Abstract
The cortical scaffolding proteins EBP50 (ERM-binding phosphoprotein-50) and E3KARP (NHE3 kinase A regulatory protein) contain two PDZ (PSD-95/DlgA/ZO-1–like) domains followed by a COOH-terminal sequence that binds to active ERM family members. Using affinity chromatography, we identified polypeptides from placental microvilli that bind the PDZ domains of EBP50. Among these are 64- and/or 65-kD differentially phosphorylated polypeptides that bind preferentially to the first PDZ domain of EBP50, as well as to E3KARP, and that we call EPI64 (EBP50–PDZ interactor of 64 kD). The gene for human EPI64 lies on chromosome 22 where nine exons specify a protein of 508 residues that contains a Tre/Bub2/Cdc16 (TBC)/rab GTPase-activating protein (GAP) domain. EPI64 terminates in DTYL, which is necessary for binding to the PDZ domains of EBP50, as a mutant ending in DTYLA no longer interacts. EPI64 colocalizes with EBP50 and ezrin in syncytiotrophoblast and cultured cell microvilli, and this localization in cultured cells is abolished by introduction of the DTYLA mutation. In addition to EPI64, immobilized EBP50 PDZ domains retain several polypeptides from placental microvilli, including an isoform of nadrin, a rhoGAP domain–containing protein implicated in regulating vesicular transport. Nadrin binds EBP50 directly, probably through its COOH-terminal STAL sequence. Thus, EBP50 appears to bind membrane proteins as well as factors potentially involved in regulating membrane traffic.
Keywords: ezrin, microfilaments, membrane traffic, NHE-RF, nadrin
Introduction
The plasma membrane and associated cortical cytoskeleton is the interface that eukaryotic cells have between the extracellular matrix, adjacent cells, and their environment. Consequently, the cell cortex is assembled into functional domains. For example, polarized epithelial cells have distinct protein and lipid compositions in the apical and basolateral domains that are separated by tight junctions (for review see Keller and Simons 1997). The cortical cytoskeleton performs many functions; it provides structural support for the plasma membrane, it participates in signaling pathways, and it participates in both endocytosis and exocytosis. Given its broad array of functions, it is not surprising that the cortical membrane organization is a complex structure of interacting proteins. As a contribution to understanding the cell cortex, we have set out to characterize the organization and function of the proteins that make up the apical cortex of epithelial cells and to elucidate their roles in regulating events at the plasma membrane.
A major component of the apical cortical cytoskeleton of epithelial cells is ezrin, an 81-kD polypeptide originally isolated from intestinal epithelial cell microvilli (Bretscher 1983). Subsequent work on ezrin identified it as a member of the ezrin/radixin/moesin (ERM) family of membrane–cytoskeleton linking proteins (for reviews see Mangeat et al. 1999; Bretscher et al. 2000). At the NH2 terminus of the ERM proteins is a ∼300-residue domain referred to as a 4.1 ERM (FERM) domain, as it is the defining feature of the 4.1-band superfamily of proteins (Chishti et al. 1998). This domain associates with membrane proteins, either directly through association with single-pass membrane proteins such as CD44, CD43, I-CAM1, 2, 3, and syndecan-2 involved in adhesion (Tsukita et al. 1994; Helander et al. 1996; Serrador et al. 1997, Serrador et al. 1998; Heiska et al. 1998; Legg and Isacke 1998; Yonemura et al. 1998), or indirectly through scaffolding proteins, such as ERM-binding phosphoprotein 50 (EBP50) and NHE3 kinase A regulatory protein (E3KARP) (Reczek et al. 1997; Reczek and Bretscher 1998; Yun et al. 1998), to membrane proteins (Hall et al. 1998a,Hall et al. 1998b; Short et al. 1998; Wang et al. 1998). In the 30 COOH-terminal residues of the ERM proteins resides an F-actin binding site (Turunen et al. 1994; Pestonjamasp et al. 1995; Berryman and Bretscher 2000), thus accounting for the view of ERM proteins as microfilament–membrane linking proteins.
