Abstract
During the life cycle of the streptomycetes, large numbers of hyphae die; the surviving ones undergo cellular differentiation and appear as chains of spores in the mature colony. Here we report that the hyphae of Streptomyces antibioticus die through an orderly process of internal cell dismantling that permits the doomed hyphae to be eliminated with minimum disruption of the colony architecture. Morphological and biochemical approaches revealed progressive disorganization of the nucleoid substructure, followed by degradation of DNA and cytoplasmic constituents with transient maintenance of plasma membrane integrity. Then the hyphae collapsed and appeared empty of cellular contents but retained an apparently intact cell wall. In addition, hyphal death occurred at specific regions and times during colony development. Analysis of DNA degradation carried out by gel electrophoresis and studies on the presence of dying hyphae within the mycelium carried out by electron microscopy revealed two rounds of hyphal death: in the substrate mycelium during emergence of the aerial hyphae, and in the aerial mycelium during formation of the spores. This suggests that hyphal death in S. antibioticus is somehow included in the developmental program of the organism.
Keywords: Streptomyces antibioticus, programmed cell death, colony development, hyphal death, multicellular development
Cell death has long been known to be a fundamental feature of animal development, but only very recently has it become a fashionable subject of general biological interest. This is because programmed cell death (PCD,1 an active physiological process of cell deletion) has been recognized as playing a relevant role in the turnover of self-renewing tissues, morphogenesis, embryonic development (Ellis et al., 1991; Cohen et al., 1992; Wyllie, 1992; Vaux et al., 1994; Sanders and Wride, 1995; Raff, 1996; Jacobson et al., 1997), and more recently in neurodegenerative diseases and cancer progression (Williams, 1991; Sen and D'Incalci, 1992; Ameisen, 1994; Barr and Tomei, 1994; Kerr et al., 1994; Cohen et al., 1996; Kusiak et al., 1996). Most studies on cell death have been carried out in animal cells. By contrast, little or no attention has been given to the process of cell death in prokaryotes, although they have been widely used as models for the study of many other basic cellular processes.
Members of the genus Streptomyces (mycelial gram-positive soil bacteria) possess two characteristics which make them attractive microorganisms for the study of cell death at the prokaryote level: first, unlike Escherichia coli (the most studied and best known single-cell prokaryote), which continuously divides by binary fission into two functionally and structurally identical daughter cells (which, therefore, are potentially immortal), the streptomycetes grow through the formation of long, multinucleoid hyphae that, with time, undergo senescence and die; second, they execute a complex developmental cycle that represents one of the probably several evolutionary attempts at multicellularity (Champness, 1988; Shapiro, 1988; Chater, 1989a; Chater and Losick, 1997). In fact, colonies of streptomycetes are now viewed as multicellular organisms containing morphologically and biochemically differentiated populations of hyphae organized into separate somatic and germ cell lineages (Chater, 1993; Champness and Chater, 1994; Bruton et al., 1995), the development of which is governed by an intricate system of intercellular communication (Horinouchi and Beppu, 1992; Kaiser and Losick, 1993; Willey et al., 1993). The colony growth cycle of the streptomycetes is initiated when a spore germinates, giving rise to one or more long multinucleoid filaments. These filaments elongate and branch repeatedly, originating a vegetative mycelium (substrate mycelium) that develops over, and into, the culture medium. As the colony ages, specialized branches emerge from the substrate mycelium and grow away from the surface of the colony, originating the reproductive aerial mycelium. Then, the aerial hyphae septate into chains of uninucleoid compartments, which finally metamorphose into thick-walled spores (McVittie, 1974; Hardisson and Manzanal, 1976; Hodgson, 1992; Chater, 1998). Along this cycle, large numbers of hyphae (including the original substrate hyphae and any portion of the aerial mycelium which does not differentiate into spores) degenerate and die. Since this phenomenon was reported for the first time (Wildermuth, 1970), autolysis has been the term most frequently used in the literature to describe the process of hyphal death which accompanies colony development in streptomycetes (Kalakoutskii and Agre, 1976; Ensign, 1978; Locci and Sharples, 1984; Méndez et al., 1985; Braña et al., 1986; Chater, 1989a,b; Hodgson, 1992; Kelemen et al., 1995).
We present studies demonstrating that autolysis (a form of cell death that follows degradation of the bacterial cell wall by the uncontrolled, lytic action of murein hydrolases) is not the predominant mechanism for hyphal death in Streptomyces antibioticus. Instead, the hyphae undergo progressive disorganization of internal cell constituents (including extensive genome digestion), preceding loss of plasma membrane integrity. Cell wall degradation, if it occurs, is just a very late event. In addition, analysis of DNA degradation carried out by gel electrophoresis revealed two rounds of hyphal death during colony development: during emergence of the aerial hyphae and during formation of the spores. We conclude that during the life cycle of S. antibioticus the mycelium does not undergo a random process of autolysis, but a highly regulated process of PCD.
Materials and Methods
Strains and Media
S. antibioticus ATCC 11891 was used in this work. The microorganism was grown as lawns on glucose/asparagine/yeast extract (GAE) medium (containing 1% glucose, 0.1% asparagine, 0.05% yeast extract, 0.05 K2HPO4, 0.05% MgSO4 · 7H2O, 0.001% FeSO4 · 7H2O, 100 mM MOPS buffer [pH 7.0], and 2% agar). Plates were inoculated by spreading confluently 0.2 ml of a spore suspension (108 spores/ml; Hardisson et al., 1978), followed by incubation at 28°C. The developmental stage of the lawns was monitored by visually observing the changes in coloration of the surface of the cultures (waxy-yellow appearance for cultures with only substrate mycelium, powder-white appearance during aerial mycelium development, and powder-gray appearance during spore formation; Wildermuth, 1970). For biochemical studies the microorganism was cultured on sterile cellophane membranes which had been overlaid previously onto the solidified culture medium. This cultivation procedure facilitates the harvesting and handling of large mycelial masses while allowing the organism to express all stages of its growth cycle (Méndez et al., 1985; Braña et al., 1986; Miguélez et al., 1994).
Microscopy
At different times of incubation, samples of the cultures (exhibiting uniformity of development) were obtained and processed for microscopy as follows. Blocks of agar containing mycelium were cut out from the culture medium and dissected into small pieces (∼3–4 mm in width and 8–10 mm in length). The pieces were fixed overnight at room temperature in 1% wt/ vol osmium tetroxide in 0.1 M veronal acetate buffer (pH 6.0), and postfixed with 0.5% wt/vol uranyl acetate in 0.1 M veronal acetate buffer (pH 6.0) for 2 h. After this, pieces were dehydrated through graded acetone solutions over a 2-h period at room temperature, embedded in Epon 812 resin and polymerized at 60°C for 36 h. Before polymerization, pieces were properly positioned to facilitate vertical sectioning of the whole mycelium. For electron microscopic observations, ultrathin sections of silver-gray interference color (thickness, 60–90 nm) were obtained with an LKB Ultramicrotome III equipped with a diamond knife and mounted on Formvar-coated copper grids. To improve contrast, ultrathin sections were poststained in the dark for 10 min on uranyl acetate drops (2% wt/ vol aqueous uranyl acetate), followed by counterstaining with lead citrate (pH 12, 1.5 min). Ultrathin sections were examined in a Philips EM300 electron microscope at an operating voltage of 60 kV, and photographed with Scientia electron microscopy film (AGFA; developed for 4 min in Kodak D19). For light microscopic observations, thin sections (∼1 μm thick) were mounted on slides, stained in Toluidine blue (0.1% wt/vol Toluidine blue in 0.1% aqueous sodium borate) for 1 min, and examined in a Nikon light microscope.
