Abstract
Laminins are heterotrimeric molecules composed of an α, a β, and a γ chain; they have broad functional roles in development and in stabilizing epithelial structures. Here, we identified a novel laminin, composed of known α and β chains but containing a novel γ chain, γ3. We have cloned gene encoding this chain, LAMC3, which maps to chromosome 9 at q31-34. Protein and cDNA analyses demonstrate that γ3 contains all the expected domains of a γ chain, including two consensus glycosylation sites and a putative nidogen-binding site. This suggests that γ3-containing laminins are likely to exist in a stable matrix.
Studies of the tissue distribution of γ3 chain show that it is broadly expressed in: skin, heart, lung, and the reproductive tracts. In skin, γ3 protein is seen within the basement membrane of the dermal-epidermal junction at points of nerve penetration. The γ3 chain is also a prominent element of the apical surface of ciliated epithelial cells of: lung, oviduct, epididymis, ductus deferens, and seminiferous tubules. The distribution of γ3-containing laminins on the apical surfaces of a variety of epithelial tissues is novel and suggests that they are not found within ultrastructurally defined basement membranes. It seems likely that these apical laminins are important in the morphogenesis and structural stability of the ciliated processes of these cells.
Keywords: laminin, testis, oviduct, lung, chromosome 9q31-34
Laminins are large glycoproteins found in all basement membranes (Ekblom, 1996). In overall appearance, laminins are cross-shaped with a single long arm arising from the coiled-coil interaction of three genetically distinct polypeptide chains and three NH2-terminal short arms, each originating from the individual polypeptide chains (Maurer, 1996). The three subunit chains are termed α, β, and γ according to the current nomenclature (Burgeson et al., 1994). The complete primary structure for each of the nine human laminin subunit chains has been elucidated: α1 (Haaparanta et al., 1991), α2, (Vuolteenaho et al., 1994), α3 (Ryan et al., 1994), α4 (Iivanainen et al., 1995a), β1 (Pikkarainen et al., 1987), β2 (Wewer et al., 1994; Iivanainen et al., 1995b), β3 (Gerecke et al., 1994), γ1 (Pikkarainen et al., 1988), and γ2 (Kallunki et al., 1992). The complete cDNA sequence for a fifth laminin α chain has been determined in mouse (Miner et al., 1995); partial cDNA sequences of human α5 (Durkin et al., 1997), and a novel chicken β chain (Ybot-Gonzalez et al., 1995) have been reported. All three chains have globular domains separated by multiple epidermal growth factor-like domains within the NH2-terminal short arms. Their long arm portions are composed of heptad repeats that are typical for α-helical coiled-coil proteins. In addition, the COOH terminus of each α chain is composed of five globular (G)1 domains (Engel, 1992).
The many functions ascribed to laminins are thought to derive from their structural and signal transduction roles, by which they contribute to the formation and stability of basement membranes, to the stability of cellular attachments to basement membranes, and to cytoskeletal rearrangements mediated by their occupancy of cell surface receptors (Ryan et al., 1994). These activities at least partially result from: (a) the binding of the COOH-terminal laminin G domains to integrins in most cells (Deutzmann et al., 1990; Drago et al., 1991; Goodman, 1992; Matter and Laurie, 1994; Rousselle et al., 1995; Chen et al., 1997), and/or to dystroglycan in muscle cells (Henry and Campbell, 1996; Pall et al., 1996; Wewer and Engvall, 1996; Cohen et al., 1997); (b) from the self-assembly of the laminins into a pericellular extracellular matrix through interactions of domains VI, which are present at the ends of the short arms of the subunit chains (Yurchenco et al., 1992; Yurchenco and Cheng, 1993, 1994); and (c) from assembly of the pericellular laminin network with a conceptually separate network of type IV collagen molecules specifically mediated by the molecule nidogen, one end of which binds the laminin γ1 chain, and the other end of which binds type IV collagen and other basement membrane matrix components (Fox et al., 1991; Battaglia et al., 1992; Aumailley et al., 1993; Reinhardt et al., 1993).
The role of laminin 5 (α3β3γ2) in stabilization of epithelial–stromal interactions is the exception to this generalized scheme. While the 5 laminin G domains of α3 bind integrins α6β4 and α3β1 on the epithelial basolateral surface (Niessen et al., 1994), the absence of domains VI on the truncated short arms of α3 and γ2 (Kallunki et al., 1992; Ryan et al., 1994), and the absence of a nidogen binding site on γ2 (Mayer et al., 1995) prevent their participation in the above-described model. Instead, the NH2 terminus of the epithelial cell–associated laminin 5 binds type VII collagen present within the subjacent stromal matrix (Rousselle et al., 1997). Thus, laminin 5 appears to play a unique role in epithelial frictional resistance, rather than a direct role in overall basement membrane structure.
The primary functions of the laminin γ1 chain in the above generalized scheme is the contribution of a domain VI which shares high amino acid sequence identity to domains VI of the laminin α and β chains and a unique sequence required for the binding of nidogen. This sequence, NIDPNAV, is present in the fourth EGF-like repeat of domain III (Mayer et al., 1979; Poschl et al., 1994). In the γ2 chain, the analogous sequence NVDPSAS is present, but does not support high affinity nidogen binding (Mayer et al., 1995).
In this report, we describe a novel human laminin γ chain. Its predicted structure indicates the presence of all the domains homologous to γ1, including a domain VI homologue and a nidogen-binding site containing a single conservative amino acid substitution. We call this chain the laminin γ3 chain. The predicted structure of this chain suggests that it should be capable of associations with other laminin chains for basement membrane assembly; however, immunolocalization studies in several tissues indicate the presence of laminin γ3 chains in regions lacking ultrastructurally identifiable basement membranes.
Materials and Methods
Isolation of a Novel Laminin 12 (α2β1γ3)
The purification of the novel laminin 12 (α2β1γ3) was carried out as follows. Human placental chorionic villi were frozen in liquid nitrogen, ground in a Waring blender, and then washed in 1 M NaCl. Unless otherwise noted, all subsequent steps were performed at 4°C. The final tissue pellet (200 g, wet weight) was suspended by stirring for 48 h in 1 liter of extraction buffer (0.5 M NaCl, 10 mM EDTA, and 625 mg/liter N-ethylmaleimide, 150 mg/liter phenylmethylsulfonyl fluoride, and 50 mM Tris-HCl, pH 7.8). The soluble fraction was collected after centrifugation (30,000 g, 60 min) and precipitated with 300 g/liter ammonium sulfate. The precipitated proteins were collected by centrifugation (30,000 g, 60 min) and dissolved in chromatography buffer (2 M urea, 25 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.8). The sample was then dialyzed against the same buffer. After dialysis, 0.5 vol of buffer-equilibrated DEAE-cellulose (DE-52; Whatman) was added and the mixture was shaken overnight. Material not bound to DEAE-cellulose was collected by filtration on a Buchner funnel (Whatman; filter 4) and precipitated by addition of 300 g/liter ammonium sulfate. The proteins were collected by centrifugation (30,000 g, 60 min), redissolved in Concanavalin A buffer (0.5 M NaCl, 5 mM CaCl2, 5 mM MgCl2, and 50 mM Tris-HCl, pH 7.8), and dialyzed against the same buffer overnight. The fraction was applied to a 2.5 × 5 cm Concanavalin A–Sepharose column (Pharmacia). Unbound material was removed by extensive washing while bound proteins were eluted by successive washing with 10 mM α-d-mannopyrannoside, 1 M α-d-glucopyrannoside, and finally 1 M α-d-mannopyrannoside (Sigma Chemical Co.). Laminins are typically recovered in the latter two fractions; each fraction was independently concentrated to 10 ml with an Amicon concentrator (30-kD membrane) and applied to a 2.5 × 100 cm Sephacryl S-500 column in 0.5 M NaCl, 50 mM Tris-HCl, pH 7.8. The fractions of interest were pooled, dialyzed against Mono-Q buffer (0.1 M NaCl and 25 mM Tris-HCl, pH 7.8), and applied to a 1 × 5 cm Mono-Q column (Pharmacia). Elution was achieved with a 60-ml 0.1–0.5-M NaCl gradient. The fraction eluted at 250 mM NaCl was taken for further study.
Protein Sequencing
Protein sequencing was performed with minor modifications of published methods (Aebersold et al., 1987). In brief, laminin 12 was resolved on a polyacrylamide gel in the presence of 2-mercaptoethanol. The bands at 205, 185, and 170 kD were excised separately, digested with trypsin, and then separated by HPLC and sequenced on an Applied Biosystem sequenator. Analysis of a trypsin digest of laminin α2 isolated from laminin 12 was performed with matrix-assisted laser desorption time-of-flight mass spectrometry performed on a Finnigan Lasermat 2000 (Chait and Kent, 1992).
cDNA Cloning
By comparison of the laminin γ1 amino acid sequence (SWISS-PROT accession number, P11047) with the dbEST database (Boguski et al., 1993; NCBI) using the program BLAST (Altschul et al., 1990) one clone (these data are available from GenBank/EMBL/DDBJ under accession number AA297192) was chosen as a possible candidate for a new laminin γ chain. To extend the cDNA, specific primers for 5′ or 3′ extension were deduced from a previously published expressed sequence tag (clone, AA297192). Nested PCR on placental Marathon-Ready cDNA (Clontech) were performed following the manufacturer's instructions using the supplied nonspecific primers with the following gene-specific primers: for the 5′ extension; in the first round, (5′-dCGCATGTGCCGTTCTCGTGGCACTGG); in the second round, (5′-dGCGGCAGGTGCACTGTCCAGTCTTGG); for the 3′ extension, in the first round, (5′-dTGCACGGGACTGCAGCCGCTGCTACCC); in the second round (5′-dGCTGCTACCCTGGCTTCTTCGACCTCC). For PCR, the Long Expand PCR Kit (Boehringer Mannheim) was used with the following conditions: denaturation, 94°C for 3 min; 10 cycles of 94°C for 30 s, 63°C (−0.5°C per cycle) for 30 s, 68°C for 4 min; 25 cycles of 94°C for 30 s, 58°C for 30 s, 68°C for 4 min (+10 s per cycle); a final extension period at 68°C of 8 min.
The PCR samples from the first round were purified (PCR Purification Kit; Qiagen) and 2% of the sample volume was used in the second round of PCR using the same PCR protocol. These PCR products were purified from an agarose gel (Gel Purification Kit; Qiagen) and either subcloned (into PCR II or PCR 2.1 vectors; Invitrogen) or directly used for sequencing. To reconfirm the nucleotide sequence and control for PCR-induced nucleotide substitutions, gene-specific primers were used to reamplify the entire γ3 cDNA. A first strand cDNA synthesis kit (Clontech) was used to synthesize cDNA from total placental RNA using oligo dT, random, or specific human laminin γ3 antisense primers following the manufacturer's protocol; PCR was used to generate overlapping clones complementary to the entire human laminin γ3 chain. Sequencing of all the obtained PCR products revealed the nucleotide sequence of laminin γ3 from what we eventually inferred as nt 297 to nt 5020. However, all further 5′ extensions failed to extend the sequence further toward the 5′ end.
