with ASF/ SF2, U1-70K, and U2AF65, but was not evident with U2AF35. These observations provide an explanation for the decreased interaction between U2AF65 and U2AF35 in sky1Δ yeast in the two-hybrid assay. The U2AF65 fusion protein expressed from the bait vector in sky1Δ yeast could not efficiently locate its target and activate transcription of the reporter gene, even though targeting of U2AF35 was unaffected by phosphorylation. Based on these results, we conclude that phosphorylation plays a direct role in the molecular targeting of RS domain–containing proteins within the nucleus.
Figure 5.
Phosphorylation mediates molecular targeting of RS domain proteins. Transcriptional repression assays using LexA fusion proteins performed in wild-type and sky1Δ yeast. Various bait plasmids were cotransformed with the reporter pJK101 (diagrammed above). Degree of reporter expression is shown as the percentage of maximal transcription achieved without LexA.
Discussion
In this report, we have taken advantage of the fact that apparently only one SR protein–specific kinase is conserved in the relatively simple eukaryote S. cerevisiae. Therefore, we were able to address the functional requirements of SR protein–specific kinases in mediating SR protein–protein interactions in vivo. An equivalent kinase deletion experiment cannot be done in mammalian cells due to the presence of at least four identified kinase activities capable of phosphorylating RS domain–containing proteins (for reviews see Fu, 1995; Misteli and Spector, 1997). The yeast model system is especially attractive due to the fact that many aspects of RNA metabolism and fundamental mechanisms of nuclear import/export are well conserved between mammalian cells and yeast. For example, this model organism was used successfully to address HIV rev function in mediating specific RNA transport in yeast, although yeast is not a natural host for this virus (Stutz and Rosbash, 1994; Stutz et al., 1995). Furthermore, the two-hybrid protein–protein interaction and transcriptional repression assays have been used widely to address the function of mammalian proteins in yeast, and often accurately reflect interactions that occur normally in mammalian cells. Our current findings illustrate multiple steps at which the SR superfamily of splicing factors may be regulated by phosphorylation and our data are consistent with published observations on the effects of phosphorylation and dephosphorylation on SR proteins in mammalian cells (see below). Although a phosphorylation-dependent nuclear localization defect was observed using mammalian SC35 expressed in yeast, a similar phenomenon also occurs with an endogenous Sky1p substrate, Npl3p (Yun, C.Y., and X.-D. Fu, manuscript submitted for publication), indicating that conserved regulatory mechanisms may operate in both yeast and mammalian cells. Most advantageously, the use of yeast has made it possible to provide quantitative measures of intracellular trafficking of SR proteins, which have been largely descriptive in previous studies.
Nuclear Import of SR Proteins
Our localization studies showed increased cytoplasmic accumulation of SR proteins in the absence of SRPK-mediated phosphorylation. The nuclear localization signal for SC35 appears to be confined to its RS domain (Cáceres et al., 1997), and, therefore, the effect of SRPK-mediated phosphorylation on SC35 localization may be readily detectable. In contrast, sequences both inside and outside of the RS domain of ASF/SF2 seem to be required for the protein to properly localize in the nucleus (Cáceres et al., 1997), and, as a result, nuclear import of ASF/SF2 may be less efficient but not abolished without SRPK-mediated phosphorylation. This possibility is consistent with the observations that ASF/SF2 mutants, including ASF/SF2-KS and RS domain–deleted ASF/SF2 (Misteli et al., 1998), were localized in both the cytoplasm and the nucleus of transfected HeLa cells. Together, these results suggest that the localization of different SR proteins may be differentially facilitated by phosphorylation, which may reflect distinct mechanisms for their nuclear import.