For high-resolution scanning electron microscopy, agar blocks containing mycelium were fixed with osmium tetroxide (1% wt/vol in 0.1 M veronal acetate buffer, pH 6.0) for 2 h, passed through increasing concentrations of acetone, and dried to critical point with a Balzers CPD-030 apparatus. The dried samples were mounted on aluminum stubs, coated with gold by vacuum evaporation (SCD-004 sputter coat; Balzers) and examined with a Jeol JSM-6100 scanning electron microscope.
Image Analysis
High-contrast photographic negatives were digitized using a Hewlett Packard 4C slide scanner. The digitized images were imported into Scion Image (Beta 2 version for Windows 95; Scion Corp.) for digital analysis. Plots of pixel intensity from zones of interest were obtained by positioning rectangular selections (452 pixels width, 600 pixels height) over such zones, or by tracing linear selections (5 pixels width) perpendicularly to the wall, passing from resin to the middle of the cells. Plots were generated by measuring pixel intensities (intensity range of each pixel 256 gray levels [8 bits]) along the linear selections (Profile Plot function) or in the rectangular selections (Surface Plot function). By using the LUT tool, pseudocolor images were generated in which the different components of the cell displayed an arbitrarily different color. All plots were obtained from hyphae considered to be cut diametrically (i.e., showing a clear-cut cell wall profile consisting of two electron-dense zones separated by a less electron-dense zone). Final images were composed and prepared for printing by using Adobe Photoshop software (4.0 version; Adobe Systems). All the images were printed using a Epson SC-800 printer.
Analytical Procedures
At various times of incubation, the mycelium from three plates (8.5 cm diam) was gently scraped from the cellophane with a plastic spatula, pooled, and suspended in 10 mM potassium phosphate buffer (pH 7.0). The suspensions were then sonicated at full power for 2 min on a MSE Soniprep 150 sonicator. Samples of disrupted mycelial suspensions (1.5 ml, by triplicate) were taken, 1.5 ml of 0.5 N perchloric acid was added, and the samples were maintained for 30 min at 0°C in an ice bath. After centrifugation, the pellets were extracted three times with 0.5 N perchloric acid at 70°C. Supernatants were pooled and assayed for RNA by the orcinol method (Schneider, 1957). Pellets were dissolved in 1.0 N NaOH and assayed for protein (Lowry et al., 1951). For dry cell weight determinations, samples (1.5 ml by triplicate) of disrupted mycelial suspensions were collected in preweighted glass vials and dried at 100°C to constant weight.
DNA Extraction and Agarose Gel Electrophoresis
Standard methods were used for DNA extraction (Hopwood et al., 1985). In brief, samples of mycelium collected from cellophane membranes were suspended in a lysing solution consisting of 5 mM Tris-HCl (pH 8.0), 25% sucrose, and lysozyme (1 mg/ml). After 1 h at 37°C, the solution was successively treated with Pronase (1 mg/ml) and 1% SDS. After phenol/chloroform extraction, the DNA solution was incubated for 1 h at 37°C in the presence of RNase (40 μg/ml) and then precipitated by adding an equal volume of 100% ethanol. The precipitated DNA was washed with 70% ethanol and suspended in TE buffer. The final DNA solution was checked spectrophotometrically for purity and concentration. Equal amounts of DNA from each sample were electrophoresed in 1% agarose gels in TBE buffer at 60–80 mA for 1–2 h. The gels were stained with ethidium bromide and photographed under UV light.
Results
Morphological Analysis of Hyphal Death
The morphological characteristics of hyphal death in S. antibioticus were analyzed by transmission and scanning electron microscopy. With this purpose, S. antibioticus was cultured at 28°C on GAE medium. Between 36 and 96 h of incubation, samples of the cultures were harvested and processed for electron microscopy as described in Materials and Methods. As observed previously (see below), during this period of incubation the cultures were particularly rich in hyphae undergoing cellular degeneration. These hyphae could be distinguished by the presence of large electron-transparent areas in the cytoplasm and by the aberrant shapes they displayed (see Figs. 1 and 3).
Figure 1.
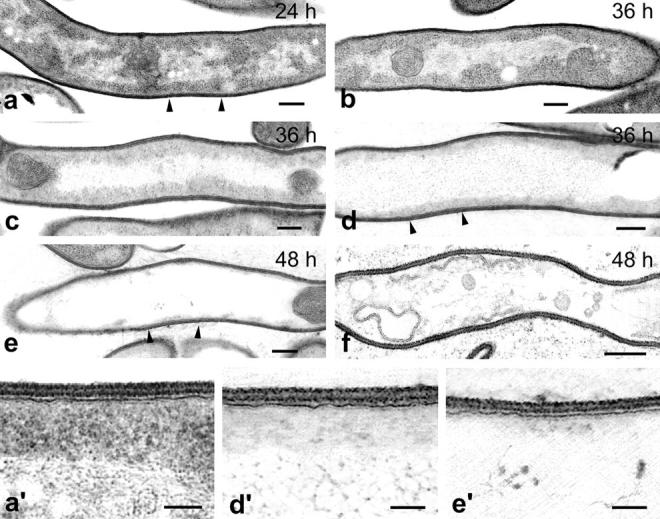
Ultrastructural features of normal and degenerating hyphae of S. antibioticus. Normal hyphae contain dark cytoplasm and various nucleoids of different sizes and shapes distributed throughout it (a). Degenerating hyphae undergo a progressive decrease in electron density in both cytoplasm and nucleoids, which condense into a long filament running along the major axis of the hypha (b and c). This is followed by dissolution of the cytoplasm and disorganization of the nucleoid, which expands dramatically into a loose network of DNA that fills most of the intracellular space of the hypha (d and e). Finally, dead hyphae appear empty of cellular contents but retain an apparently intact cell wall (f). (a′, d′, and e′) Higher magnification views of the zones marked by arrows in a, d, and e, respectively. Note that the fibrillar substructure of the nucleoid appears less electron dense and more disorganized in d′, and that this fibrillar substructure has almost completely disappeared in e′. Also note that the cell wall and the plasma membrane appear intact at all stages of cellular degeneration. Numbers indicate the age of the cultures. Bars: (a–f) 0.2 μm; (a′, d′, and e′) 0.1 μm.
