Abstract
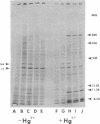
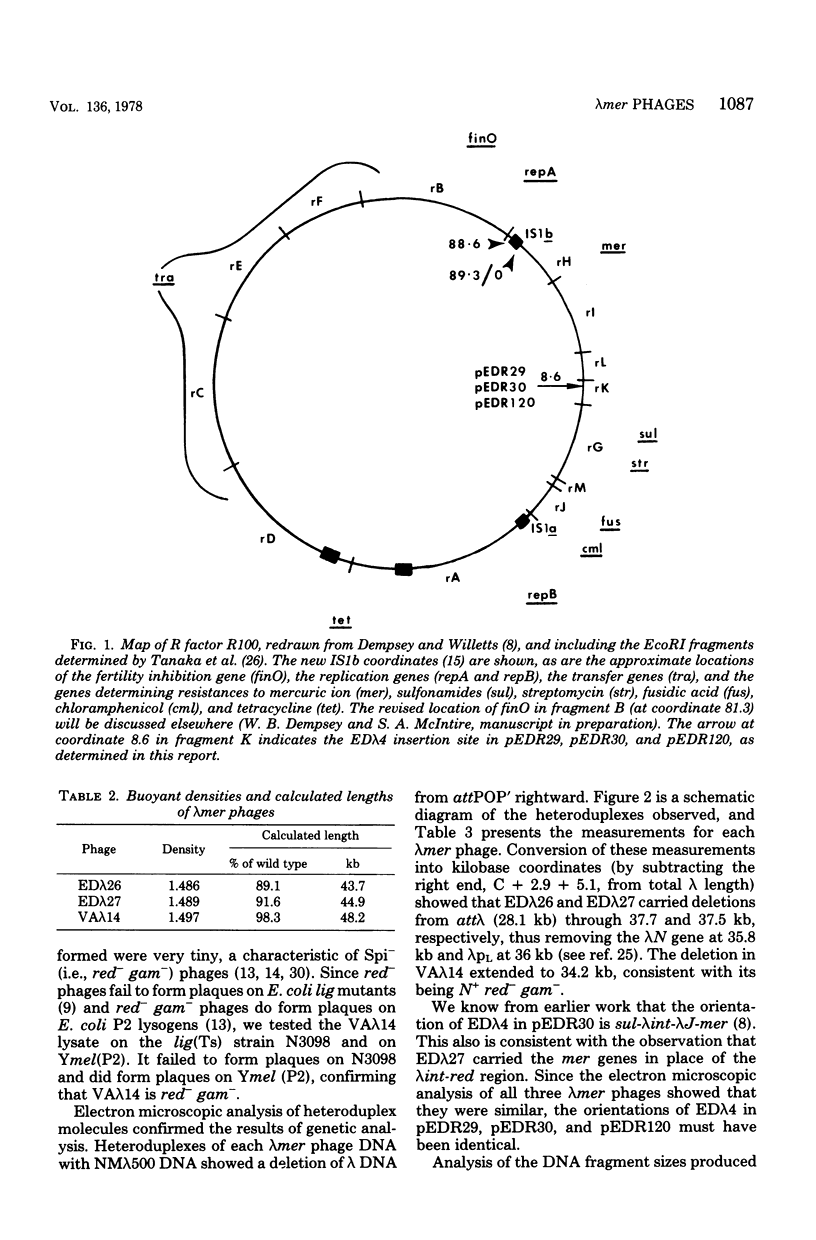
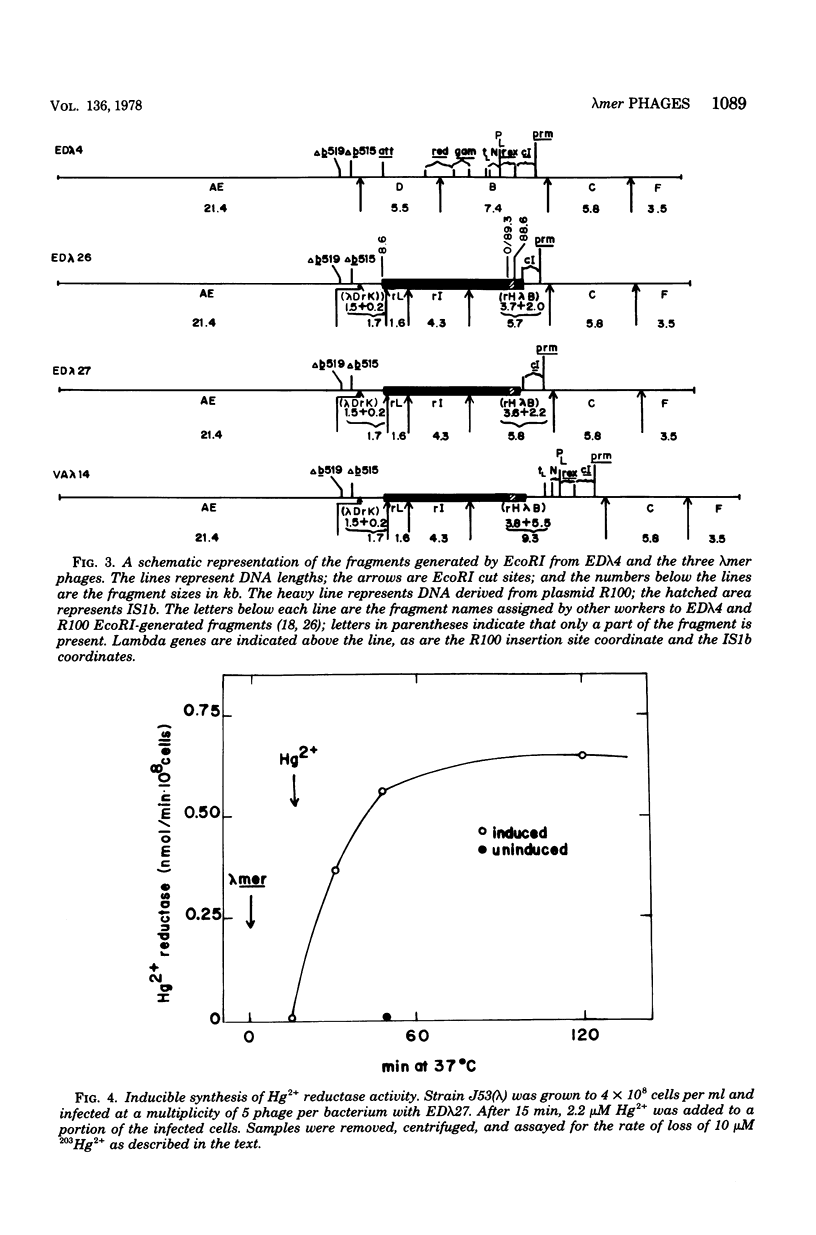
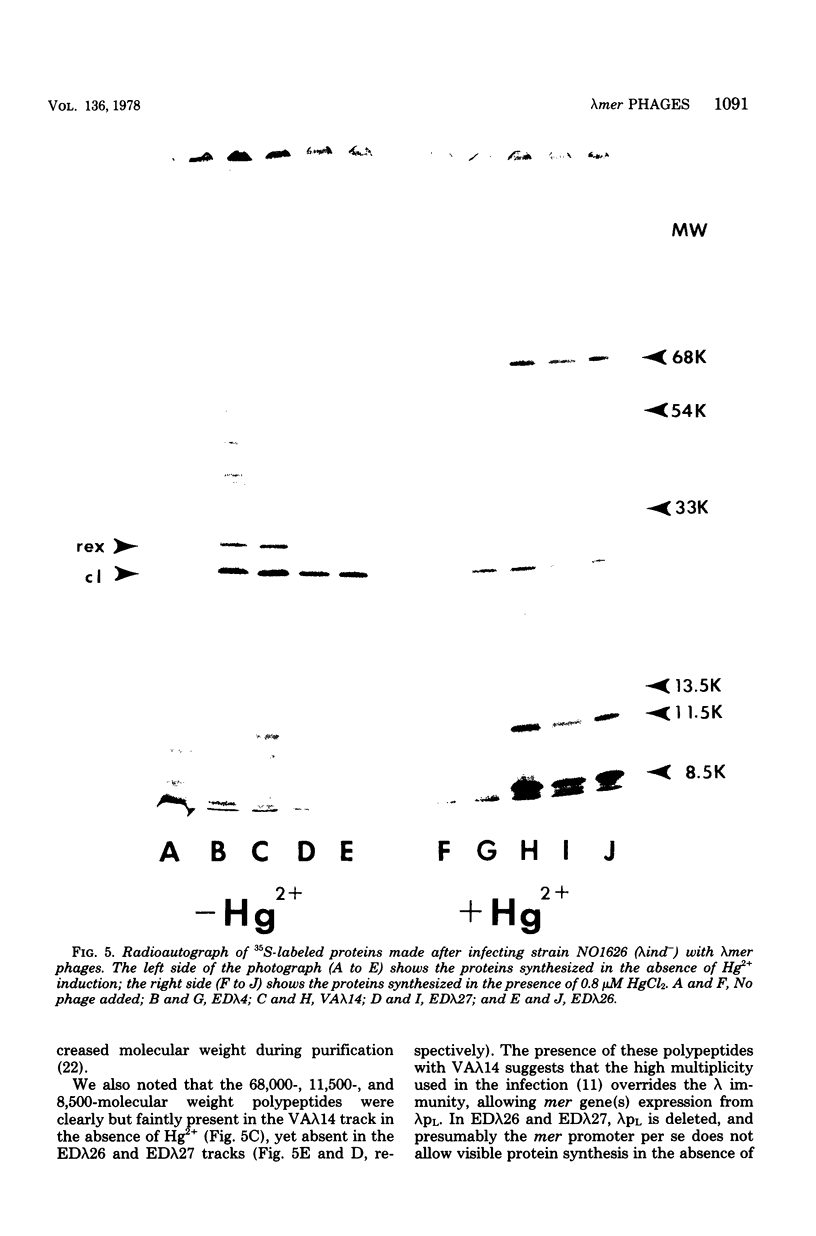
Three lambdamer (resistance to Hg2+ and mercurials) transducing phages were prepared from three independent cointegrate isolates of bacteriophage lambda and plasmid R100. DNA heteroduplex and restriction nuclease analyses of the lambdamer DNA showed that all three phages had resulted from lambda insertion at kilobase coordinate 8.6 of plasmid R100, followed by loss of different lengths of lambda DNA and replacement with different lengths of R100 DNA. Two of the lambdamer phages were defective, containing deletions from lambdaatt through the lambdaN gene and into the lambdarex gene; the third, VAlambda14, was an N+ Spi- plaque-forming phage. With VAlambda14, N-dependent transcription of R100 mer from the lambdapL promoter suggested that transcription of mer proceeded in the direction from IS1b toward the sulfonamide resistance determinant (i.e., from a plasmid promoter in restriction nuclease fragment EcoRI-H toward fragment EcoRI-I). Phage-directed protein synthesis in a UV-irradiated lambdaind- lysogen showed the Hg2+-inducible synthesis of three major polypeptides of molecular weights 68,000, 11,500, and 8,500 and three minor ones of molecular weights 54,000, 33,000, and 13,500. The largest of the major polypeptides