The linking function of ERM proteins is subject to conformational regulation. In the dormant state, ERM proteins are unable to interact with at least some of their ligands, including EBP50 and F-actin, because the binding sites are masked (Gary and Bretscher 1995; Reczek and Bretscher 1998). Masking is achieved by the association of the ∼100 COOH-terminal residues with the NH2-terminal FERM domain (Gary and Bretscher 1995; Magendantz et al. 1995). Since the NH2-terminal domain of any ERM member can asssociate with the COOH-terminal domain of any member, these regions were called N-ERM association domains (ERMADs) and C-ERMADs. Recent structural studies have provided a view of how the masking is achieved: the N-ERMAD consists of a relatively compact domain to which the C-ERMAD binds as a highly extended inhibitory peptide masking a remarkable amount of the surfaces of both the N- and C-ERMAD (Pearson et al. 2000). Activation is believed to result in dissociation of the C-ERMAD from the N-ERMAD to reveal the interface containing membrane and F-actin binding sites. Currently, activation is believed to be a two-step process involving phosphatidylinositol 4,5-bisphosphate and phosphorylation of a conserved threonine 20 residues from the COOH terminus (Nakamura et al. 1995, Nakamura et al. 1999; Niggli et al. 1995; Hirao et al. 1996; Heiska et al. 1998; Matsui et al. 1998; Pietromonaco et al. 1998; Huang et al. 1999; Barret et al. 2000). In support of this mechanism, Hamada et al. 2000 have suggested that binding of phosphatidylinositol 4,5-bisphosphate to the FERM domain results in the movement of a peptide loop in the FERM domain that is likely to reduce its affinity for the C-ERMAD, and Pearson et al. 2000 have suggested that phosphorylation of the conserved threonine might destabilize the N-/C-ERMAD interaction.
Based on our model for ezrin activation (Gary and Bretscher 1995), we previously sought to identify proteins that might bind to the isolated, conformationally activated NH2-terminal domain of ezrin and moesin. Although several adhesion proteins had been shown to interact with this domain, we reasoned that in epithelial cells, where the ezrin-rich apical surface is not attached to the extracellular matrix, other molecules might associate with ezrin. Since ezrin is highly enriched in human placenta (Berryman et al. 1995), we used this tissue as starting material for the identification of proteins that bound immobilized NH2-terminal domains of ezrin and moesin. This approach led to the isolation and characterization of EBP50 (Reczek et al. 1997; Reczek and Bretscher 1998). EBP50 is the human homologue of rabbit Na+/H+ exchanger regulatory factor described previously (for review see Weinman et al. 2000). Human EBP50 is a 358-residue protein that is significantly enriched in microvilli and contains two related postsynaptic density (PSD)-95/DlgA/ZO-1–like (PDZ) domains in its NH2-terminal half and an ERM binding sequence in its COOH-terminal 30 residues. PDZ domains frequently associate with specific COOH-terminal sequences of transmembrane proteins (Songyang et al. 1997) (for reviews see Saras and Heldin 1996; Kornau et al. 1997), and, indeed, the first PDZ domain of EBP50 has been shown to have specificity for the sequence D-(S/T)-(R/Y)-L (Hall et al. 1998a; Wang et al. 1998) and to bind to the cystic fibrosis transmembrane conductance regulator (CFTR) (through -DTRL) (Yang and Tonks 1991; Short et al. 1998), the β2-adrenergic receptor (β2AR) (through -DSLL) (Hall et al. 1998a,Hall et al. 1998b; Cao et al. 1999), and the PDGF receptor (through -DSFL) (Maudsley et al. 2000). These interactions may be involved in retaining membrane proteins at the apical surface, as seems to be the case for the CFTR (Short et al. 1998), or in regulating the availability of EBP50 to bind to Na+/H+ exchanger isoform 3 (NHE3) (Hall et al. 1998b) or, as has been shown for the β2AR, in trafficking endocytosed receptor back to the plasma membrane (Cao et al. 1999). EBP50 has also been shown to interact with the cytoplasmic proteins YAP65 (Mohler et al. 1999) and G protein–coupled receptor kinase 6A (GRK6A) (Hall et al. 1999).
In addition to binding the ERM family members, EBP50 was independently identified as a protein that binds the NH2-terminal domain of merlin (Murthy et al. 1998), the product of the neurofibromatosis II tumor suppressor gene (Rouleau et al. 1993; Trofatter et al. 1993). Another protein, E3KARP, has been identified that, like EBP50, has closely related PDZ domains and a COOH-terminal ERM binding domain. However, although EBP50 binds both ERM proteins and merlin with high affinity, E3KARP binds much more readily to ERM proteins than to merlin (Yun et al. 1997, Yun et al. 1998; Reczek and Bretscher 1998; Nguyen et al. 2001).