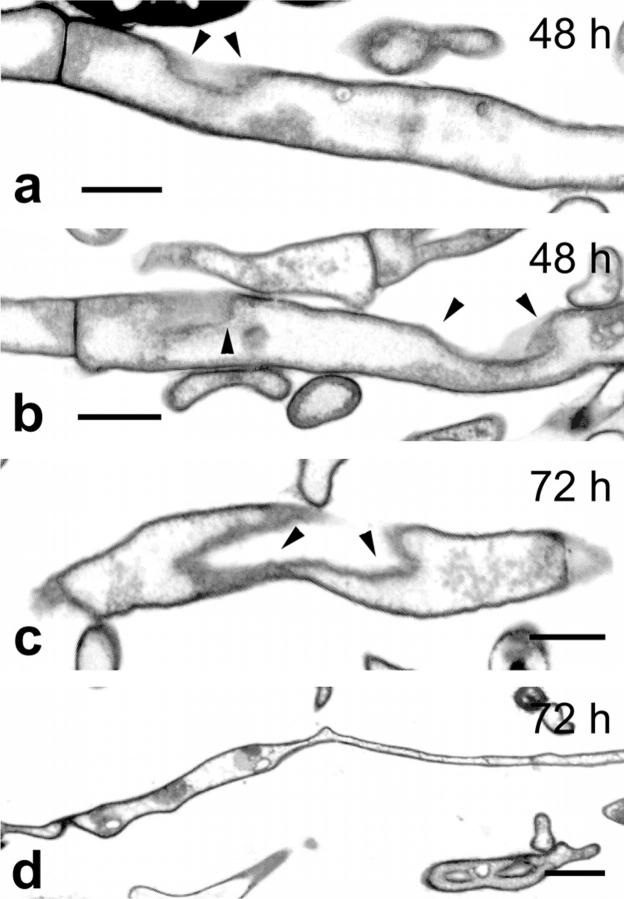
Figure 3.
Transmission electron micrographs of degenerating hyphae of S. antibioticus showing changes in hyphal morphology. (a and b) Longitudinal sections of hyphae showing zones where the wall appears collapsed (arrowheads). (c) Longitudinal section of a hypha showing a cross-sectioned, deeply collapsed zone of the cell wall (arrowheads). (d) Collapsed hypha displaying aberrant shapes. Numbers indicate the age of the cultures. Bars: (a–c) 0.5 μm; (d) 1 μm.
From inspection of numerous hyphae at different stages of cellular degeneration, a logical progression of cytological changes for hyphal death could be deduced (Fig. 1). The first recognizable cytological change characteristic of hyphal death was a disorganization of the nucleoid which loses its condensed, axially disposed form and expands into a large, open network of DNA. During this process, the fibrillar substructure of the nucleoid becomes progressively less electron-dense and more disorganized. As the degree of disorganization of the nucleoid increases, the cytoplasm becomes less electron-dense. Such a clearing of the cytoplasm probably results from degradation of ribosomes and other macromolecular constituents, as is suggested by the marked reduction in the RNA and protein contents which the colony undergoes during this period of development (see Fig. 9). Finally, what remains of the nucleoid is a large electron-translucent area that shows no signs of fibrillar substructure and fills most of the intracellular space of the hypha. Only small electron-dense aggregates of unknown chemical nature were present in that region of the cytoplasm.
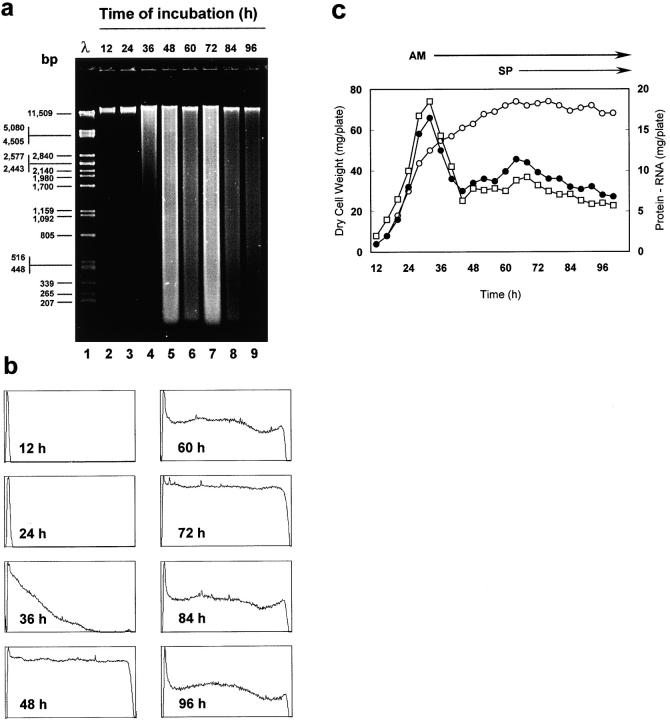
Figure 9.
(a and b) DNA degradation during colony development in S. antibioticus. (a) Agarose gel electrophoresis of S. antibioticus DNA from different time points during colony development. Samples of mycelium were harvested at the times indicated above each lane. Total DNA was then isolated from the samples and subjected to electrophoresis in a 1% agarose gel. All the lanes were loaded with equal amounts of DNA. Lane 1 contains PstI-digested λ phage DNA as a molecular size marker. (b) Pixel intensity traces corresponding to lanes 2–9. A digitized image of the gel was transferred to Scion Image, an image analysis program. Lanes were analyzed by tracing linear selections (5 pixels, width) and measuring pixel intensities along the trajectory of each lane with the help of the Profile Plot function of the software. Y-axis: pixel intensity (0–250, arbitrary values). X-axis: lane trajectory. (c) Changes in dry cell weight and macromolecular content during colony development in S. antibioticus. The microorganism was cultured on cellophane-covered GAE medium. At different times of incubation, the mycelium was removed from the cellophane and the dry cell weight (open circles) and the total RNA (filled circles) and protein contents (open boxes) were determined. The plotted values are the mean of three determinations. Arrows at the top of the figure indicate the developmental stage of the cultures. AM, period of aerial mycelium development; SP, period of spore formation.
While all this is occurring, the plasma membrane shows no signs of ultrastructural disorganization and remains as a continuous, well-stained structure close to the inner face of the wall (Fig. 1, d′ and e′). Only after the cytoplasm has completely cleared and the fibrillar components of the nucleoid have almost disappeared does the plasma membrane retract from the wall and dissociate into a number of vesicles (Fig. 1 f). Moreover, an undamaged, triple-layered cell wall was persistently seen at all stages of cellular degeneration. All of these ultrastructural changes were particularly evident in the plots of pixel intensity obtained by digital analysis of the images. As Fig. 2 shows, all the plots generated from hyphae undergoing cellular degeneration revealed progressive disorganization of the nucleoid and loss of cytoplasmic electron density with maintenance of plasma membrane and cell wall integrity.
Figure 2.