Increased cytoplasmic localization of hypophosphorylated SR proteins may be due to inefficient nuclear import or accelerated export. Recently, it was reported that overexpression of Clk/Sty or SRPK2 in mammalian cells increases cytoplasmic localization of ASF/SF2 (Cáceres et al., 1998; Kuroyanagi et al., 1998; our unpublished observations), indicating that SR protein–specific kinases may play an active role in facilitating nuclear export of SR proteins. Alternatively, the dissolution of nuclear speckles by these kinases elevates the nucleoplasmic pool of SR proteins, which may indirectly lead to an increase in detectable cytoplasmic protein levels, as previously suggested (Cáceres et al., 1998). In either case, the observation that over-phosphorylation appears to stimulate SR protein export is incompatible with the idea that the lack of phosphorylation also accelerates nuclear export of SC35 in sky1Δ yeast. Therefore, we favor the interpretation that SKY1-mediated phosphorylation functions at the nuclear import step, which provides a rationale for our earlier findings that all SRPK family members from yeast to humans are largely, but not exclusively, localized in the cytoplasm (Takeuchi and Yanagida, 1993; Wang et al., 1998; Siebel et al., 1999). Together, these observations suggest that SRPKs may catalyze phosphorylation of their substrates in the cytoplasm and facilitate their nuclear import.
Molecular Targeting to Nascent Transcripts
In yeast, we observed that unphosphorylated SR proteins appear to distribute uniformly in the nucleoplasm, yet their efficient targeting to appropriate molecular targets (in this case, the LexA binding site) requires phosphorylation mediated by Sky1p. These data are consistent with the recent report on phosphorylation-dependent recruitment of SR proteins to nascent transcripts (Misteli et al., 1998). In addition, the transcriptional repression assay provided a quantitative measure of recruitment, and, therefore, allowed the comparison of different RS domain–containing splicing factors. For example, we observed that targeting of the SC35 fusion protein is more dependent on phosphorylation than that of ASF/SF2 and other RS domain–containing splicing factors, which may result from the phosphorylation dependence of SC35 for both efficient nuclear import and intranuclear movement. The mechanism for phosphorylation-dependent targeting is not entirely clear, but may be explained by the possibility that an unphosphorylated RS domain may interact with other nuclear constituents. For example, it was suggested, based on in vitro experiments, that phosphorylation is necessary for sequence-specific binding of SR proteins to their RNA targets (Tacke et al., 1997; Xiao and Manley, 1997).
In addition to increasing protein–nucleic acid binding specificity, phosphorylation can modulate protein–protein interactions among RS domains in vitro (Xiao and Manley, 1997; Wang et al., 1998). Our finding that phosphorylation increases ASF/SF2-U1-70K interactions, but has little effect on ASF/SF2-U2AF35 interactions, suggests that phosphorylation can differentially modulate protein–protein interactions depending on the protein pairs. Therefore, phosphorylation may regulate molecular targeting of RS domain–containing proteins to their appropriate protein partners after being recruited to actively transcribing regions in the nucleus. This phosphorylation-dependent selectivity may be crucial for an orderly assembly of splicing factors on specific transcripts at specific stages of the splicing reaction.
Dynamic Distribution of Splicing Factors in the Nucleus
In mammalian cells, splicing factors are believed to be “stored” in nuclear speckles (for reviews, see Spector, 1993; Singer and Green, 1997) and recruited to nascent transcripts to carry out the splicing reaction. The exchange of SR proteins between the nucleoplasm and nuclear speckles is highly dynamic, as illustrated by monitoring the movement of GFP-ASF/SF2 in living cells (Misteli et al., 1997, 1998). Earlier studies have shown that increased phosphorylation releases SR proteins to the nucleoplasm (Gui et al., 1994a; Colwill et al., 1996a; Misteli et al., 1997; Duncan et al., 1998; Wang et al., 1998), making them generally available to be recruited to the sites of transcription and splicing, whereas dephosphorylation appears to be essential for the resolution of spliceosomes (Mermoud et al., 1994), and as a result, contributes to the accumulation of SR proteins in nuclear speckles (Misteli and Spector, 1997). In the current study, SR proteins in sky1Δ yeast remained soluble, suggesting that unphosphorylated SR proteins may not form nonspecific aggregates resembling inclusion bodies in yeast, yet they do not interact with each other efficiently. Interestingly, we observed that removal of the RS domains from both U2AF65 and U2AF35 allowed them to interact very efficiently in both wild-type and sky1Δ yeast, even better than the interaction between the full-length proteins in wild-type yeast. This result implies that even in wild-type cells, some proteins may not be sufficiently phosphorylated for efficient interaction. Because we detect U2AF65/35 in large complexes in both wild-type and sky1Δ yeast, it is possible that phosphorylated and unphosphorylated proteins may interact with different sets of proteins.