Genomic DNA
The sequence of the 5′ end of the cDNA was determined from the human genomic P1 clone DMPC-HFF#1-1461F2, which was obtained from a PCR-based library screen performed at Genome Systems, Inc. The oligonucleotide primers that were provided to Genome Systems specifically amplify exon 2 of the human γ3 gene (sense primer, 5′-dCCCCGCAGGGGAAGGCGGGTCCTG; antisense primer, 5′-dGGCTTATGAGATCACGTATGTGAG). To obtain the sequence of the missing 5′ end, the genomic clone was sequenced (in 4% DMSO) with gene-specific antisense primers. The sequence of the laminin γ3 5′ untranslated region was confirmed by RT-PCR from placental RNA in 4% DMSO, (sense primer, 5′-dCGCGCGGCGTCGGTGCCCTTGACC; antisense primer 5′-dGCTTGTAGATGGCAAAGCTCTCAGG).
Nucleotide Sequencing
Nucleotide sequences were determined with a Thermo Sequenase cycle sequencing kit and 33P-ddNTP (Amersham Pharmacia) using either the M13 forward or reverse primers or gene-specific primers synthesized in our laboratory. A 1:1 ratio of inosine to guanosine was included in the sequencing mix. Sequence data were assembled and manipulated using Genetyx-Max 8.0 and Genestream-1 at http://genome.eerie.fr/home.html (Software Development Co., Ltd.). The signal peptide cleavage site was predicted using http://genome.cbs.dtu.dk/services/SignalP/ (Nielsen et al., 1997).
Northern Blot Analysis
A 956-bp PCR product (nt 1316–2271) was generated (Long Expand PCR Kit; Boehringer Mannheim) from placental cDNA, purified (PCR purification kit; Qiagen), and labeled with [33P]dCTP (NEN) using the rediprime DNA labeling system (Amersham). Without further purification, the probe was denatured in the same buffer containing 1/10 (vol/vol) human Cot-1 DNA (Boehringer Mannheim), and 1/10 (vol/vol) sheared salmon testes DNA (GIBCO BRL) at 94°C for 5 min then chilled before use. Northern blots (Clontech) were prehybridized in 50% formamide, 5× SSPE, 1× Denhardt's, 1% SDS, 10% Dextran-sulfate, 0.1 mg/ml salmon sperm DNA (GIBCO BRL) at 42°C for 2 h the probe was added and hybridized for 20 h. The blot was washed three times in 2× SSC, 1% SDS at 42°C and two times in 0.1× SSC, 1% SDS at 42°C. Blots were placed on a BioMax MR film (Kodak) with a BioMax TranScreen-LE intensifying screen (Kodak) for 20 h at −70°C.
Recombinant Expression of Domain I of the γ3 Chain
A cDNA encoding the COOH terminus of human γ3 was cloned into the HisTrx and pPEP-T vectors (kindly provided by Richard Kammerer, Biozentrum, Basel, Switzerland; based on the pET system; Novagen). The HisTrx vector has a histidine-tagged bacterial thioredoxin cDNA as a carrier in front of the cloning site; pPEP-T has a piece of the coiled-coil domain of mouse β1 in front of the cloning site. The γ3 cDNA fragment used was amplified by PCR from human placenta cDNA (see cDNA cloning) using primers that include the EcoRI adapters: forward, 5′-GCGGATC CGAGGAAGCTGAGCGGGTGGGTGCTG-3′; reverse, 5′-GCGAA TTCTTACTGCCAGCTGGCACAGTTCTCGGG-3′. The resultant plasmids were transformed into BL21(DE3) pLysS bacteria (Novagen) and fusion proteins were isolated according to the pET System manual (Novagen). A recombinant fragment containing only histidine-tagged thioredoxin was similarly expressed and purified.
Antibody Production
The 170-kD band (i.e., γ3 chain) was excised from the reducing SDS-PAGE gel described above and injected into a rabbit for antibody production following standard procedures (Harlow and Lane, 1988). The resulting serum (R16) was evaluated by Western analysis and shown to react with the 170-kD γ3 chain, and showed minor cross-reactivity with other laminin chains at high antibody concentrations. All antibody-related studies presented in this communication were conducted at concentrations well below those where cross-reactivity was observed. The histidine-tagged, thioredoxin-γ3 fusion protein was used for the production of a second rabbit antiserum (R21) which reacted with a single band in Western blots of placental extracts.
Affinity Purification
The R16 antiserum was affinity-purified by binding to gel-purified γ3 that had been transferred to nitrocellulose and then eluted with 1 M acetic acid followed by immediate neutralization. The R21 serum was purified by binding to the histidine-tagged, mouse β1-human γ3 fusion protein coupled to activated CNBr-Sepharose; the bound antibodies were eluted with 2 M urea in PBS, or with 1 M acetic acid which was immediately neutralized. The immunofluorescent patterns produced by these two affinity-purified antibody pools were indistinguishable, and were similar to whole R21 serum with reduced background staining. Only affinity-purified preparations of R21 serum were used for these studies. Antibodies made against histidine-tagged thioredoxin were similarly isolated by affinity chromatography from R21 serum; immunofluorescent patterns with these controls were blank.
Immunofluorescent Analyses
Most tissues were obtained from various colleagues using specimens for other purposes: these include tissues from male and female rats; from normal human tissues discarded after surgery; and from rhesus monkey, Macaca mulatta. Bovine tissues were purchased from a local slaughterhouse. Dissected and blocked tissues were placed directly in embedding compound (O.C.T.; Sakura Finetek) and frozen by immersion in liquid nitrogen-cooled isopentane. 10-μm sections were made on a Leica CM 3000 or 3050 and collected on Superfrost slides (Fisher Scientific). Sections were air dried and stored at −20°C until use. Just before use, sections were immersed in acetone at −20°C and then rinsed three times in PBS at room temperature. Sections were incubated with primary antibodies diluted in PBS containing: 2% normal goat serum, 0.25% sodium azide, and 0.1% Triton X-100. Sections were incubated overnight at 4°C; they were washed in three changes of PBS (5 min per wash) and then incubated for 45–60 min with secondary antibody coupled to either Cy3, FITC, or Texas red. After incubation, sections were washed and coverslipped in Prolong (Molecular Probes). The sections were imaged on a Leica confocal laser scanning microscope (Leica TCS-NT). The gain was adjusted in each channel of the confocal to assure that there was no bleeding across the channels; this adjustment is performed at the outset of each confocal session. Images were transferred to Adobe Photoshop and cropped for reproduction. The brightness and contrast were adjusted to make printed images similar to that obtained on the microscope monitor.
Other primary laminin reactive primaries used were: polyclonal anti-EHS-laminin-1 (Sigma Chemical Co.); monoclonal anti-laminin α2 chain (mAb 1922, Chemicon); polyclonal anti-laminin α4 (Miner et al., 1997); polyclonal anti-laminin α5 chain (Miner et al., 1995); two monoclonal anti-laminin β1 chain (545, Marinkovich et al., 1992; clone C21, Green et al., 1992); monoclonal anti-laminin γ2 chain (Verrando et al., 1987). Monoclonal anti-PGP 9.5 (Ultraclone, Ltd.) was used to identify nerves in skin. Secondary antibodies used were: goat anti-rabbit FITC (ICN Pharmaceuticals); goat anti–rabbit-Cy3 (Jackson ImmunoResearch Laboratories).
In Situ Hybridizations
Paraffin sections were processed for in situ hybridizations as previously described in detail (Libby et al., 1997). In brief, cRNA probes for the laminin γ3 chain were generated from human γ3 clones; cRNAs were labeled during transcription by the incorporation of digoxigenin-UTP (Boehringer Mannheim); ∼1 μg/ml of cRNA was used for hybridization; hybridizations were performed at high stringency (50% formamide and 5× SSC, 60°C; see Libby et al., 1997 for complete details). After overnight hybridization, sections were washed (50% formamide, 1× SSC, for 30 min at 60°C) and the unhybridized probe was destroyed by RNase A. The hybrids were detected with an anti-digoxigenin antibody coupled to alkaline phosphatase (Boehringer Mannheim). Sections were incubated overnight with anti-digoxigenin diluted 1:1,000 in blocking solution (Boehringer Mannheim). After washing to remove unbound antibody, endogenous alkaline phosphatase activity was blocked by washing in levamisole for 10 min; the alkaline phosphatase reaction was carried out overnight at room temperature.
Other Methods
SDS-PAGE (Laemmli, 1970) and electrophoretic transfer of proteins to nitrocellulose with immunoblot analysis were performed essentially as previously described (Lunstrum et al., 1986). For the FISH analysis, a 1217 bp cDNA fragment was generated by RT-PCR from placental RNA, using the sense primer 5′-dAGTGCCACTATAACGGCACATGCG and antisense primer 5′-dCTCGTGTCTGCAAGGAGTCTGTCA. The gel band was purified and subcloned (PCR II vector; Invitrogen). After the sequence of the fragment was verified, the resultant plasmid was used for the fluorescent in situ localization of the LAMC3 gene (SeeDNA Biotech, Inc.).
Results
Characterization of Laminin 12 (α2β1γ3)
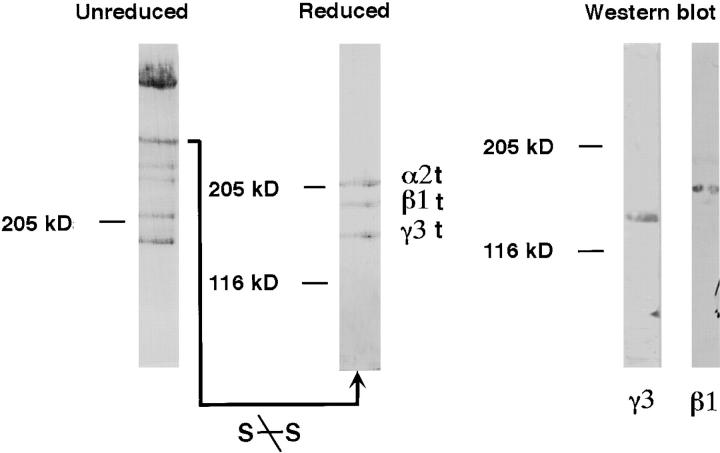
Laminin 12 was extracted from human chorionic villi using EDTA and partially purified by a combination of DEAE-cellulose, Concanavalin A, Sephacryl S-500, and Mono-Q chromatography (see Materials and Methods). The final fraction of interest resulting from the above protocol contains multiple laminins. Laminin 12 was resolved from this mixture by SDS-PAGE (3–5% polyacrylamide) under nonreducing conditions. Six bands were resolved (Fig. 1, Unreduced). Only the bands at ∼560 kD and at the top of the gel were reactive with a polyclonal anti-laminin antiserum (Sigma Chemical Co.; not shown). Therefore, the resolved band at 560 kD was excised, reduced in 10% 2-mercaptoethanol SDS-PAGE sample buffer, and resolved by 5% SDS-PAGE. Three bands were observed with masses of ∼205, 185, and 170 kD (Fig. 1, Reduced). The band at 185 kD reacted with a monoclonal antibody (clone 545; Marinkovich et al., 1992) specific to the laminin β1 chain (Fig. 1, Western blot). Each of the three bands was digested with trypsin, the peptides were resolved by HPLC, and selected resolved peptides were sequenced. The sequences obtained are shown in Table I. The 205-kD chain contained three peptides sequence identical to human laminin α2 (published residues 536, 70, 1367; Vuolteenaho et al., 1994). On that basis, the band was identified as human laminin α2, despite our observation that the 205-kD band did not react with anti-α2 mAb (mAb 1922; Chemicon). The band at 185 kD produced two peptides identical to human β1, and was thereby confirmed as human β1.