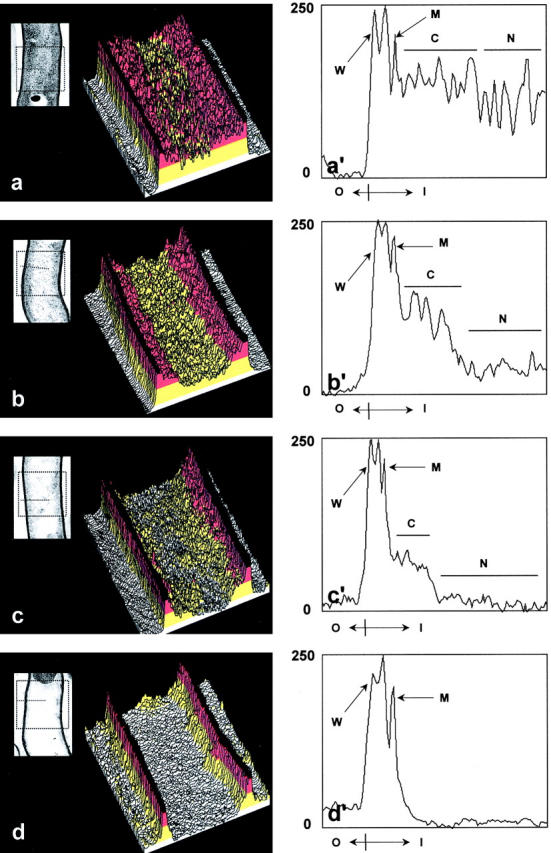
Pixel intensity scan analysis of normal and degenerating hyphae of S. antibioticus. Electron micrographs of a normal hypha (a, inset) and of degenerating hyphae at different representative stages of nucleoid disorganization (b–d, insets) were digitized and analyzed by measuring pixel intensities in zones of interest (see Materials and Methods). By using the Scion Image software, rectangular and linear selections were traced on the images and positioned such that the zones of interest could be scanned (a–d, insets, dashed lines). Pixel intensity values obtained from rectangular selections are plotted as oblique-viewed, three-dimensional representations (a–d). For highlighting particular regions of the cell, three-dimensional representations were pseudocolored such that the nucleoid appears yellow, the cytoplasm red, the cell wall black, and the resin that surrounds the hypha white. Pixel intensity values obtained from linear selections are plotted as two-dimensional representations (a′–d′). Y-axis: pixel intensity (inverted). X-axis: O, outside cell; I, inside cell. Traces corresponding to the cell wall (which gives two peaks of density) and the plasma membrane are marked by arrows (W and M, respectively), and those corresponding to the cytoplasm and the nucleoid by lines (C and N, respectively).
In the upper zone of the substrate mycelium and throughout the aerial mycelium, dying hyphae undergo a series of drastic changes in their morphology. As Fig. 3 shows, accompanying degeneration of the nucleoid, the hyphae collapse and undergo distortion of the hyphal shape (Fig. 3, a–c). The end result is an aberrant hyphal structure, empty of cellular contents but still retaining a continuous, apparently intact cell wall (Fig. 3 d). These morphological changes were also examined by using scanning electron microscopy. As Fig. 4 a shows, normal hyphae typically appear as long, cylindrical cells the surfaces of which are totally smooth. In degenerating hyphae, however, the wall intrudes and forms small depressions at many sites along the hypha (Fig. 4, b and c). Then, the wall collapses and the hyphae progressively shrink, acquiring an irregular, tubular-deflated appearance (Fig. 4, d–g). These aberrant, but otherwise characteristic, hyphal shapes probably arise as a consequence of the loss of water and cellular contents in degenerating hyphae or, more likely, of their displacement towards the growing parts of the colony (see below).
Figure 4.
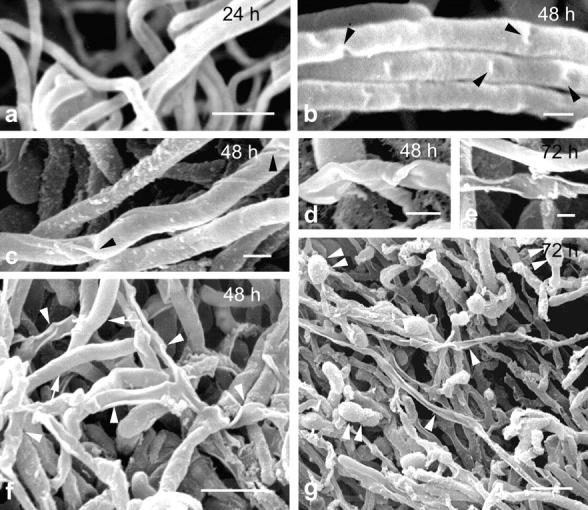
Changes in surface morphology accompanying hyphal death. Scanning electron micrographs of normal (a) and degenerating hyphae (b–g) of S. antibioticus. Degenerating hyphae initially display a number of discrete depressions along the wall (b and c, arrowheads). As the process continues, the hyphae shrink and collapse (d). Finally, dead hyphae appear as irregular, tubular-deflated structures (e). (f and g) Lower-magnification pictures showing normal hyphae (f, arrows), dead hyphae (f and g, arrowheads), and spores (g, arrowhead pairs). Numbers indicate the age of the cultures. Bars: (a, f, and g) 2 μm; (b–e) 0.5 μm.
Time Course of Hyphal Death during Colony Development
A series of light and electron microscopic studies was performed to examine the time course of hyphal death during colony development in S. antibioticus. Accordingly, the microorganism was grown in GAE medium. At different times during growth, samples of the cultures were harvested, fixed, and processed for microscopy (Materials and Methods). Large semithin sections were used to examine the general organization of the colony in the light microscope. Ultrathin sections obtained from areas of interest were used for the ultrastructural characterization of the process in the electron microscope. The results obtained are shown in Figs. 5–8.
Figure 5.

(Left) Vertical sections of colonies of S. antibioticus grown for 12 h (a) and 24 h (b and c). (a and b) Semithin sections showing the general organization of the colonies. Colonies are formed by a thin, compact layer of substrate hyphae that develop above the culture medium and by a large, hyphal network that develops within it. (c) Ultrathin section throughout the substrate mycelium. The hyphae are all similar in ultrastructural appearance (a granular, electron-dense cytoplasm with nucleoids of different size and shape distributed throughout it) and do not display symptoms of cellular disorganization. (c′) Enlarged detail of the hypha pointed to in c (arrow). (a–c) Arrowheads point to the surface of the culture medium. (Right) Vertical sections of a colony of S. antibioticus grown for 36 h. (a) Semithin section showing formation of aerial hyphae in the upper region of the colony. (b) Ultrathin section extending from the upper region of the colony to the surface of the culture medium. Hyphae with evident symptoms of cellular degeneration are visible near the boundary with the culture medium. (b′) Enlarged detail of the hypha pointed to in b (arrow) showing the thin sheath (arrowhead) that surrounds it. Arrowheads in b indicate the agar surface. Bars in left panels: (a and b) 20 μm; (c) 5 μm; (c′) 0.2 μm. Bars in right panels: (a) 15 μm; (b) 5 μm; (b′) 0.1 μm.
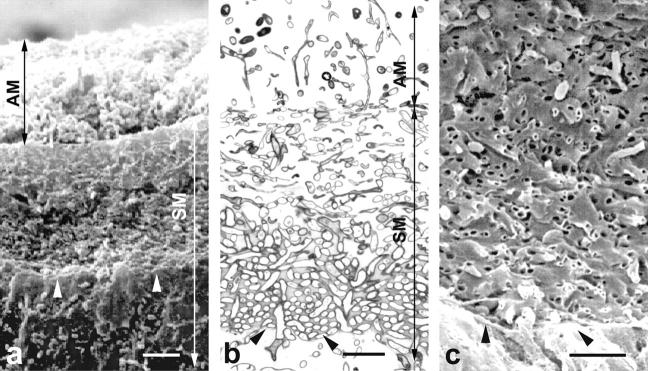
Figure 8.