In fact, interactions involving hypophosphorylated RS domain–containing splicing factors may resemble those in nuclear speckles where a series of rearranged protein–protein and protein–RNA interactions have been triggered by phosphatases during splicing. To some extent, these dephosphorylation-induced rearrangements may cause the coalescence of splicing complexes on nascent transcripts to give rise to the appearance of speckles in the nucleus. Therefore, one might imagine that nuclear speckles are postsplicing structures for recycling of splicing factors, rather than storage sites where splicing factors are randomly accumulated. This scenario may best explain the observations that nuclear speckles contain essentially all splicing factors as well as poly (A)+ mRNA (Xing et al., 1993, 1995), but are not sites for the majority of nascent transcripts detected by BrUTP pulse labeling (Jackson et al., 1993).
A Model for Phosphorylation-dependent Movement of Splicing Factors in Cells
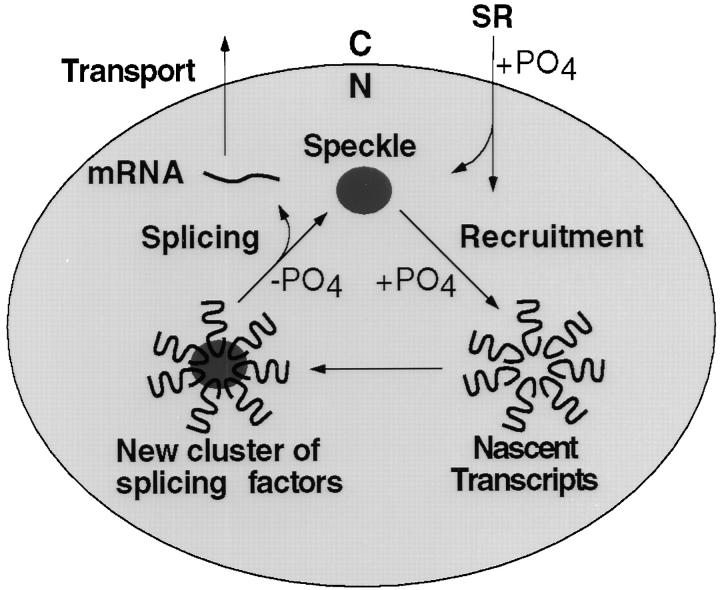
A synthesis of our results with published reports is presented in a model (Fig. 6), depicting the dependence of SR protein trafficking on phosphorylation. In this model, nuclear import of newly translated or shuttling RS domain–containing proteins is facilitated by phosphorylation. Newly imported SR proteins are then partitioned between speckles and the nucleoplasm based in part on the extent of phosphorylation. A large body of evidence suggests that mRNA splicing takes place cotranscriptionally in the nucleoplasm whereas nuclear speckles have been described as “storage sites” for splicing factors. Release of SR proteins from speckles is facilitated by phosphorylation (Gui et al., 1994a; Colwill et al., 1996a), which is a prerequisite for their subsequent recruitment to nascent transcripts (Misteli et al., 1998). We show in the current study that specific targeting of nucleoplasmic SR proteins to their appropriate RNA and protein partners are also affected by phosphorylation. Once assembled on nascent transcripts, the resolution of splicing complexes during the splicing reaction appears to require dephosphorylation of SR proteins (Mermoud et al., 1994), which may accompany a series of rearrangements involving both RNAs and proteins in the spliceosome. As a result, hypophosphorylated SR proteins may be engaged in interactions with a distinct set of proteins, causing them to remain in speckles, as mature transcripts proceed to nuclear export.