Figure 1.
Identification of laminin 12 isolated from human placenta. A partially purified preparation of laminins isolated from an EDTA extract of placenta was subjected to 3–5% SDS-PAGE (Unreduced). The gel band in the indicated position (bar) was excised from the gel and resolved by 5% SDS-PAGE after disulfide-bond reduction. Three bands were seen from the single unreduced band (Reduced). The 185-kD band was recognized by an anti-β1 antibody, while the 170-kD band was specifically recognized by a polyclonal antibody (R16; which used the same band as an antigen; Western blot). Each of the three bands (Reduced gel) was subsequently identified as laminin α2, β1, and γ3 (see text for details). As they have faster electrophoretic mobilities than expected for these chains they are marked α2t, β1t, and γ3t to indicate that they are truncated.
Table I.
Comparison of Peptide Sequences Obtained from Placental Laminin 12 205- and 185-kD Bands and Published Sequences of α2 and β1 Residues
| Human α2* res. no. 536 | KIQMSGWYL | |
| Peptide from 205-kD band | IQMSG-YL | |
| Human α2* res. no. 70 | KLVEHVPGQPVR | |
| Peptide from 205-kD band | LVEHVPGQPVR | |
| Human α2* res. no. 1367 | RGTTMTPPADLIEK | |
| Peptide from 205-kD band | GTTMTPPADLIEK | |
| Human β1‡ no. 171 | RYSDIEPSTEGEVIFR | |
| Peptide from 185-kD band | YSDIEPSTEGEVIFR | |
| Human β1‡ res. no. 558 | RIPSWTGAGFVR | |
| Peptide from 185-kD band | IPSWTGAGFVR |
In contrast to the easy identification of the other two bands, the band at 170 kD contained three sequences not contained within any known laminin chain. The NH2-terminal sequence of the 170-kD chain was determined, and it also was novel; i.e., nonidentical to known laminin sequences. As these four sequences from the 170-kD band were derived from an unknown laminin and we had identified the laminin α and β chains, we assumed these sequences were derived from a novel laminin γ chain that we call γ3.
The apparent molecular masses for the 205- and 185-kD bands are not consistent with the literature values published for the α2 and β1 chains, respectively. Thus, these bands are indicated in Fig. 1 as α2t, β1t, and γ3t to indicate that they have been processed (truncated). Laminin 2 and laminin 4 were also present in these preparations; when characterized by similar procedures (not described here in detail) they showed molecular masses consistent with literature predictions, suggesting that our preparations were not extensively and nonspecifically degraded. Together these observations suggest that the truncations observed for the γ3-containing molecules may be physiologically relevant.
Characterization of the γ3 cDNA
The cDNA sequences of human γ1 and γ2 were used to probe the National Center for Biomedical Information (NCBI) expressed-sequence-tag database (dbEST), and a clone was identified that was homologous, but not identical, to γ1 and γ2. The sequence of this clone was used to design PCR primers for extensions at 3′ and 5′ ends (see Materials and Methods) using human placental cDNA, and additional sequence information was obtained by a combination of genomic DNA and placental cDNA sequencing. The resulting sequence is shown in Fig. 2. The deduced amino acid sequence contains regions with 100% identity to all three of the peptide sequences obtained from the 170-kD band (underlined in Fig. 2). The nucleotide sequence reported in this paper has been submitted to GenBank/EMBL Data Bank with the accession number AF041835.
Figure 2.
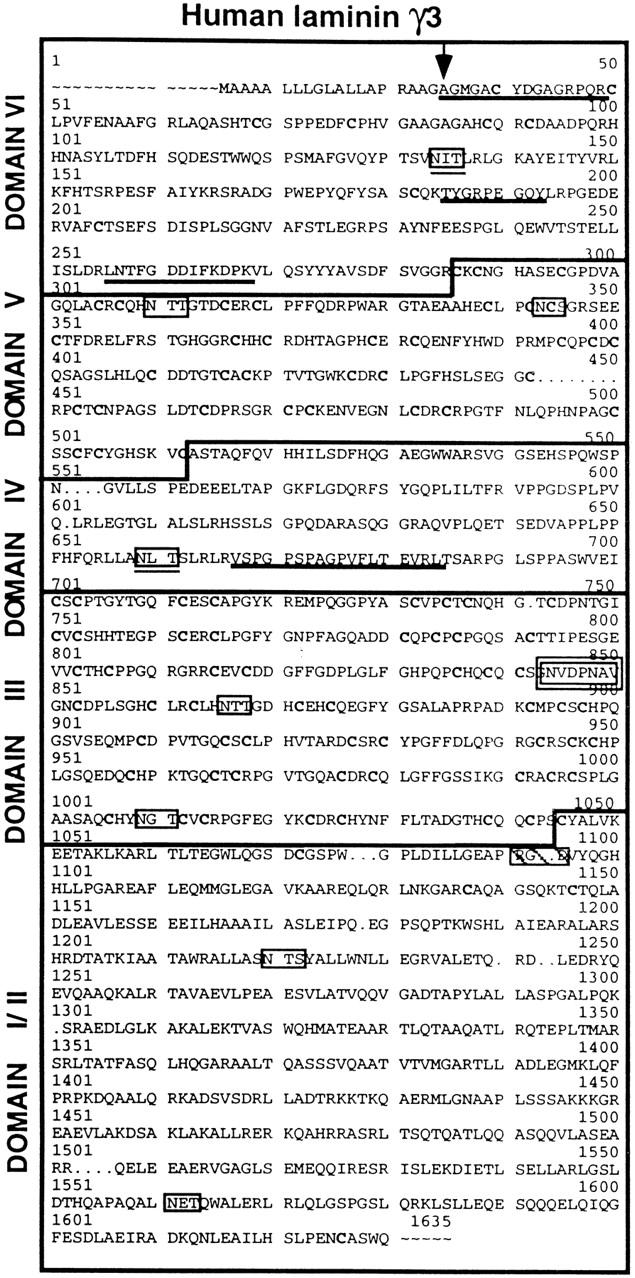
The complete amino acid sequence of human γ3 as predicted from the corresponding cDNA sequence. The amino acid positions are numbered in accordance to the homologous locations within laminin γ1 (Pikkarainen et al., 1988). A 19–amino acid signal sequence precedes the γ3 NH2 terminus (arrow marks the signal peptide cleavage site). Peptide sequences obtained from Edman analyses of the fragmented γ3t gel band (Fig. 1, Reduced) are underlined. Potential glycosylation sites are boxed, and those conserved between γ1 and γ3 are also underlined. The nidogen binding consensus sequence is boxed in bold type. The sequence RGD is boxed and hatched.
The DNA sequence contains an open reading frame predicting 1620 amino acids, including a 19–amino acid-long putative signal peptide that closely meets the criteria described by Nielsen et al. (1997). The predicted cleavage site was confirmed by protein sequencing of the γ3 NH2 terminus; this sequence exactly matched the predicted amino acid sequence following the signal peptide. The overall sequence of γ3 is most similar to that of γ1, sharing 52% amino acid similarity with human γ1 (Pikkarainen et al., 1988). In addition, the amino acid sequence predicted by the γ3 cDNA contains a domain distribution most like that of the γ1 chain. All six domains are represented.
Overall, the γ3 chain has 43.6% amino acid identity with the γ1 chain and 34% identity with the γ2 chain. The highest conservation is seen between domains γ1VI and γ3VI (Fig. 3). Domains γ3V and III also show considerable similarity to domains γ1V and III and γ2V and III.
Figure 3.
Comparison of the predicted domain structures and the percent amino acid sequence identity of γ3 with γ1 and with γ2. The γ3 cDNA predicts a full-length laminin chain where all the domains found in γ1 are represented. On the left side a diagram of the domain structure of both γ1 and γ3 is given; on the right, the domain structure of γ2. Above each diagram is noted by domain the percent amino acid identity between γ1 or γ2 and γ3. The degree of amino acid identity of γ3 to γ1 is higher than that of γ3 to γ2, suggesting that γ3 has a closer evolutionary relationship with γ1 than γ2.
The predicted γ3 sequence contains nine potential glycosylation sites (Fig. 2, boxed), only two of which (Fig. 2, boxed and underlined) are conserved in both human and mouse γ1. As these conserved sites are contained within the globular domains IV and VI, it is likely that these sites are used physiologically. There is a single RGD sequence (boxed, hatched) within domain II, but this site is not conserved in either human or mouse γ1 and γ2 proteins. The sequence NVDPNAV (Fig. 2, double boxed) occurs within the fourth EGF-like repeat of domain III and is a homologue of the nidogen binding site (NIDPNAV) within the same domain of γ1. These sequences differ by only a single conservative amino acid substitution.
LAMC3 Maps to Chromosome 9q31-q34
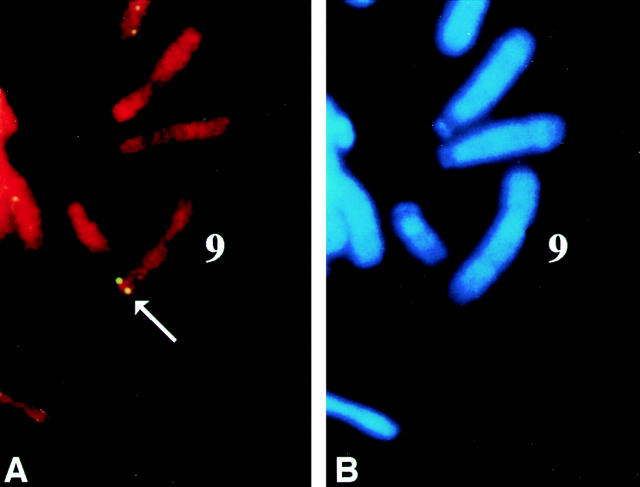
The γ3 chromosomal location was determined by searching the NCBI Human Genomic Sequencing Index data base with the γ3 cDNA sequence. The sequence is identical to a database, Sequence Tagged Sites (clone WI-14302), that has been localized to chromosome 9q33-q34. A 1.2-kb γ3 cDNA probe within domains I and II of the predicted protein, the regions of least homology among the γ chains, was used to localize LAMC3 by fluorescent in situ hybridization (FISH) analysis (SeeDNA Biotech, Inc.). The results confirm the localization to chromosome 9q31-q34 (Fig. 4).