Scanning (a and c) and transmission (b) electron micrographs of a 5-d-old colony of S. antibioticus. (a) A general view of the colony (vertical cut) showing the aerial (AM) and the substrate mycelium (SM). (b) Ultrathin section extending from the basal zone of the aerial mycelium to the surface of the culture medium. This region consists of a dense mass of empty dead hyphae (only the walls remain) surrounded by amorphous, moderately electron-dense material. Note that the dead hyphae appear as empty, tubular structures near the boundary with the culture medium, whereas in the aerial mycelium and in the upper zone of the substrate mycelium they appear collapsed. (c) Detail of the substrate mycelium near the surface of the culture medium viewed by scanning electron microscopy. The arrowheads mark the position of the culture medium. Bars: (a) 10 μm; (b and c) 5 μm.
During the first 24 h of incubation (Fig. 5, left), the substrate mycelium grew on the surface of the culture medium by forming a thin compact layer of hyphae. Within it, however, the hyphae penetrated to depths of >60 μm. Throughout these colonies, both above and within the culture medium, the hyphae were indistinguishable ultrastructurally: they appeared very electron-dense and showed no apparent symptoms of cellular degeneration (Fig. 5, c and c′). The general organization of the colonies was basically similar after 36 h of incubation (Fig. 5, right). However, the presence of hyphae surrounded by a thin sheath growing upwards from the substrate mycelium revealed that the aerial mycelium begins form at this stage of development. Ultrathin sections corresponding to these colonies revealed few hyphae with symptoms of degeneration, and these appeared at or near the boundary with the culture medium (Fig. 5 b).
After 48 h of incubation, the colonies contained a well-developed aerial mycelium (Fig. 6, left). In semithin sections, the aerial mycelium appeared as a loose network of hyphae that develop upwards into the air (Fig. 6 a). Ultrathin sections through representative zones of these colonies, illustrating the vertical distribution of hyphae undergoing cell degeneration, are shown in Fig. 6, b–d. No hyphae with symptoms of cellular degeneration were encountered in the aerial mycelium at this stage of development, where a majority of the hyphae showed a dense, heavily stained cytoplasm (Fig. 6 b). Below this zone and near the boundary with the culture medium (not only above, but also within it), the substrate mycelium appeared as an intricate network of hyphae in different stages of cellular degeneration, ranging from hyphae with electron-transparent areas in their cytoplasm to hyphae in which the cytoplasmic components had completely disappeared (Fig. 6, c and d). At the bottom of the colony (∼130 μm below the surface of the culture medium), a minority of the hyphae displayed a quite different morphological form of cell death, which was identifiable in 5–10% of the hyphae present in that region (Fig. 6, right). These hyphae did not collapse nor did they undergo progressive disorganization of the nucleoid and cytoplasm with maintenance of cell wall integrity. Instead, there was an early rupture of the wall and plasma membrane followed by rapid release of cellular contents into the surrounding medium, as suggested by the almost total absence of hyphae exhibiting stages of cellular degeneration later than those shown in Fig. 6, b and c. Only very few hyphae showing the initial stages of cell wall degradation could be examined and all these hyphae showed lightly stained cytoplasm with nucleoids of various sizes and shapes irregularly distributed through it.
Figure 6.
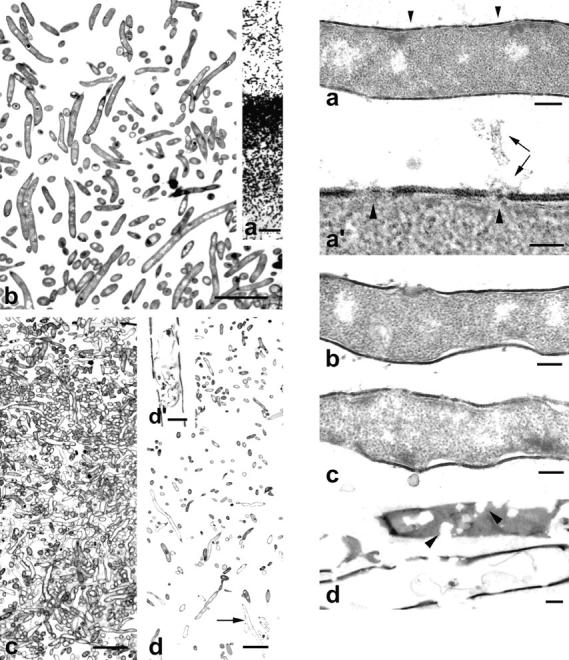
(Left) Vertical sections of a colony of S. antibioticus grown for 48 h. (a) Semithin section of the colony showing a well-developed aerial mycelium. b–d are ultrathin sections through the following distinct regions of the colony: (b) aerial mycelium; (c) substrate mycelium above the surface of the culture medium; (d) substrate mycelium below the surface of the culture medium. Note the presence of dead hyphae throughout the substrate mycelium and its absence in the aerial mycelium. (d′) Enlarged detail of the hypha pointed to in d (arrow). Note the rupture of the cell wall. (Right) Ultrastructural features of degenerating hyphae at the bottom of the colony. (a) Early stage of cellular degeneration. Alterations in both cell wall and plasma membrane are evident (see also a′). However, there is no evidence for a process of nucleoid disorganization. (a′) Enlarged detail of the zone marked by arrowheads showing small holes in the wall, rupture of the plasma membrane (arrowheads), and leakage of cytoplasmic contents (arrows). (b and c) Intermediate stages of cellular degeneration. Note that the cell wall appears severely damaged and that the plasma membrane had detached from the wall. (d) Final stage of cellular degeneration. Arrowheads point to zones of wall rupture (sagittal section). All these stages of cellular degeneration appeared in cultures after 48 h of incubation. Bars in left panels: (a) 15 μm; (b–d) 5 μm; (d′) 0.5 μm. Bars in right panels: (a–d) 0.2 μm; (a′) 0.1 μm.
After 72 h of incubation, the colonies displayed abundant sporulation in the aerial mycelium (Fig. 7). Ultrathin sections of these colonies did not reveal changes in the substrate mycelium, which appeared composed almost entirely of empty dead hyphae. In the aerial mycelium, however, a fraction of the hyphal population metamorphoses into chains of spores, while the remainder (nonsporulating hyphae) degenerates and dies. Dead hyphae appeared throughout the aerial mycelium, but they were maximally abundant towards the boundary with the substrate mycelium (data not shown).
Figure 7.