is identified as the subunit of the mercuric reductase enzyme. The functions of the smaller polypeptides are not known. Hg2+ reductase enzyme assays confirmed the regulation of mer synthesis during phage infection.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Clark D. L., Weiss A. A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977 Oct;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey W. B., Willetts N. S. Plasmid co-integrates of prophage lambda and R factor R100. J Bacteriol. 1976 Apr;126(1):166–176. doi: 10.1128/jb.126.1.166-176.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. M., Hicks M. L., Gellert M. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol. 1973 Jul 15;77(4):531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- Hirsch H. J., Saedler H., Starlinger P. Insertion mutations in the control region of the galactose operon of E. coli. II. Physical characterization of the mutations. Mol Gen Genet. 1972;115(3):266–276. doi: 10.1007/BF00268890. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Lindahl L., Nomura M. Specialized transducing phages for ribosomal protein genes of Escherichia coli. Proc Natl Acad Sci U S A. 1975 Jan;72(1):6–10. doi: 10.1073/pnas.72.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Chandler M. Mapping of the drug resistance genes carried by the r-determinant of the R100.1 plasmid. Mol Gen Genet. 1977 Nov 29;157(1):17–23. doi: 10.1007/BF00268682. [DOI] [PubMed] [Google Scholar]
- Lindahl G., Sironi G., Bialy H., Calendar R. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc Natl Acad Sci U S A. 1970 Jul;66(3):587–594. doi: 10.1073/pnas.66.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Chattoraj D. K. The role of Chi mutations in the Spi- phenotype of phage lambda: lack of evidence for a gene delta. Mol Gen Genet. 1975 Dec 30;143(1):35–41. doi: 10.1007/BF00269418. [DOI] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Murray K. Manipulation of restriction targets in phage lambda to form receptor chromosomes for DNA fragments. Nature. 1974 Oct 11;251(5475):476–481. doi: 10.1038/251476a0. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IV. The F sequences in F14. J Mol Biol. 1974 Nov 15;89(4):565–584. doi: 10.1016/0022-2836(74)90036-9. [DOI] [PubMed] [Google Scholar]
- Reyes O., Gottesman M., Adhya S. Suppression of polarity of insertion mutations in the gal operon and N mutations in bacteriophage lambda. J Bacteriol. 1976 Jun;126(3):1108–1112. doi: 10.1128/jb.126.3.1108-1112.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. Physical mapping of the att-N region of coliphage lambda: apparent oversaturation of coding capacity in the gam-ral segment. Biochimie. 1974;56(11-12):1497–1503. doi: 10.1016/s0300-9084(75)80272-0. [DOI] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]