Figure 4.
Localization of LAMC3 to chromosome 9, band q31-q34 by FISH. The position of LAMC3 was probed using a 1.3-kb cDNA probe within predicted protein domains I and II of γ3. The FISH signal (A) was superimposed over the DAPI-banded chromosomes (B) to identify the location of the γ3 gene. Both alleles of the γ3 gene are labeled (A, arrow) at the end of the long arm of chromosome 9 (B, 9).
Laminin γ3 Associates with α2β1 to Form Laminin 12 which, in Placenta, Is Lacking Part of the I/II Domains of All Three Chains
To determine the domains present within α2t, the 205-kD gel band, purified from placenta, was fragmented with trypsin and the resulting peptides were fractionated by HPLC; the masses of the eluted peptides were determined by mass spectroscopy. The ion chromatograms were then evaluated relative to the masses predicted from the published amino acid sequence for α2 in order to determine the NH2- and COOH-terminal peptides present within the digest. The results identified a number of tryptic peptides; among these, the peptide LVEHVPGQP(VR), beginning at residue 70 within domain VI, was the most NH2-terminal; the peptide GTTMTPPADLIEK, beginning at residue 1367 within domain III, was the most COOH-terminal. These results indicate that α2t is a fragment containing the short arm of the laminin α2 chain. This conclusion is consistent with the observation that the initial peptide sequence identified from α2t was within the short arm domains (above).
β1t and γ3t are also short arm fragments, as all the peptide sequences determined for both species are present within the short arm domains. However, the masses of α2t, β1t, and γ3t are greater than predicted for the short arms alone. In addition, α2t, β1t, and γ3t are not separable by gel electrophoresis without the reduction of disulfide bonds. Therefore, this truncated laminin 12 molecule is very likely to contain portions of domain II of all three chains, as the interchain disulfide bonds should lie between these domains. It is of interest to note that domain II of γ3 contains three cysteinyl residues whose bonding partners are not readily identified and are not present in domain II of other laminin chains. These three cysteinyl residues are conserved in mouse γ3 (Albus, A., and R.E. Burgeson, unpublished observation). Whether these cysteinyl residues could form intrachain, interchain, or intermolecular disulfide bonds that in some way contribute to the cleavage of γ3 chain-containing laminins is unknown.
Tissue Distribution of γ3 Expression by Northern Analysis
Tissue RNA blots and Master Blot dot blots (Clontech) were probed with a γ3 nucleotide probe (nt 1316 to 2277). A single major transcript of ∼5 kb, consistent in size with other laminin γ chains, is present in several of the tissues examined (Fig. 5 A). A small amount of a second larger transcript can also be detected. This larger transcript is most likely due to differences in polyadelylation or due to inefficient splicing. The γ3 chain RNA is abundant in spleen, testis, placenta, lung, and liver; lesser amounts are seen in kidney and ovary (Fig. 5 A). The predominance of a single transcript allowed use of the RNA Master Blot (Clontech) to determine expression in a large number of other tissues. On this dot blot, tissue RNA concentrations have been normalized to housekeeping genes. The Master Blot (Fig. 5 B) confirms the abundant presence of γ3 transcripts in placenta, adrenal gland, testis, lung, and fetal kidney, but also shows detectable levels of γ3 transcripts in numerous additional tissues, including brain and skeletal muscle.
Figure 5.
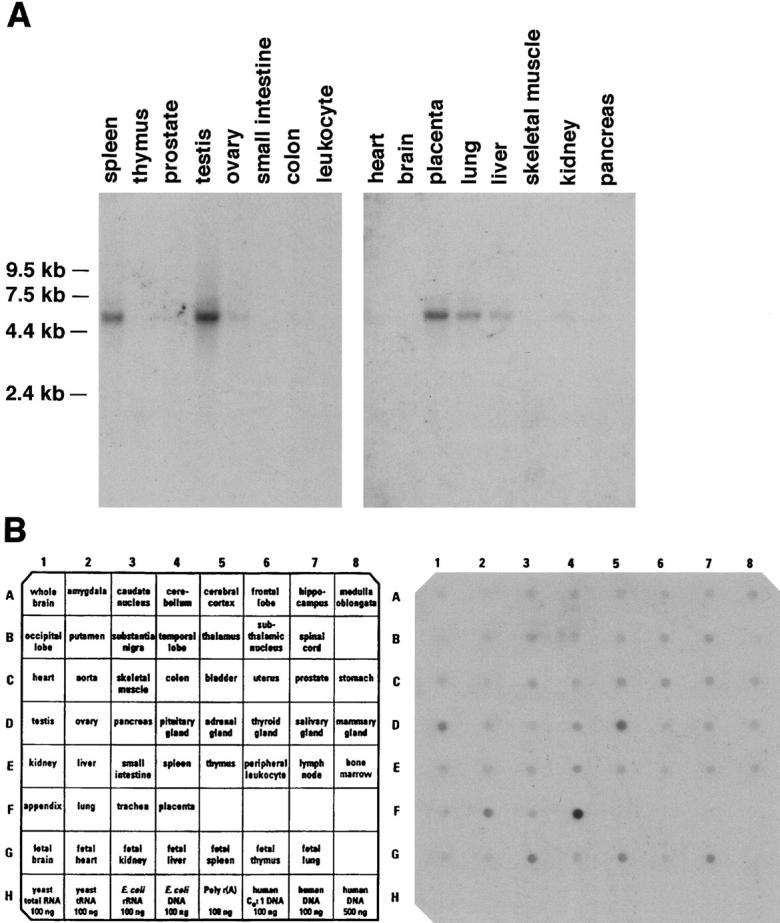
Laminin γ3 chain is expressed widely in human tissues. The same cDNA probe described in Fig. 4 was used to probe tissue blots (A) and a dot blot (B). Only a single major RNA species is detected on the tissue blots; strong signals are obtained from testis, spleen, placenta, and lung. Less intense signals are obtained from ovary, kidney, liver, and skeletal muscle. Dot blots indicate a more widespread tissue distribution, including thyroid, gut, and some fetal tissues.
Characterization of the Immunospecificity of Anti-Laminin γ3 (R16; R21)
A polyclonal antiserum, R16, was made in a rabbit to the γ3 chain excised from a reduced SDS-PAGE gel similar to that shown in Fig. 1. Another, R21, was made to recombinant γ3 protein (see Methods). The R16 antiserum recognizes the γ3 chain on immunoblots of placental extracts, but at very high antibody concentrations, it shows some reactivity with the β1 and γ1 chains as well. Thus, as a control, human neonatal foreskin was immunostained with anti-laminin γ1 (polyclonal anti-laminin 1; Sigma Chemical Co.), anti-laminin γ2 (GB3, Verrando et al., 1987), and with anti-laminin γ3 (R16). Crisp, brilliant fluorescence was observed along the dermal-epidermal junction, and around capillaries with the anti-γ1 antibodies (data not shown), and in the basement membrane at the dermal-epidermal junction with anti-γ2 (data not shown); in contrast, no signal above background was detected using the anti-γ3 reagent (R16) when it was applied at dilutions of 1:250 or more (data not shown). The antigen could not be unmasked by treatment of the cryosections with 2, 4, or 6 M urea, or with 2 M guanidinium-HCl (data not shown). As all known laminin chains have been detected in skin within either the epithelial basement membranes or the vascular basement membranes, these results indicate that the cross-reactivity detected by Western blot analyses using the polyclonal anti-γ3 (R16) antibody was either not apparent by immunohistochemistry, or was below detection at the antibody concentrations used. For the subsequent anatomical experiments (below), R16 was diluted 1:250 or greater to assure no cross-reactivity was occurring. R21, the affinity-purified antiserum to recombinant γ3, was also tested on sections of neonatal foreskin. As with R16, no immunoreactivity was seen (data not shown); thus we conclude that this antiserum has no cross-reactivity with other known γ chains. Neither R21 nor R16 antiserums label the blood vessel basement membranes (see below) consistent with a lack of cross-reactivity to other γ chains.
γ3-containing Laminins Are Localized to Peripheral Nerves and to Ciliated Epithelial Apical Surfaces
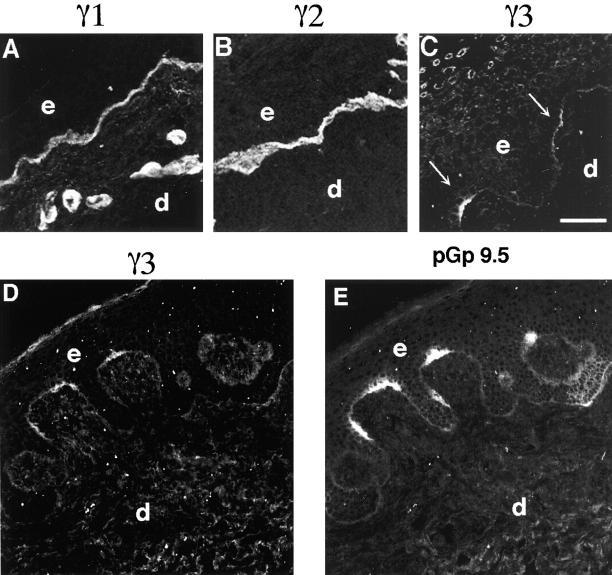
Unlike the lack of anti-laminin γ3 chain immunoreactivity seen in neonatal foreskin, laminin γ3 chain immunoreactivity was detected in human leg skin. As shown in Fig. 6 A, and consistent with published results, laminin γ1 chain reactivity is seen at the dermal-epidermal junction and within the basement membranes of the vasculature, while laminin γ2 chain immunoreactivity is restricted to the dermal-epidermal junction (Fig. 6 B). The laminin γ3 chain immunoreactivity is further restricted to distinct patches widely spaced along the dermal-epidermal junction (Fig. 6 C). In experiments not shown, the immunoreactivity did not correlate positively or negatively with sites of cell proliferation, nor did it correlate with fixed positions relative to the rete ridges. However, there is a direct correlation of the laminin γ3 chain immunoreactivity (Fig. 6 D) with sites where nerves cross the dermal-epidermal junction as detected by an antibody to the neuronal marker PGP9.5 (Fig. 6 E), which reacts with ubiquitin COOH-terminal hydrolase (Day et al., 1990). The results in skin suggest that γ3-containing laminins may be deposited into the dermal– epidermal junction by nerve or nerve associated cells, or that its expression by epithelial cells is induced by the adjacent nerve.