Figure 6.
A model for phosphorylation-dependent SR protein targeting/recycling in the dynamics of nuclear speckles. Different structures in the illustration reflect progressive time points in speckle assembly and disassembly rather than specific locations of these structures in the nucleus. In addition, the illustrated structure is intended to represent a speckle or a portion of it because the speckled nuclear domain is highly dynamic in living cells as documented by Spector and colleagues (Misteli et al., 1997).
In summary, the novel system developed in this study enabled us to dissect this complex process in a relatively simple genetic background. The data presented in this report illustrate that SR protein kinases can affect translocation of RS domain–containing proteins from the cytoplasm to the nucleus as well as molecular targeting within the nucleus, in addition to modulation of RS domain affinities observed in vitro. Defects in combinations of these processes provide a probable explanation for the critical dependence of RS domain interactions on SRPK-mediated phosphorylation in cells. These phosphorylation-dependent steps provide multiple avenues for pre-mRNA splicing regulation in response to internal or external signaling.
Acknowledgments
We thank Mira Guzijan, Harold Fisk, and Jane Wu for assistance and expert advice; and Lana Feng, Jiwu Wang, Chi Yun, Brent Gravely, and Tom Maniatis for critical comments. T3R and NCoR control plasmids were kindly provided by M.G. Rosenfeld.
Abbreviation used in this paper
- snRNPs
small nuclear ribonucleoprotein particles
Footnotes
J.M. Yeakley is the recipient of a National Institutes of Health (NIH) postdoctoral fellowship. H. Tronchère was supported by the Human Frontiers Science Program. J.A. Dyck is funded by the Damon Runyon-Walter Winchell Cancer Research Foundation. X.-D. Fu is a Leukemia Scholar. This work was supported by an NIH grant to X.-D. Fu.
References
- Cáceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor ASF/SF2 structural domains. EMBO (Eur Mol Biol Organ) J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intracellular distribution. EMBO (Eur Mol Biol Organ) J. 1996a;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Feng L, Yeakley JM, Gish GD, Cáceres JF, Pawson T, Fu X-D. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996b;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Bell JC. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;17:5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241:300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994a;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Tronchère H, Chandler SD, Fu X-D. Purification and characterization of a kinase specific for the serine and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia MA, Blanco, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewert LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AE. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO (Eur Mol Biol Organ) J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucl Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing. In RNA Structure and Function. R. Simons and M. Grunberg-Manago, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 279–307.
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/ U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol. 1998;18:1765–1773. doi: 10.1128/mcb.18.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel, C., L. Feng, C. Guthrie, and X.-D. Fu. 1999. Conservation of yeast protein kinase specific for the SR family of splicing factors. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed]
- Singer RH, Green MR. Compartmentalization of eukaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. A functional interaction between Rev and yeast pre-mRNA is related to splicing complex formation. EMBO (Eur Mol Biol Organ) J. 1994;13:4096–4104. doi: 10.1002/j.1460-2075.1994.tb06727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Yanagida M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle. Mol Biol Cell. 1993;4:247–260. doi: 10.1091/mbc.4.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunel C, Lührmann R. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature. 1993;363:283–286. doi: 10.1038/363283a0. [DOI] [PubMed] [Google Scholar]
- Tronchère H, Wang J, Fu X-D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR–based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wang H-Y, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu X-D. SRPK2: a differentially expressed SR protein–specific kinase involved in mediating the interaction and localization of pre-mRNA splicing in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dong Z, Bell LR. Sex-lethal interactions with protein and RNA. Roles of glycine-rich and RNA binding domains. J Biol Chem. 1997;272:22227–22235. doi: 10.1074/jbc.272.35.22227. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao S-H, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xiao S-H, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO (Eur Mol Biol Organ) J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC35 domain. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]