Since many of the proteins documented to bind the PDZ domains of EBP50 are not expected to be enriched in the apical aspect of all epithelial cells that contain EBP50, we used affinity chromatography to identify proteins from isolated placental microvilli that bind the PDZ domains of EBP50. We have identified several binding candidates. In particular, the placental form of nadrin (Harada et al. 2000), a protein having a rhoGAP domain, binds the PDZ domains of EBP50 as does a Tre/Bub2/Cdc16 (TBC)/rabGAP domain–containing protein that we refer to as EPI64. The focus of this report is on the isolation and characterization of EPI64, the identification of which adds further support for a role of EBP50 in regulating membrane traffic.
Materials and Methods
Human placenta was obtained from consenting patients at Tompkins Community Hospital, Ithaca, NY. Adult female CD-1 mice were provided by Dr. M. Salpeter and M. Strang (Cornell University). Restriction enzymes and other reagents for molecular biology were purchased from GIBCO BRL and New England Biolabs, Inc.
Production and Purification of Recombinant Proteins
The design and production of the ezrin 1–296 (ezrin NH2-terminal domain), and glutathione S-transferase (GST)–EBP50 constructs 1–358 (full-length EBP50), 1–97 (PDZ-1), 1–138 (PDZ-1+), 1–248 (PDZ-1+ PDZ-2), and 241–358 (COOH-terminal half) have been described previously (Reczek et al. 1997; Reczek and Bretscher 1998). GST–EBP50 138–248 (PDZ-2) was generated by subcloning the 0.35-kb NruI–EcoRI fragment of GST–EBP50 1–248 into the SmaI–EcoRI sites of the pGEX-2T vector (Amersham Pharmacia Biotech). To make the untagged EPI64 construct, the cDNA sequence encoding residues 1–508 was amplified by PCR using primers that generated EcoRI and HindIII sites at the ends. This product was subcloned into the pQE16 expression vector (QIAGEN). The GST–EPI64 constructs were made similarly, using PCR to amplify the cDNA sequence for residues 1–508 with primers generating BamHI and EcoRI sites at the ends, except the 3′ primer used for the DTYLA mutant encoded an additional alanine residue after the wild-type sequence. These products were then subcloned into the pGEX-2T vector. The Xpress epitope-tagged EPI64 constructs were derived from the GST–EPI64 constructs by subcloning the appropriate BamHI–EcoRI EPI64 coding fragments into the EpiTag vector (Invitrogen). All recombinant sequences were determined to be free of PCR errors by nucleotide sequence analysis. MBP-E3KARP was a gift from Dr. C. Yun (Johns Hopkins University, Baltimore, MD).
The expression of the ezrin NH2-terminal and GST–EBP50 constructs has been described (Reczek et al. 1997; Reczek and Bretscher 1998), and the expression of GST–EBP50 138–248, and GST–EPI64 was carried out similarly here. For the expression of untagged EPI64, the plasmid encoding this construct was transformed into the Escherichia coli strain M15 [pRep4] (QIAGEN). Saturated overnight cultures were inoculated at 1:20 dilution in LB medium containing 100 μg/ml ampicillin and 25 μg/ml kanamycin and grown for 90 min at 37°C. Isopropyl β-d-thiogalactopyranoside was added to 2 mM, and cells were grown for an additional 180 min. Cells were harvested by centrifugation at 8,000 g for 15 min.
The purification of ezrin 1–296 has been described (Reczek et al. 1997). The GST fusion proteins were purified according to the manufacturer's protocol supplied with the vectors (Amersham Pharmacia Biotech). To purify bacterially expressed untagged EPI64, induced cells from 50 ml of culture were resuspended in Tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 7.4, at 4°C) containing 50 μg/ml phenylmethylsulfonyl fluoride and 75 μg/ml benzamidine and lysed by sonication (Branson Ultrasonics Corp.). The resulting extract was clarified by centrifugation at 48,000 g for 10 min, and the soluble supernatant was mixed with 50 μl of a 25% slurry of PDZ-1 + PDZ-2 GST–EBP50–agarose beads for 45 min. The beads were washed five times in 1 ml Tris-buffered saline, and bound protein was eluted by boiling 2 min in SDS sample buffer (Laemmli 1970) or by the addition of Tris-buffered saline made up to 2 M NaCl.