Figure 6.
Laminin γ3 chain is expressed in restricted sites, correlated with nerve terminations, in human skin. Laminin γ1, γ2, and γ3 chains and nerve-specific ubiquitin COOH-terminal hydrolase (PGP 9.5) in human leg skin were localized by immunohistochemistry and confocal laser microscopy. Laminin γ1 (A) immunoreactivity is present throughout the dermal-epidermal junctional basement membrane, as well as in the basement membranes of the glands and hair follicles. Laminin γ2 immunoreactivity (B), on the other hand, is restricted to the basement membrane of the dermal-epidermal junction and that underlying the epithelial cells of the outer root sheath of the hair follicles. Finally, laminin γ3 immunoreactivity (C) is most restricted and is found in distinct patches within the dermal-epidermal junction basement membrane. Laminin γ3 immunofluorescence (D) and a nerve-specific marker (E, anti-PGP 9.5) are colocalized when both primaries are applied to the same section.
Laminin γ3 is also expressed in the neural retina at the apical surface of the retina and in the outer synaptic layer (Libby, R.T., Y. Xu, E.P. Gibbons, M.-F. Champliaud, M. Koch, R.E. Burgeson, D.D. Hunter, and W.J. Brunken, manuscript submitted for publication); in the retina, the γ3 chain is coexpressed with the α4, α3, and β2 chains. Native γ3-containing laminins have not been isolated as yet from the retina; however, they have from another region of the central nervous system, the cerebellum, from which we have obtained two novel laminins, α3β2γ3 and α4β2γ3 (Champliaud, M.-F., unpublished observations). In addition, anatomical methods (immunohistochemistry and in situ hybridization), demonstrate the expression of γ3 in cerebellum and forebrain structures (Brunken, W.J., unpublished observations). It seems likely that γ3-containing laminins will be a general feature of the matrix in the CNS.
The Northern analysis indicated that the laminin γ3 chain was most strongly expressed in placenta, testis, lung, liver, spleen, and ovary. Therefore, we examined the localization of γ3 chains within testis, lung, and ovary. The reactivity within the epididymis and the fallopian tube were particularly striking. Thus, the distribution of γ3 in these tissues was extensively studied. In the female reproductive system, the oviduct was strongly reactive. Cryosections of the bovine (Fig. 7, A–F) or rat (Fig. 7, G–I) ampulla reacted for γ3 using R16 (Fig. 7, A, D, and G–I), or R21 (Fig. 7, B and E) showed brilliant immunoreactivity at the apical surfaces of the tubal mucosa. Double immunofluorescent studies performed with laminin γ3 and either laminin α2 (Fig. 7, A and D) or laminin α5 antibodies (Fig. 7 E) demonstrated that both of these α chains are restricted to the basement membranes of the tubal epithelial and the subjacent endothelium whereas γ3 is expressed at the apical surface. The pre-immune serum from rabbit 21 (Fig. 7 C) was negative, as was the reactivity of anti-thioredoxin antibodies purified from the R21 serum by immunoaffinity (Fig. 7 F).
Figure 7.
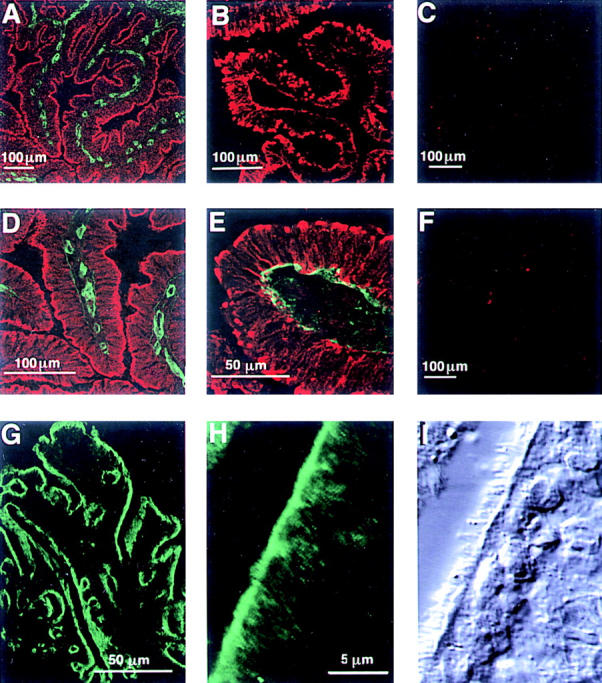
Laminin γ3 is expressed on the apical surface of the ciliated epithelium of the bovine and rat fallopian tube. Confocal laser microscope images of freshly frozen tissue sections of fallopian tube; in each image, five optical sections were superimposed. The laminin γ3-reactive antiserum R16 (A and D, red) and anti-laminin α2 chain (A and D, green) were applied to bovine tissue together; γ3 immunoreactivity is confined to the apical surface of the epithelium, whereas α2 is present in the basement membrane; D is a higher magnification of one of the folia in A. Application of the γ3-reactive R21 serum alone (B) also labels the apical surface of the bovine epithelium, but in plaque-like structures. R21 pre-immune serum is nonreactive (C). Simultaneous visualization of R21 immunoreactivity (E, red) and α5 immunoreactivity (E, green) shows that the γ3 is found on the apical side of the epithelium and in cytoplasmic stores in the long processes of these columnar tubal cells whereas α5-immunoreactivity is confined to the basement membrane. Antibodies purified from the R21 serum with the carrier protein, His-thioredoxin are negative (F). γ3 shows a similar distribution in rat fallopian tube (G and H): R16 reactivity is present at the apical surface over the whole of the ampulla (G) and in a higher power view of the epithelium (H); comparison of the fluorescent image in H with the differential interference contrast image in I demonstrates that γ3 immunoreactivity is associated with the ciliated surface of the epithelium.
The pattern of immunoreactivity for R16 in the rat oviduct was identical to that seen in bovine tissue (Fig. 7 G). Higher magnification micrographs of the epithelial apical surface of the rat ampulla (Fig. 7, H and I) show the γ3 chain to be localized to the apical surface of the epithelial cells at the base of the cilia.
It should be noted that the labeling pattern of R21 differed somewhat from that of R16. In general, the pattern with R21 was somewhat punctate, showing large deposits of immunoreactivity at the apical surface, and increased cytoplasmic labeling of the tubal epithelium, whereas the R16 immunoreactivity was more restricted to the apical extracellular surface. These observations suggest that the R21 antiserum, made to recombinant domain I, may recognize the unfolded γ3 chain better than the R16 antiserum, which should recognize primarily short arm domains.
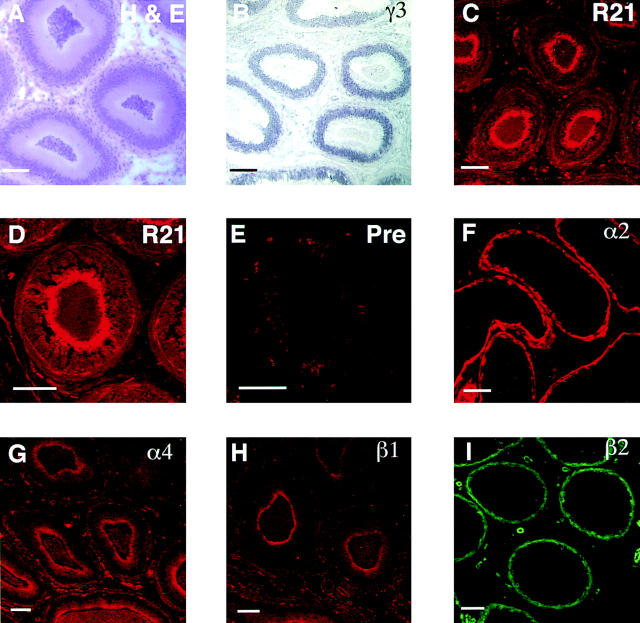
The male monkey reproductive tract was examined also. Like the fallopian tube, the epithelium in the epididymis is a single columnar epithelium (Fig. 8, A, H, and E). In situ hybridization performed on adjacent sections of the monkey epididymis (Fig. 8 B; γ3) localized transcripts for the γ3 chain to the apical region of the epithelial cells (compare Fig. 8, A with B). R16 (data not shown) and R21 (Fig. 8, C and D, R21) sera gave similar patterns, reacting with both the basal and apical surfaces of the epithelial cells. The R21 antiserum reacted with apparently intracellular stores of γ3, as was seen in the bovine fallopian tube. The preimmune control serum from R21 showed only punctate autofluorescence (Fig. 8 E, Pre).
Figure 8.
Laminin γ3 is expressed on the surface of male reproductive epithelium. Light (A and B) and confocal (C–I) images of the epididymis from monkey (rhesus). In A, a section was fixed and stained with H&E for orientation; the epididymis is composed of a columnar epithelium, with considerable interstitial matrix between adjacent folds. (B) By in situ hybridization, a γ3-specific probe is localized to the epithelium; the reaction product (dark blue) is concentrated above the nuclear region of the cell; this section is not counter-stained. (C and D) γ3 immunoreactivity (R21) is present at the surface of the epithelium; (E) pre-immune (Pre) serum is nonreactive in this tissue. (F–I) Immunolocalization of other laminin chains; both α2 and β2 are confined to the basement membranes of the epithelium and blood vessels, whereas both α4 and β1 are seen at the apical surface of the epithelium; α4 is also expressed in the interstitial matrix and the basement membrane. Bars, 100 μm.
Potential chains partners were explored by examination of the same tissue with antibodies specific for a variety of other laminin chains: α2 (Fig. 8 F), α4 (G), β1 (H), and β2 (I). We used two monoclonal antibodies to test for the presence of β1 at the apical surface (clones 545; and C21) both gave the same pattern of immunolabeling; only the results with clone 545 are shown. As can be seen readily, α2 and β2 were restricted to the basal surface of the epithelial cells, while staining for α4 and β1 were also seen at the apical surface. Thus, in contrast to the results from placental extracts, α4 (and not α2) appears to be a candidate chain partner for γ3 in the epididymis. These observations suggest that a wide variety of γ3-containing laminins will be expressed in a tissue-specific pattern.
Expression of laminin γ3 chain was examined in the rat as well and the tissue distribution of γ3 in the rat epididymis was similar to that described for the monkey (data not shown); namely, γ3 immunoreactivity was localized to the apical surface of the epithelium. We also studied other regions of the rat reproductive system. Unlike laminin-1 immunoreactivity, which is localized to the basement membrane of the seminiferous tubules (Fig. 9 A), γ3 immunoreactivity is not present within the basement membrane of the seminiferous tubules nor is it found around the interstitial cells (Fig. 9 A, arrows; Fig. 9 B, asterisk). Within the seminiferous tubules, only the occasional tubule reacted strongly with the laminin γ3 reactive serum (R16, Fig. 9 B); it was our impression that those tubules identified by the antibody contained nearly mature spermatids. Further along the male reproductive system, in the ductus deferens, laminin-1 immunoreactivity (Fig. 9 C; arrows mark the apical surface of the epithelium) was seen along the epithelial basement membrane, in the lamina propria and ensheathing the smooth muscle cells of the muscular layer. In contrast, γ3 immunoreactivity (R16) was found at the apical and basal surfaces of the epithelial cells, as well as intracellularly (Fig. 9 D).