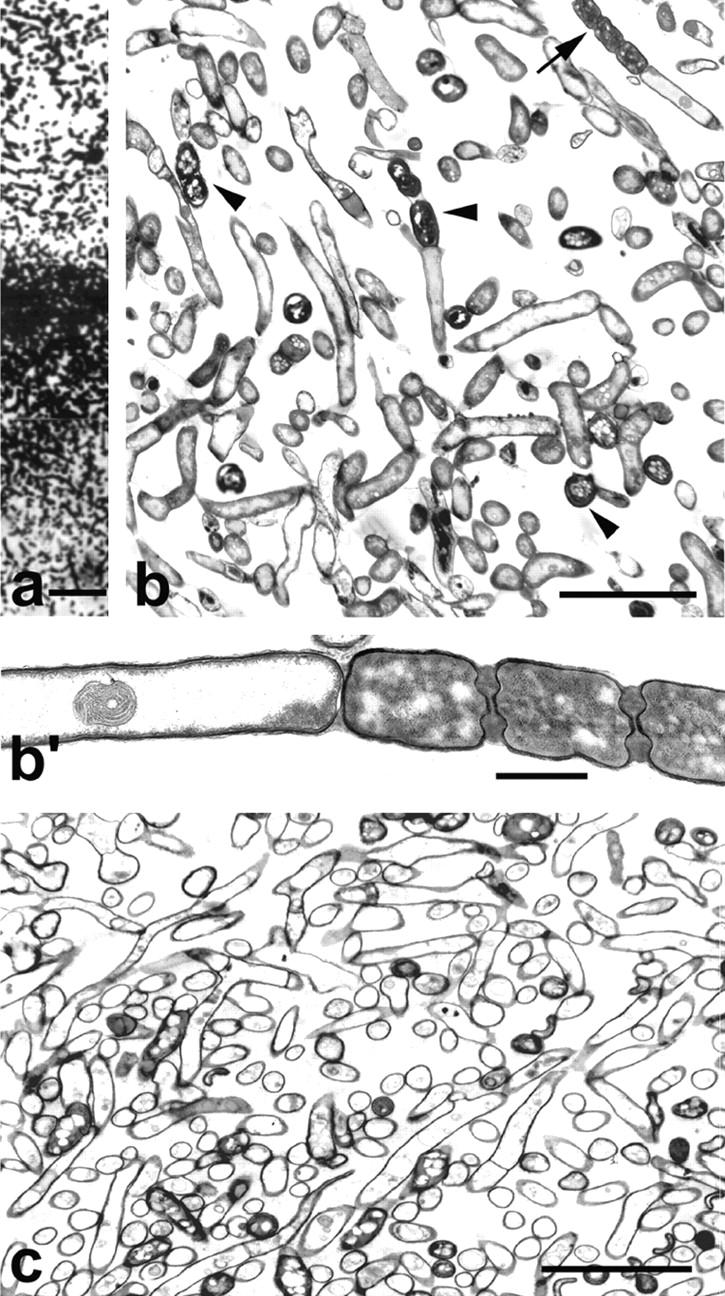
Sections of a colony of S. antibioticus grown for 72 h. (a) Vertical semithin section through the aerial and the substrate mycelium. (b) Vertical ultrathin section through a part of the aerial mycelium. Note the presence of spores (arrowheads) and hyphae at different stages of cellular degeneration. (b′) Higher magnification of the sporulating hypha pointed to in b (arrow) showing cytoplasmic degeneration in the nonsporulating segment. (c) Vertical ultrathin section through a part of the substrate mycelium. Bars: (a) 15 μm; (b and c) 5 μm; (b′) 1 μm.
The oldest colonies examined, 5 d old, consisted entirely of dead hyphae and mature spores (Fig. 8). All that remained in the older parts of these colonies was an intricate, pseudo-skeletal structure formed by the walls of the dead hyphae. Many such hyphae displayed aberrant morphologies, but near to the boundary with the culture medium a majority of dead hyphae retained their original tubular shape. This is probably because in such a zone of the colony the mycelium was less affected by the emergence of the aerial hyphae. Since the aerial mycelium completely depends upon translocation of water and nutrients from the substrate mycelium (a part of which is provided by dying hyphae themselves; Méndez et al., 1985), its development will draw fluids from the nearest subjacent hyphae which, as a consequence of this, will undergo collapse and distortion of shape.
Hyphal Death and Associated Biochemical Changes
S. antibioticus was cultured on sterile cellophane membranes overlaid on solid GAE medium. At different times during growth, samples of mycelium were carefully removed from the cellophane membranes and used either to examine the state of DNA by agarose gel electrophoresis or to estimate changes in the macromolecular content of the hyphae.
Changes in DNA Integrity.
As Fig. 9 a shows, DNA extracted from samples collected during the first 24 h of incubation (lanes 2 and 3) did not shows signs of degradation. However, after 36 h of incubation and continuing through later time points (Fig. 9 a, lanes 4–9), DNA degradation was apparent as a smear beneath a band of high molecular weight DNA. As can be seen, DNA degradation coincided in time with the presence in the colony of hyphae undergoing nuclear degeneration (see Figs. 5–7). On the other hand, there were two time points at which DNA degradation reached a maximum (Fig. 9 b, 48 and 72 h of incubation). These corresponded to the same time points at which the colonies were found to form aerial hyphae and to differentiate into spores, respectively (see Figs. 6 and 7).
Changes in Dry Cell Weight and Macromolecular Content.
As Fig. 9 c shows, over the period 12–32 h after inoculation the cultures entered a phase of rapid growth during which all the growth parameters increased steadily. This was followed by a long period of slow biomass accumulation (which began with the emergence of the aerial mycelium and extended throughout its development), during which the total contents of RNA and protein in the mycelium varied significantly: there was a marked decrease during emergence of the aerial hyphae (between 32 and 44 h of incubation), followed by a slight increase between 44 and 64 h of incubation and by a moderate but persistent decrease during the period of spore formation.
Discussion
As in most, if not all, bacterial systems exhibiting multicellularity (Mendelson et al., 1997; Shapiro, 1997; Shimkets and Dworkin, 1997), colony development in streptomycetes is maintained by a tight balance between cell proliferation and cell death processes. Much of what we currently know about hyphal death in streptomycetes comes from an early study on colony development in Streptomyces coelicolor, carried out by Wildermuth almost 30 years ago (Wildermuth, 1970). In that work, it was reported that the mycelium undergoes extensive breakdown during the life cycle of the colony and that this process is accompanied by the presence of vacuole-like spaces in the cytoplasm (later identified as deposits of some fatty storage material; Olukoshi and Packter, 1994; Plaskitt and Chater, 1995), condensation of the nucleoid, and the rupture of the plasma membrane. However, this study was limited in two relevant aspects: these cytological alterations were not precisely placed within the time framework of the overall death process; and there was no information on early cytological changes and, therefore, no clear picture of the sequence of morphological events leading to hyphal death could be deduced from these observations. Since that time, hyphal death has been reported repeatedly to occur as a normal part of colony development in streptomycetes (Kalakoutskii and Agre, 1976; Ensign, 1978; Locci and Sharples, 1984; Chater, 1989a, 1993; Hodgson, 1992), but no attempts have been made to elucidate the mechanisms underlying this process. Moreover, despite decades of extensive morphological studies on colony development in streptomycetes, it is remarkable that we still know so little concerning the ultrastructural aspects of hyphal death.