Affinity Chromatography and Binding Assays
To prepare affinity resins, GST fusion proteins that had been purified according to the manufacturer's protocol were dialyzed into PBS and then re-bound to glutathione–agarose at ∼130 μM. Ezrin 1–296 beads were made as described previously (Reczek et al. 1997).
Human placental microvilli were prepared as described by Berryman et al. 1995. For extracts, fresh microvilli in saline, at ∼10 mg/ml total protein concentration, were pelleted by centrifugation at 20,000 g for 30 min and then resuspended and lysed in the same starting volume of ice-cold extraction buffer (50 mM Tris, 300 mM NaCl, 1% Triton X-100, 50 μg/ml phenylmethylsulfonyl fluoride, 75 μg/ml benzamidine, pH 7.4, at 4°C). The extract was clarified by centrifugation at 100,000 g for 30 min at 4°C, and the resulting supernatant (final concentration ∼5 mg/ml) was used to set up binding reactions.
Affinity binding assay reactions were carried out by mixing 50 μl of 25% slurry of protein-coupled beads with 1 ml of soluble microvillar extract at 4°C for 90 min. The beads were washed three times in extract buffer, followed by two washes in extract buffer without Triton X-100. Bound extract proteins were eluted from the beads by two sequential elutions in three-bed volumes of extract buffer made up to 2 M NaCl and lacking Triton X-100. Eluates from the same reaction were pooled and then boiled in SDS sample buffer.
Phosphatase Assays
Phosphatase treatment of EPI64 isolated from human placental microvilli was performed according to methods described previously (Reczek et al. 1997).
Antibodies
Polyclonal antibodies to EPI64 were elicited in rabbits and affinity purified as described (Bretscher 1983) using purified recombinant GST–EPI64 as antigen. Monoclonal antibodies to the Xpress epitope tag were purchased from Invitrogen.
SDS-PAGE, Blot Overlays, and Immunoblots
SDS-PAGE was performed according to (Laemmli 1970). For some experiments gels were stained with Coomasssie brilliant blue R-250 or were silver stained (Oakley et al. 1980). For blots, proteins were transferred from gels to polyvinylidene fluoride membranes using a semidry electroblotter (Integrated Biosystems). All blots were developed using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech).
Blot overlays and probe biotinylation were carried out according to methods described previously (Gary and Bretscher 1993). For control experiments, the biotinyl probes were omitted or, in the case of the MBP–E3KARP fusion protein, biotinyl MBP alone was used.
Immunoblots were blocked in 10% nonfat dry milk and then probed with 0.1 μg/ml affinity-purified EPI64 antibodies in 1% milk, followed by 0.1 μg/ml peroxidase-conjugated goat anti–rabbit IgG in 1% milk. Primary antibodies were omitted for control blots.
Murine tissue samples were obtained from adult female CD-1 mice. Total SDS-soluble tissue and cell lysates were prepared as described previously (Reczek et al. 1997). Tissue and cell lysate samples analyzed by SDS-PAGE were normalized according to total protein concentration.
Sequence Analysis
Affinity chromatography from extracts of human placental microvilli was used on a large scale to isolate sufficient amounts of the 120-, 65-, and 64-kD candidates for peptide microsequencing. Eluates were resolved by preparative SDS-PAGE, the resultant gels were stained briefly with Coomassie brilliant blue R-250, and regions containing the desired bands were excised and then washed in 50% acetonitrile. These samples were sent to Harvard Microchem, where sequence analysis was performed by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ quadrupole ion trap mass spectrometer.
The Institute for Genomic Research human cDNA database was searched using the EPI64 peptide sequences. cDNA clones that matched these query sequences were obtained from Genome Systems Inc. The insert sizes were determined by restriction endonuclease digestion using enzymes appropriate for the cloning sites in each library parent vector. The cDNA insert of a clone from a human prostate library was sequenced in its entirety using sets of primers that yielded overlapping sequence information. All nucleotide sequencing was done using an automated cycle sequencer (model 373A; Applied Biosystems).