Figure 9.

Laminin γ3 is expressed on the surface of rat ciliated epithelia in testis and lung. Various tissues from the rat male reproductive system (A–D) were incubated with either laminin-1 (α1/β1/γ1) antiserum (left column) or γ3 antiserum (right column). (A and B) testis; (C and D) ductus deferens. The laminin-1 immunoreactivity is present in the basement membrane in these structures (left column); arrows denote interstitial cells in A, and the apical borders of the epithelium in C; the basal lamina surrounding the smooth muscle is also easily distinguished (C). In contrast, laminin γ3 expression is distinctly not in the basement membrane of the testis (B) nor of the ductus deferens (D). γ3 expression is limited to the seminiferous tubules themselves and does not surround the interstitial cells (B, asterisk). Within the seminiferous tubules, γ3 expression appears to change with the state of maturation of sperm (see text for details). In the ductus deferens, γ3 expression (D) is limited to the surface of epithelial cells (basolateral and apical). No γ3 expression is seen in the smooth muscle of the ductus deferens. In lung (E and F), laminin γ3 is expressed on the apical surface of nonrespiratory airways. γ3 immunoreactivity is limited to the apical surface of the epithelium (E; compare to the differential interference contrast image of the same section in F). Scale bars are given for each pair of figures (A and B, C and D, E and F).
The apical distribution of the γ3 chain is not confined to the reproductive system; in rat lung, the ciliated epithelial cells lining the bronchi were also strongly reactive with the anti-laminin γ3 antiserum, R16 (Fig. 9 E). Again, the fluorescence was apparent along the apical surface, as determined by differential interference contrast microscopy (Fig. 9 F). No γ3-immunoreactivity was seen in respiratory epithelium nor in the pulmonary capillary bed (not shown).
Discussion
The laminin γ3 chain described here is the eleventh laminin subunit to be identified. The predicted primary and secondary structure of this chain suggests that γ3 is more closely related to human γ1 than γ2. Unlike γ2, the γ3 cDNA sequence predicts a laminin subunit without the short-arm truncations predicted for γ2. Perhaps more significantly, γ3 contains a γ1-like nidogen binding motif with only a single conservative amino acid substitution, suggesting that γ3-containing laminins should be capable of associating with other basement membrane molecules through nidogen interactions (Mayer et al., 1995; Poschl et al., 1996). In addition, domain VI of γ3 shares the highest sequence identity with domain VI of the γ1 chain. As this latter domain has been shown to support laminin self-assembly (Yurchenco and Cheng, 1994), it seems reasonable to suggest that domain VI of the γ3 chain may also support self-assembly.
Two of the predicted glycosylation sites in the γ3 chain are also found in human and in mouse γ1; these are within short-arm globular domains, i.e., VI and IV. Interestingly, the glycosylation site in domain IV is also found in human (Kallunki et al., 1992) and in mouse (Sugiyama et al., 1995) γ2. This remarkable conservation of glycosylation sites among these three chains and between these species suggests that these sites are indeed important and glycosylated; they are likely to be critical in the folding of this region or may play another important function.
The RGD sequence within domain II of the γ3 chain is found in neither γ1 nor in γ2. Moreover, it seems likely that this sequence is not functional within native γ3-containing laminins as it is located within the coiled-coil region of γ3. However, it very well may promote integrin-mediated recognition of non-native molecules or of protein fragments.
In placenta, the γ3 chain can combine with the laminin α2 and β1 chains. This observation suggests that, unlike γ2 which pairs preferentially with β3, γ3 may pair with any β chain, with the possible exception of β3, and with any of the known α chains. This prediction suggests the existence of an additional 10 laminins with the following chain compositions: α1β1γ3, α1β2γ3, α2β1γ3, α2β2γ3, α3β1γ3, α3β2γ3, α4β1γ3, α4β2γ3, α5β1γ3, and α5β2γ3. In both the epididymis and the fallopian tube, γ3 is not combined with α2. In the epididymis, the α4 and β1 chains appear to be potential partners. Given that the total number of human laminins is not known, at least one additional β chain has been identified in chicken (Ybot-Gonzalez et al., 1995; Liu et al., 1998) and in mammals (Olson, P.F., unpublished observations), assigning a final laminin numerical identifier to these laminins is premature. However, as we have shown α2β1γ3 to be the twelfth laminin to be identified, we provisionally call this heterotrimer, laminin 12.
The masses of the chains of laminin 12 as approximated by electrophoretic mobility are considerably less than predicted by the amino acid sequences and from prior experience with the α2 and β1 chains. They are also less than the α2, β1, and β2 chains present in laminins 2 and 4 obtained from the same preparations. The reason for these more rapid electrophoretic migration rates appears to be proteolysis within the domains II of the chains comprising this molecule. In placenta, this proteolysis may be physiological, since laminins 2 and 4 isolated from the same preparations are apparently intact. The significance of this observation awaits considerable additional experimentation before it is understood. However, we have observed three cysteinyl residues within domain II of the γ3 chain that are not present in other human α, β, or γ chains. It is possible that a disulfide bond between two of these residues distorts the coiled-coil conformation making molecules containing this chain more susceptible to proteolysis. At this time, we do not know if truncation of laminins containing the γ3 chain can be generalized to tissues other than placenta; however, this seems unlikely in that our isolation of γ3-containing laminins from the CNS do not show the same truncation (Champliaud, unpublished observations). The COOH-terminal truncation of α2t explains its lack of reactivity with an anti-α2 antibody, mAb 1922, which is specific for the α2 G domain (Engvall et al., 1990).
The antiserum to the recombinant γ3 domain I fusion protein (R21) was originally made to evaluate this potential processing, since epitopes contained within this domain should be absent from the processed molecule. In this regard, the immunohistochemical data is not definitive. Like R16, R21 immunoreactivity is seen at the apical surface; however, the apical reactivity is distinctly different than that for R16. Specifically, R21 immunoreactivity appears as a plaque-like structure at the cell surface, with some reactivity within the cells. Thus, it seems possible that R21 epitopes are entirely intracellular but it is also possible that some of the R21 epitopes are present at the apical surface of these epithelial cells. Further experimentation beyond the scope of this report is required to address this question. However, laminins containing γ3 chains have been immunoisolated from the medium of A204 cells derived from a human rhabdosarcoma (Champliaud, M.-F., unpublished data), indicating that γ3-containing laminins are capable of being secreted in vitro. These γ3-containing laminins from A204 are a mixture of processed (truncated) and unprocessed (untruncated) molecules.
Reports of laminins in tissue locations not identified as basement membranes are increasingly frequent. In the brain, laminins have been observed not only within the basement membrane of capillaries, but also at other sites not conceptualized as basement membranes (Higuchi et al., 1991; Jucker et al., 1992, 1996a,b; Mori et al., 1992; Tian et al., 1996, 1997; Hagg et al., 1997; Yamamoto et al., 1997). In the eye, the laminin β2 chain has been identified in both basement membrane and non-basement membrane locations (Hunter et al., 1992; Libby et al., 1996; Libby et al., 1997; Toti et al., 1997). Laminins have also been observed in cartilage (Durr et al., 1996).
Intriguingly, the laminin γ3 chain appears most commonly to be associated with non-basement membrane structures. In the cerebellum, γ3 chains are detected in the pericellular nets surrounding both neurons and glia (Brunken, W.J., unpublished observations). Reported elsewhere (Libby et al., manuscript submitted for publication, see above), γ3 is present within the neural retina at two extracellular sites: between the outer segments of the photoreceptors, and at the synapses of the photoreceptors with the bipolar and horizontal cells. In the retina, these γ3-containing molecules are the products of the Müller glial cells which, like the tubal epithelium, contain a considerable store of intracellular γ3 chain. The laminin α3 and β2 chains are also present at these sites, whereas the γ1 and γ2 chains are absent. The functions fulfilled by these laminins are unclear, but possibilities include: stabilization of neural architecture; induction and stabilization of differentiated neural phenotypes (Hunter et al., 1992b; Libby et al., 1996; Hunter and Brunken, 1997); and stabilization of synaptic junctions.
However, the most abundant expression of γ3 as detected by Northern analyses is not within neural tissues, but rather is in the testis, the placenta, the spleen, the lung, and the ovary. γ3 immunoreactivity is present at the bases of the epithelial cilia of the epididymis, the trachea, the bronchi, and the oviduct. There are no structures resembling basement membrane at these sites. However, γ3 chains may be present within the basement membranes along the basolateral surfaces of some of these epithelia. The chain partners for the γ3 chain in these apical laminins are not yet known with certainty. However, in epididymis, the laminin α2 chain does not colocalize with γ3 at the apical epithelial surface; rather, the α4 and β1 chains are present at that location and, thereby, are potential chain partners. Thus, it seems likely that γ3 will be as promiscuous as γ1 with respect to partner choice during laminin assembly.
The presence of laminins along ciliated epithelial surfaces was unexpected and their functions there are unknown. Perhaps a modified basement membrane containing at least laminin helps organize or stabilize the specialized cytoskeleton of the cilia. Laminins at these apical surfaces may also participate in the anchorage of mucins to the surface. Alternatively, laminins might stabilize the outfoldings of the plasma membranes of the cilia. Similar functions for laminins have been postulated to stabilize the junctional folds beneath synapses at neuromuscular junctions (Noakes et al., 1995), and contribute to the organization of epithelial hemidesmosomes (Langhofer et al., 1993; Baker et al., 1996). Laminins expressed at the apical surface of the retina are thought to play a role in photoreceptor morphogenesis, specifically outer segment formation and synapse development (Libby et al., 1997, 1998).
Consistent with the above speculations regarding an essential function for γ3 containing laminins in neural tissues, the chromosomal locus of LAMC3 at 9q31-q34 is shared with four diseases having various degrees of neural dysfunction in common: Fukuyama congenital muscular dystrophy (FCMD); muscle-eye-brain syndrome; Walker-Warburg syndrome; and retinitis pigmentosa-21 with deafness (RP-21). The genetic cause of the latter three of these conditions is unclear. While γ3 expression may be affected in FCMD, the LAMC3 cannot be the genetic cause of the problem as a retrotransposal insertion in a different gene has recently been identified in 87% of the FCMD alleles (Kobayashi et al., 1998). However, LAMC3 is an excellent candidate for mutations underlying one or more of remaining syndromes in this cluster of human diseases, particularly RP-21.