Therefore, the main objectives of this study were to characterize the sequence of ultrastructural changes leading to hyphal death in S. antibioticus, to identify the zones of the colony where they occur, and to determine the developmental time at which they appear. Our electron microscopy study has revealed some previously unrecognized aspects of hyphal death in streptomycetes. We have discerned that nucleoid degradation is a relatively early event in the hyphal death process and that this degradation clearly precedes the rupture of the plasma membrane. Moreover, we also observed that nucleoid degradation is accompanied by progressive digestion of cytoplasmic contents and distortion of the hyphal shape, and that all this occurs with maintenance of cell wall integrity and without disturbing the general organization of the colony.
In addition, we have observed that the hyphae die in specific zones and at specific times during the colony life cycle. Studies on the presence and distribution of degenerating hyphae within the mycelium carried out by electron microscopy and analysis of DNA degradation carried out by gel electrophoresis revealed two rounds of hyphal death during colony development in S. antibioticus. The first round coincided with development of the aerial mycelium. It caused massive death in the substrate mycelium but had no apparent effect on the emergent aerial hyphae. The second round was not triggered until sporulation had been initiated and was more selective, since it only affected the basal, nonsporulating parts of the aerial hyphae. These observations are also interesting because they suggest that hyphal death is somehow included into the developmental program of the colony.
All together, the results obtained in this study demonstrate that the hyphae of S. antibioticus do not die via autolysis (see Introduction). Instead, they undergo an orderly process of internal cell dismantling (including extensive genome digestion), followed by shrinkage and distortion of the hyphal shape that resembles PCD in animal development. However, there are some aspects of the cytology and functions of hyphal death in S. antibioticus that distinguish it from PCD in higher organisms. First, dying hyphae do not display features such as reduction in nuclear size, condensation of chromatin, and internucleosomal cleavage of DNA (which gives a characteristic ladder when analyzed by electrophoresis), as is characteristically seen in eukaryotic cells undergoing PCD via apoptosis. This is probably a consequence of the quite different ultrastructural organization of the prokaryotic nucleoid, which lacks a nuclear membrane and contains an extensively folded DNA molecule not arranged into nucleosomes (Kellenberger and Arnold-Schulz-Gahmen, 1992; Robinow and Kellenberger, 1994). Second, dead hyphae do not completely disappear, but form part of the colony structure where they still could potentially perform two, nonmutually exclusive roles: they could provide a mechanical support for aerial hyphae to develop far from the surface of the culture medium, and they could serve as a conducting system for passage of water and solutes within the colony (a process of considerable importance for the aerial hyphae which develop in the absence of a surrounding liquid medium). Moreover, as dying hyphae provide nutrient support for development of the aerial mycelium in S. antibioticus (Méndez et al., 1985), maintenance of cell wall integrity would allow the cytoplasmic contents of these hyphae to be degraded and reused for growth without disturbing the general architecture of the colony.
In conclusion, our study has provided morphological evidence for the existence of a process of PCD in a prokaryotic organism. This adds new support to the hypothesis that the basic structure of the cell death processes has been preserved and extended throughout evolution (Vaux et al., 1994; Vaux and Strasser, 1996; Hochman, 1997). Establishing the mechanisms and signals that regulate such a process will be a major challenge for the future. In this respect, it is important to note that several bacterial plasmids carrying genes capable of killing their host have been reported recently to be responsible for the death of specific subpopulations of bacterial cells (Bugge and Gerdes, 1995; Naito et al., 1995; Yarmolinsky, 1995; Chaloupka and Vinter, 1996; Franch and Gerdes, 1996; Holcík and Lyer, 1997). Interestingly, two such plasmid-encoded killer systems seem to be present in Streptomyces spp. (Holcík and Lyer, 1997, and references therein). Finally, the colony growth cycle of the streptomycetes provides a useful prokaryotic system for the study of the mechanism and role of cell death in development. Such studies may provide insights into the role of cell death in more complex eukaryotic systems and may also provide insights into the evolution of this important phenomenon.
Acknowledgments
We thank A. Quintana for his help with the scanning microscopy and A.M. Nistal for helping with digital imaging.
This work was supported by grants BIO94-1025 and PM97-0197 from the CICYT, Spain.
Abbreviation used in this paper
- PCD
programmed cell death
References
- Ameisen JC. Programmed cell death (apoptosis) and cell survival regulation: relevance to AIDS and cancer. AIDS. 1994;8:1197–1213. doi: 10.1097/00002030-199409000-00001. [DOI] [PubMed] [Google Scholar]
- Barr PJ, Tomei LD. Apoptosis and its role in human disease. Biotechnology. 1994;12:487–493. doi: 10.1038/nbt0594-487. [DOI] [PubMed] [Google Scholar]
- Braña AF, Méndez C, Díaz LA, Manzanal MB, Hardisson C. Glycogen and trehalose accumulation during colony development in Streptomyces antibioticus. . J Gen Microbiol. 1986;132:1319–1326. doi: 10.1099/00221287-132-5-1319. [DOI] [PubMed] [Google Scholar]
- Bruton CJ, Plaskitt KA, Chater KF. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolorA3(2) Mol Microbiol. 1995;18:89–99. doi: 10.1111/j.1365-2958.1995.mmi_18010089.x. [DOI] [PubMed] [Google Scholar]
- Bugge R, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- Chaloupka J, Vinter V. Programmed cell death in bacteria. Folia Microbiol. 1996;41:451–464. [Google Scholar]
- Champness WC. New loci required for Streptomyces coelicolormorphological and physiological differentiation. J Bacteriol. 1988;170:1168–1174. doi: 10.1128/jb.170.3.1168-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champness, W.C., and K.F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp. In Regulation of Bacterial Development. P.J. Piggot, C.P. Morgan, and P. Youngman, editors. American Society for Microbiology, Washington, DC. 61–93.
- Chater, K.F. 1989a. Aspects of multicellular differentiation in Streptomyces coelicolor A3(2). In Genetics and Molecular Biology of Industrial Microorganisms. CL. Hershberger, S.W. Queener, and G. Hegeman, editors. American Society for Microbiology, Washington, DC. 99–107.
- Chater KF. Multilevel regulation of Streptomycesdifferentiation. Trends Genet. 1989b;5:372–376. doi: 10.1016/0168-9525(89)90172-8. [DOI] [PubMed] [Google Scholar]
- Chater KF. Genetics of differentiation in Streptomyces. . Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- Chater KF. Taking a genetic scalpel to the Streptomycescolony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- Chater, K.F., and R. Losick. 1997. The mycelial life-style of Streptomyces coelicolor A3(2) and its relatives. In Bacteria as Multicellular Organisms. J.H. Shapiro and M. Dworkin, editors. Oxford University Press, New York. 149–182.
- Cohen JJ, Duke RC, Fadok VA, Sellins K. Apoptosis and programmed cell death in immunity. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- Cohen, L.D., N.F. Theuvenet, and J.L. Noebels. 1996. Programmed cell death in neurologic disease. In Current Neurology. Vol. 16. S.H. Appel, editor. Mosby, St. Louis. 253–280.