We have identified the laminin γ3 chain together with the α2 and β1 chains within laminin 12 from human placenta, but multiple other combinations are possible in other tissues. It would be of particular interest if γ3 were to associate with the β2 chain and well as with the α2 chain, as both these chains have been reported to show neural and muscle-associated expression and function. The γ3-containing laminins are likely to be the subject of considerable interest as they constitute a novel class of laminin molecules distributed outside of the traditional basement membrane. The identification of the function of this diverse family of laminins remains to be elucidated by future experiments.
Acknowledgments
This work was supported by the U.S. Public Health Service grants AR35689 (R.E. Burgeson), EY12037 (D.D. Hunter); the E. Matilda Ziegler Foundation (W.J. Brunken); and additional support from the Cutaneous Biology Research Center, Massachusetts General Hospital.
Abbreviations used in this paper
- FCMD
Fukuyama congenital muscular dystrophy
- FISH
fluorescent in situ hybridization
- G
globular
Footnotes
The authors gratefully acknowledge the excellent technical support provided by Ms. Carol Milbury, and the expert assistance of Dr. Yimin Ge with confocal microscopy. The authors also thank Dr. Richard Kammerer for the HisTrx and a pPEP-T vector.
Dr. Brunken is on leave from the Department of Biology, Boston College.
References
- Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SBH. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aumailley M, Battaglia C, Mayer U, Reinhardt D, Nischt R, Timpl R, Fox JW. Nidogen mediates the formation of ternary complexes of basement membrane components. Kidney Int. 1993;43:7–12. doi: 10.1038/ki.1993.3. [DOI] [PubMed] [Google Scholar]
- Baker S, Hopkinson S, Fitchmun M, Andreason G, Frasier F, Plopper G, Quaranta V, Jones J. Laminin-5 and hemidesmosomes: role of the alpha 3 chain subunit in hemidesmosome stability and assembly. J Cell Sci. 1996;109:2509–2520. doi: 10.1242/jcs.109.10.2509. [DOI] [PubMed] [Google Scholar]
- Battaglia C, Mayer U, Aumailley M, Timpl R. Basement-membrane heparan sulfate proteoglycan binds to laminin by its heparan sulfate chains and to nidogen by sites in the protein core. Eur J Biochem. 1992;208:359–366. doi: 10.1111/j.1432-1033.1992.tb17195.x. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST-database for “expressed sequence tags.” . Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Burgeson RE, Chiquet M, Deutzmann R, Ekblom P, Engel J, Kleinman H, Martin GR, Meneguzzi G, Paulsson M, Sanes J, et al. A new nomenclature for the laminins. Matrix Biol. 1994;14:209–211. doi: 10.1016/0945-053x(94)90184-8. [DOI] [PubMed] [Google Scholar]
- Chait BT, Kent SB. Weighing naked proteins: practical, high-accuracy mass measurement of peptides and proteins. Science. 1992;257:1885–1894. doi: 10.1126/science.1411504. [DOI] [PubMed] [Google Scholar]
- Chen L, Shick V, Matter M, Laurie S, Ogle R, Laurie G. Laminin E8 alveolarization site: heparin sensitivity, cell surface receptors, and role in cell spreading. Am J Physiol. 1997;272:L494–L503. doi: 10.1152/ajplung.1997.272.3.L494. [DOI] [PubMed] [Google Scholar]
- Cohen M, Jacobson C, Yurchenco P, Morris G, Carbonetto S. Laminin-induced clustering of dystroglycan on embryonic muscle cells: comparison with agrin-induced clustering. J Cell Biol. 1997;136:1047–1058. doi: 10.1083/jcb.136.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day IN, Hinks LJ, Thompson RJ. The structure of the human gene encoding protein gene product 9.5 (PGP9.5), a neuron-specific ubiquitin C-terminal hydrolase. Biochem J. 1990;268:521–524. doi: 10.1042/bj2680521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutzmann R, Aumailley M, Wiedemann H, Pysny W, Timpl R, Edgar D. Cell adhesion, spreading and neurite stimulation by laminin fragment E8 depends on maintenance of secondary and tertiary structure in its rod and globular domain. Eur J Biochem. 1990;191:513–522. doi: 10.1111/j.1432-1033.1990.tb19151.x. [DOI] [PubMed] [Google Scholar]
- Drago J, Nurcombe V, Bartlett PF. Laminin through its long arm E8 fragment promotes the proliferation and differentiation of murine neuroepithelial cells in vitro. Exp Cell Res. 1991;192:256–265. doi: 10.1016/0014-4827(91)90184-v. [DOI] [PubMed] [Google Scholar]
- Durkin M, Loechel F, Mattei M, Gilpin B, Albrechtsen R, Wewer U. Tissue-specific expression of the human laminin alpha5-chain, and mapping of the gene to human chromosome 20q13.2-13.3 and to distal mouse chromosome 2 near the locus for the ragged (Ra) mutation. FEBS Lett. 1997;411:296–300. doi: 10.1016/s0014-5793(97)00686-8. [DOI] [PubMed] [Google Scholar]
- Durr J, Lammi P, Goodman SL, Aigner T, von der Mark K. Identification and immunolocalization of laminin in cartilage. Exp Cell Res. 1996;222:225–233. doi: 10.1006/excr.1996.0028. [DOI] [PubMed] [Google Scholar]
- Ekblom, P., and R. Timpl. 1996. The Laminins. In Cell Adhesion and Communication. Vol. 2. C. Goridis, editor. Harwood Academic Publishers, The Netherlands. 321 pp.
- Engel J. Laminins and other strange proteins. Biochemistry. 1992;31:10643–10651. doi: 10.1021/bi00159a001. [DOI] [PubMed] [Google Scholar]
- Engvall E. Laminin variants: why, where and when? . Kidney Int. 1993;43:2–6. doi: 10.1038/ki.1993.2. [DOI] [PubMed] [Google Scholar]
- Engvall E, Earwicker D, Haaparanta T, Ruoslahti E, Sanes JR. Distribution and isolation of four laminin variants; tissue restricted distribution of heterotrimers assembled from five different subunits. Cell Regul. 1990;1:731–740. doi: 10.1091/mbc.1.10.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Mayer U, Nischt R, Aumailley M, Reinhardt D, Wiedemann H, Mann K, Timpl R, Krieg T, Engel J, et al. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO (Eur Mol Biol Organ) J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke DR, Wagman DW, Champliaud MF, Burgeson RE. The complete primary structure for a novel laminin chain, the laminin B1k chain. J Biol Chem. 1994;269:11073–11080. [PubMed] [Google Scholar]
- Goodman SL. Alpha 6 beta 1 integrin and laminin E8: an increasingly complex simple story. Kidney Int. 1992;41:650–656. doi: 10.1038/ki.1992.100. [DOI] [PubMed] [Google Scholar]
- Green TL, Hunter DD, Chan W, Merlie JP, Sanes JR. Synthesis and assembly of synaptic cleft protein s-laminin by cultured cells. J Biol Chem. 1992;267:2014–2022. [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Haaparanta T, Uitto J, Ruoslahti E, Engvall E. Molecular cloning of the cDNA encoding human laminin A chain. Matrix. 1991;11:151–160. doi: 10.1016/s0934-8832(11)80153-8. [DOI] [PubMed] [Google Scholar]
- Hagg T, Portera-Cailliau C, Jucker M, Engvall E. Laminins of the adult mammalian CNS; laminin-alpha2 (merosin M-) chain immunoreactivity is associated with neuronal processes. Brain Res. 1997;764:17–27. doi: 10.1016/s0006-8993(97)00419-8. [DOI] [PubMed] [Google Scholar]
- Heimann P, Menke A, Rothkegel B, Jockusch H. Overshooting production of satellite cells in murine skeletal muscle affected by the mutation “muscular dystrophy with myositis” (mdm, Chr 2) Cell Tissue Res. 1996;283:435–441. doi: 10.1007/s004410050554. [DOI] [PubMed] [Google Scholar]
- Henry MD, Campbell KP. Dystroglycan: an extracellular matrix receptor linked to the cytoskeleton. Curr Opin Cell Biol. 1996;8:625–631. doi: 10.1016/s0955-0674(96)80103-7. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Ohnishi T, Arita N, Hiraga S, Iwasaki H, Mori S, Hayakawa T. Immunohistochemical localization of fibronectin, laminin and fibronectin-receptor in human malignant gliomas—in relation to tumor invasion. No to Shinkei. Brain Nerve (Tokyo) 1991;43:17–23. [PubMed] [Google Scholar]
- Hunter DD, Brunken WJ. Beta 2 laminins modulate neuronal phenotype in the rat retina. Mol Cell Neurosci. 1997;10:7–15. doi: 10.1006/mcne.1997.0632. [DOI] [PubMed] [Google Scholar]
- Hunter DD, Llinas R, Ard M, Merlie JP, Sanes JR. Expression of s-laminin and laminin in the developing rat central nervous system. J Comp Neurol. 1992a;323:238–251. doi: 10.1002/cne.903230208. [DOI] [PubMed] [Google Scholar]
- Hunter DD, Murphy MD, Olsson CV, Brunken WJ. S-laminin expression in adult and developing retinae: a potential cue for photoreceptor morphogenesis. Neuron. 1992b;8:399–413. doi: 10.1016/0896-6273(92)90269-j. [DOI] [PubMed] [Google Scholar]
- Iivanainen A, Sainio K, Sariola H, Tryggvason K. Primary structure and expression of a novel human laminin a4 chain. FEBS Lett. 1995a;365:183–188. doi: 10.1016/0014-5793(95)00462-i. [DOI] [PubMed] [Google Scholar]
- Iivanainen A, Vuolteenaho R, Sainio K, Eddy R, Shows TB, Sariola H, Tryggvason K. The human laminin beta 2 chain (S-laminin): structure, expression in fetal tissues and chromosomal assignment of the LAMB2 gene. Matrix Biol. 1995b;14:489–497. doi: 10.1016/0945-053x(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Jucker M, Bialobok P, Hagg T, Ingram DK. Laminin immunohistochemistry in brain is dependent on method of tissue fixation. Brain Res. 1992;586:166–170. doi: 10.1016/0006-8993(92)91390-z. [DOI] [PubMed] [Google Scholar]
- Jucker M, Tian M, Ingram D. Laminins in the adult and aged brain. Mol Chem Neuropathol. 1996a;28:209–218. doi: 10.1007/BF02815224. [DOI] [PubMed] [Google Scholar]
- Jucker M, Tian M, Norton D, Sherman C, Kusiak J. Laminin alpha 2 is a component of brain capillary basement membrane: reduced expression in dystrophic dy mice. Neuroscience. 1996b;71:1153–1161. doi: 10.1016/0306-4522(95)00496-3. [DOI] [PubMed] [Google Scholar]
- Kallunki P, Sainio K, Eddy R, Byers M, Kallunki T, Sariola H, Beck K, Hirvonen H, Shows TB, Tryggvason K. A truncated laminin chain homologous to the B2 chain: structure, spatial expression, and chromosomal assignment. J Cell Biol. 1992;119:679–693. doi: 10.1083/jcb.119.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;398:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langhofer M, Hopkinson SB, Jones JC. The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. J Cell Sci. 1993;105:753–764. doi: 10.1242/jcs.105.3.753. [DOI] [PubMed] [Google Scholar]
- Libby RT, Hunter DD, Brunken WJ. Developmental expression of laminin beta 2 in rat retina. Further support for a role in rod morphogenesis. Investig Ophthalmol Vis Sci. 1996;37:1651–1661. [PubMed] [Google Scholar]
- Libby RT, Xu Y, Selfors LM, Brunken WJ, Hunter DD. Identification and cellular source of laminin β2 in adult and developing vertebrate retinae. J Comp Neurol. 1997;389:655–667. doi: 10.1002/(sici)1096-9861(19971229)389:4<655::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Libby RT, Lavalee C, Balkema GW, Brunken WJ, Hunter DD. Characterization of laminin β2 in retinal function. Investig Ophthalmol Vis Sci. 1998;39:S574. [Google Scholar]
- Liu J, Swasdison S, Xie W, Brewton RG, Mayne R. Primary structure and expression of a chicken laminin beta chain: evidence for four beta chains in birds. Matrix Biol. 1998;16:471–481. doi: 10.1016/s0945-053x(98)90018-x. [DOI] [PubMed] [Google Scholar]
- Lunstrum GP, Sakai LY, Keene DR, Morris NP, Burgeson RE. Large complex globular domains of type VII procollagen contribute to the structure of anchoring fibrils. J Biol Chem. 1986;261:9042–9048. [PubMed] [Google Scholar]
- Marinkovich MP, Lunstrum GP, Keene DR, Burgeson RE. The dermal-epidermal junction of human skin contains a novel laminin variant. J Cell Biol. 1992;119:695–703. doi: 10.1083/jcb.119.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter ML, Laurie GW. A novel laminin E8 cell adhesion site required for lung alveolar formation in vitro. J Cell Biol. 1994;124:1083–1090. doi: 10.1083/jcb.124.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer, P., and J. Engel. 1996. Structure of laminins and their chain assembly. In The Laminins. Vol. 2. P. Ekblom and R. Timpl, editor. Harwood Academic Publishers, The Netherlands. 27–50.
- Mayer U, Nischt R, Poschl E, Mann K, Fukuda K, Gerl M, Yamada Y, Timpl R. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO (Eur Mol Biol Organ) J. 1979;12:1879–1885. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Poschl E, Gerecke DR, Wagman DW, Burgeson RE, Timpl R. Low nidogen affinity of laminin-5 can be attributed to two serine residues in EGF-like motif gamma 2III4. FEBS Lett. 1995;365:129–132. doi: 10.1016/0014-5793(95)00438-f. [DOI] [PubMed] [Google Scholar]
- Miner JH, Patton BL, Lentz SI, Gilbert DJ, Snider WD, Jenkins NA, Copeland NG, Sanes JR. The laminin a chains: expression, developmental transitions, and chromosomal locations of a1-5, identification of heterotrimeric laminins 8-11 and cloning a novel a3 isoform. J Cell Biol. 1997;137:685–701. doi: 10.1083/jcb.137.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Lewis RM, Sanes JR. Molecular cloning of a novel laminin chain, alpha 5, and widespread expression in adult mouse tissues. J Biol Chem. 1995;270:28523–28526. doi: 10.1074/jbc.270.48.28523. [DOI] [PubMed] [Google Scholar]
- Mori S, Sternberger NH, Herman MM, Sternberger LA. Variability of laminin immunoreactivity in human autopsy brain. Histochemistry. 1992;97:237–241. doi: 10.1007/BF00267633. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hogervorst F, Jaspars LH, de Melkar AA, Delwel GO, Hulsman EH, Kuikman I, Sonnenberg A. The alpha 6 beta 4 integrin is a receptor for both laminin and kalinin. Exp Cell Res. 1994;211:360–367. doi: 10.1006/excr.1994.1099. [DOI] [PubMed] [Google Scholar]
- Noakes PG, Gautam M, Mudd J, Sanes JR, Merlie JP. Aberrant differentiation of neuromuscular junctions in mice lacking s-laminin/laminin beta 2. Nature. 1995;374:258–262. doi: 10.1038/374258a0. [DOI] [PubMed] [Google Scholar]
- Pall E, Bolton K, Ervasti J. Differential heparin inhibition of skeletal muscle alpha-dystroglycan binding to laminins. J Biol Chem. 1996;271:3817–3821. doi: 10.1074/jbc.271.7.3817. [DOI] [PubMed] [Google Scholar]
- Pikkarainen T, Eddy R, Fukushima Y, Byers M, Shows T, Pihlajaniemi T, Saraste M, Tryggvason K. Human laminin B1 chain. A multidomain protein with gene (LAMB1) locus in the q22 region of chromosome 7. J Biol Chem. 1987;262:10454–10462. [PubMed] [Google Scholar]
- Pikkarainen T, Kallunki T, Tryggvason K. Human laminin B2 chain. Comparison of the complete amino acid sequence with the B1 chain reveals variability in sequence homology between different structural domains. J Biol Chem. 1988;263:6751–6758. [PubMed] [Google Scholar]
- Poschl E, Fox JW, Block D, Mayer U, Timpl R. Two non-contiguous regions contribute to nidogen binding to a single EGF-like motif of the laminin gamma 1 chain. EMBO (Eur Mol Biol Organ) J. 1994;13:3741–3747. doi: 10.1002/j.1460-2075.1994.tb06683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poschl E, Mayer U, Stetefeld J, Baumgartner R, Holak T, Huber R, Timpl R. Site-directed mutagenesis and structural interpretation of the nidogen binding site of the laminin gamma1 chain. EMBO (Eur Mol Biol Organ) J. 1996;15:5154–5159. [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mann K, Nischt R, Fox JW, Chu ML, Krieg T, Timpl R. Mapping of nidogen binding sites for collagen type IV, heparan sulfate proteoglycan, and zinc. J Biol Chem. 1993;268:10881–10887. [PubMed] [Google Scholar]
- Rousselle P, Golbik R, van der Rest M, Aumailley M. Structural requirement for cell adhesion to kalinin (laminin-5) J Biol Chem. 1995;270:13766–13770. doi: 10.1074/jbc.270.23.13766. [DOI] [PubMed] [Google Scholar]
- Rousselle P, Keene DR, Ruggiero F, Champliaud MF, Rest M, Burgeson RE. Laminin 5 binds the NC-1 domain of type VII collagen. J Cell Biol. 1997;138:719–728. doi: 10.1083/jcb.138.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Tizard R, VanDevanter DR, Carter WG. Cloning of the LamA3 gene encoding the alpha 3 chain of the adhesive ligand epiligrin. Expression in wound repair. J Biol Chem. 1994;269:22779–22787. [PubMed] [Google Scholar]
- Sugiyama S, Utani A, Yamada S, Kozak CA, Yamada Y. Cloning and expression of the mouse laminin gamma 2 (B2t) chain, a subunit of epithelial cell laminin. Eur J Biochem. 1995;228:120–128. doi: 10.1111/j.1432-1033.1995.tb20239.x. [DOI] [PubMed] [Google Scholar]
- Tian M, Hagg T, Denisova N, Knusel B, Engvall E, Jucker M. Laminin-alpha2 chain-like antigens in CNS dendritic spines. Brain Res. 1997;764:28–38. doi: 10.1016/s0006-8993(97)00420-4. [DOI] [PubMed] [Google Scholar]
- Tian M, Jacobson C, Gee SH, Campbell KP, Carbonetto S, Jucker M. Dystroglycan in the cerebellum is a laminin alpha 2-chain binding protein at the glial-vascular interface and is expressed in Purkinje cells. Eur J Neurosci. 1996;8:2739–2747. doi: 10.1111/j.1460-9568.1996.tb01568.x. [DOI] [PubMed] [Google Scholar]
- Toti P, De Felice C, Malandrini A, Megha T, Cardone C, Villanova M. Localization of laminin chains in the human retina: possible implications for congenital muscular dystrophy associated with alpha 2-chain of laminin deficiency. Neuromuscular Disorders. 1997;7:21–25. doi: 10.1016/s0960-8966(96)00399-9. [DOI] [PubMed] [Google Scholar]
- Verrando P, Hsi BL, Yeh CJ, Pisani A, Serieys N, Ortonne JP. Monoclonal antibody GB3, a new probe for the study of human basement membranes and hemidesmosomes. Exp Cell Res. 1987;170:116–128. doi: 10.1016/0014-4827(87)90121-2. [DOI] [PubMed] [Google Scholar]
- Vuolteenaho R, Nissinen M, Sainio K, Byers M, Eddy R, Hirvonen H, Shows TB, Sariola H, Engvall E, Tryggvason K. Human laminin M chain (merosin): complete primary structure, chromosomal assignment, and expression of the M and A chain in human fetal tissues. J Cell Biol. 1994;124:381–394. doi: 10.1083/jcb.124.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wewer U, Engvall E. Merosin/laminin-2 and muscular dystrophy. Neuromuscular Disorders. 1996;6:409–418. doi: 10.1016/s0960-8966(96)00384-7. [DOI] [PubMed] [Google Scholar]
- Wewer UM, Gerecke DR, Durkin ME, Kurtz KS, Mattei MG, Champliaud MF, Burgeson RE, Albrechtsen R. Human beta 2 chain of laminin (formerly S chain): cDNA cloning, chromosomal localization, and expression in carcinomas. Genomics. 1994;24:243–252. doi: 10.1006/geno.1994.1612. [DOI] [PubMed] [Google Scholar]
- Xu H, Wu XR, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin alpha 2 (Lama2) gene. Nat Genet. 1994;8:297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Shibata N, Kanazawa M, Kobayashi M, Komori T, Ikeya K, Kondo E, Saito K, Osawa M. Localization of laminin subunits in the central nervous system in Fukuyama congenital muscular dystrophy: an immunohistochemical investigation. Acta Neuropathol. 1997;94:173–179. doi: 10.1007/s004010050690. [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P, Runswick S, Smyth N, Edgar D. Regulated expression of a novel laminin beta subunit during the development of the chick embryo. Differentiation. 1995;59:215–223. doi: 10.1046/j.1432-0436.1995.5940215.x. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Cheng YS. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J Biol Chem. 1993;268:17286–17299. [PubMed] [Google Scholar]
- Yurchenco PD, Cheng YS. Laminin self-assembly: a three-arm interaction hypothesis for the formation of a network in basement membranes. Contrib Nephrol. 1994;107:47–56. doi: 10.1159/000422960. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Cheng YS, Colognato H. Laminin forms an independent network in basement membranes [published erratum appears in J. Cell Biol.118:493] J Cell Biol. 1992;117:1119–1133. doi: 10.1083/jcb.117.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]