- Ellis RE, Yuan JY, Horvitz HR. Mechanisms and functions of cell death. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- Ensign JC. Formation, properties and germination of actinomycete spores. Annu Rev Microbiol. 1978;32:185–219. doi: 10.1146/annurev.mi.32.100178.001153. [DOI] [PubMed] [Google Scholar]
- Franch T, Gerdes K. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol Microbiol. 1996;21:1049–1060. doi: 10.1046/j.1365-2958.1996.771431.x. [DOI] [PubMed] [Google Scholar]
- Hardisson C, Manzanal MB. Ultrastructural studies of sporulation in Streptomyces. . J Bacteriol. 1976;127:1443–1454. doi: 10.1128/jb.127.3.1443-1454.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisson C, Manzanal MB, Salas JA, Suarez JE. Fine structure and biochemistry of arthrospore germination in Streptomyces antibioticus. . J Gen Microbiol. 1978;105:203–214. doi: 10.1099/00221287-105-2-203. [DOI] [PubMed] [Google Scholar]
- Hochman A. Programmed cell death in prokaryotes. Crit Rev Microbiol. 1997;23:207–214. doi: 10.3109/10408419709115136. [DOI] [PubMed] [Google Scholar]
- Hodgson, D.A. 1992. Differentiation in Actinomycetes. In Prokaryotic Structure and Function: A New Perspective. Society for General Microbiology Symposium. S. Mohan, C. Dow, and J.A. Cole, editors. Cambridge University Press, Cambridge. Vol. 47. 407–440.
- Holcík M, Lyer VN. Conditionally lethal genes associated with bacterial plasmids. Microbiology. 1997;143:3403–3416. doi: 10.1099/00221287-143-11-3403. [DOI] [PubMed] [Google Scholar]
- Hopwood, D.A., M.J. Bibb, K.F. Chater, T. Kieser, C.J. Bruton, H.M. Kiesser, D.J. Lydiate, C.P. Smith, J.M. Ward, and H. Schrempf. 1985. Methods for isolating Streptomyces “total” DNA. In Genetic Manipulation of Streptomyces: A Laboratory Manual. John Innes Foundation, Norwich, United Kingdom. 71–80.
- Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–875. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- Kalakoutskii LV, Agre NS. Comparative aspects of development and differentiation in actinomycetes. Bacteriol Rev. 1976;40:469–524. doi: 10.1128/br.40.2.469-524.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen GH, Plaskitt KA, Lewins CG, Findlay KC, Buttner MJ. Deletion of DNA lying close to the glk A locus induces ectopic sporulation in Streptomyces coelicolorA3(2) Mol Microbiol. 1995;17:221–230. doi: 10.1111/j.1365-2958.1995.mmi_17020221.x. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Arnold-Schulz-Gahmen B. Chromatins of lower protein content: special features of their compaction and condensation. FEMS (Fed Eur Microbiol Soc) Lett. 1992;100:361–370. doi: 10.1111/j.1574-6968.1992.tb14064.x. [DOI] [PubMed] [Google Scholar]
- Kerr JFR, Winterford CM, Harmon BV. Apoptosis: its significance in cancer and cancer therapy. Cancer. 1994;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kusiak JW, Izzo JA, Zhao B. Neurodegeneration in Alzheimer disease. Is apoptosis involved? . Mol Chem Neuropathol. 1996;28:153–162. doi: 10.1007/BF02815217. [DOI] [PubMed] [Google Scholar]
- Locci, R., and G.P. Sharples. 1984. Morphology. In The Biology of the Actinomycetes. M. Goodfellow, M.M. Mordarski, and S.T. Williams, editors. Academic Press, London. 165–199.
- Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- McVittie A. Ultrastructural studies on sporulation in wild-type and white colony mutants of Streptomyces coelicolor. . J Gen Microbiol. 1974;81:291–302. doi: 10.1099/00221287-81-2-291. [DOI] [PubMed] [Google Scholar]
- Mendelson, N.H., B. Salhi, and C. Li. 1997. Physical and genetic consequences of multicellularity in Bacillus subtilis. In Bacteria as Multicellular Organisms. J.H. Shapiro and M. Dworkin, editors. Oxford University Press, New York. 326–349.
- Méndez C, Braña AF, Manzanal MB, Hardisson C. Role of substrate mycelium in colony development in Streptomyces. . Can J Microbiol. 1985;31:446–450. doi: 10.1139/m85-083. [DOI] [PubMed] [Google Scholar]
- Miguélez EM, García M, Hardisson C, Manzanal MB. Autoradiographic study of hyphal growth during aerial mycelium development in Streptomyces antibioticus. . J Bacteriol. 1994;176:2105–2107. doi: 10.1128/jb.176.7.2105-2107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- Olukoshi ER, Packter NM. Importance of stored triacylglycerols in Streptomyces: possible carbon source for antibiotics. Microbiology. 1994;140:931–943. doi: 10.1099/00221287-140-4-931. [DOI] [PubMed] [Google Scholar]
- Plaskitt KA, Chater KF. Influences of developmental genes on localized glycogen deposition in colonies of a mycelial prokaryote, Streptomyces coelicolorA3(2): a possible interface between metabolism and morphogenesis. Phil Trans R Soc Lond. 1995;347:105–121. [Google Scholar]
- Raff MC. Size control: the regulation of cell numbers in animal development. Cell. 1996;86:173–175. doi: 10.1016/s0092-8674(00)80087-2. [DOI] [PubMed] [Google Scholar]
- Robinow C, Kellenberger E. The bacterial nucleoid revisited. Microbiol Rev. 1994;58:211–232. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders EJ, Wride MA. Programmed cell death in development. Int Rev Cytol. 1995;163:105–173. doi: 10.1016/s0074-7696(08)62210-x. [DOI] [PubMed] [Google Scholar]
- Schneider WC. Determination of nucleic acids in tissues by pentose analysis. Methods Enzymol. 1957;3:680–684. [Google Scholar]
- Sen S, D'Incalci M. Apoptosis, biochemical events and relevance to cancer chemotherapy. FEBS Lett. 1992;307:122–127. doi: 10.1016/0014-5793(92)80914-3. [DOI] [PubMed] [Google Scholar]
- Shapiro JA. Bacteria as multicellular organisms. Sci Am. 1988;256:82–89. [Google Scholar]
- Shapiro, J.A. 1997. Multicellularity: the rule, not the exception: lessons from Escherichia coli colonies. In Bacteria as Multicellular Organisms. J.H. Shapiro and M. Dworkin, editors. Oxford University Press, New York. 14–49.
- Shimkets, L.J., and M. Dworkin. 1997. Mycobacterial multicellularity. In Bacteria as Multicellular Organisms. J.H. Shapiro and M. Dworkin, editors. Oxford University Press, New York. 220–244.
- Vaux DL, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Haecker G, Strasser A. An evolutionary perspective on apoptosis. Cell. 1994;76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- Wildermuth H. Development and organization of the aerial mycelium in Streptomyces coelicolor. . J Gen Microbiol. 1970;60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. . Genes Dev. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- Williams GT. Programmed cell death: apoptosis and oncogenes. Cell. 1991;65:1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]
- Wyllie AH. Apoptosis and the regulation of cell number in normal and neoplastic tissue: an overview. Cancer Res. 1992;48:6691–6696. doi: 10.1007/BF00048057. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]