Protein and DNA sequence alignments and editing were done using the software programs MEGALIGN and EDITSEQ (DNASTAR Inc.), respectively.
Immunofluorescence Microscopy and Cell Transfection
Cryosections of human placenta were prepared and stained as described previously (Berryman et al. 1993). Affinity-purified EPI64 antibodies were used at ∼3 μg/ml.
JEG-3 cultured cells were grown on glass coverslips in MEM supplemented with 10% FCS. Cells were transfected using Genejammer transfection reagent (Stratagene) according to the manufacturer's protocol. 24–48 h after transfection, cells were fixed and stained for microscopy as described by Franck et al. 1993 using a 1:400 dilution of anti-Xpress monoclonal antibody (Invitrogen). In cases where cells or tissues were double labeled, ∼3 μg/ml affinity-purified EPI64, EBP50, or ezrin antibodies were also used. Actin was stained with a 1:200 dilution of rhodamine phalloidin (Molecular Probes, Inc.).
Cells and tissue sections were viewed with an Axiovert 100-TV fluorescence microscope (Carl Zeiss Inc.), and images were acquired using Metamorph imaging software (Universal Imaging Corp.).
Results
Identification of EBP50 PDZ Domain–binding Proteins
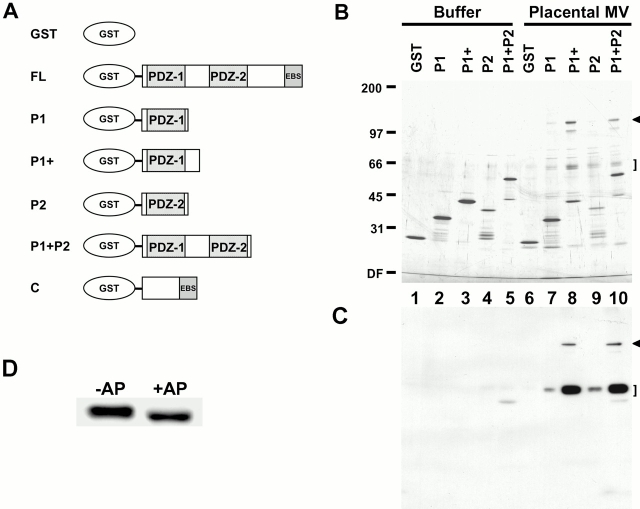
Both ezrin and EBP50 are highly enriched in human placental microvilli, so we used these isolated structures as starting material in an attempt to identify physiologically relevant proteins that bind the PDZ domains of EBP50. A series of GST fusion proteins containing the first, the second, or both PDZ domains of EBP50 (Fig. 1 A) were expressed as soluble proteins in bacteria. After purification, they were re-bound to glutathione–agarose beads at equimolar concentrations. The beads were incubated with total detergent soluble extracts of human placental microvilli and then washed extensively, and bound proteins were eluted in high salt buffer and resolved by SDS-PAGE (Fig. 1 B). Beads with immobilized GST alone, or the fusion proteins incubated with just buffer, served as controls. In analyzing these results, it is necessary to identify those bands that are derived from the fusions proteins (lanes 1–5) to determine which polypeptides originate from the placental microvillar extract (lanes 6–10). Such an analysis reveals that a distinct group of polypeptides at ∼120, ∼65, and ∼64 kD bound specifically to all fusion proteins containing PDZ-1 (Fig. 1 B, lanes 7–10). However, these proteins clearly bound better to a construct of PDZ-1 with additional COOH-terminal sequences (P1+), rather than to the minimal PDZ-1 domain (P1), and bound to a lesser extent to just PDZ-2 (P2). No binding was seen to the control GST beads (lane 6). In addition to the 120-, 65-, and 64-kD bands, additional specific polypeptides were also recovered in lower abundance on the various PDZ domain beads, and these are discussed below.
Figure 1.
Identification of EBP50 PDZ domain–binding candidates. (A) Summary of GST–EBP50 fusion proteins used for affinity chromatography. The cDNA sequences encoding human EBP50 residues 1–358 (FL), residues 1–97 (P1), residues 1–138 (P1+), residues 138–248 (P2), residues 1–248 (P1 + P2), and residues 241–358 (C) were expressed as soluble recombinant proteins fused to GST. These proteins were purified, immobilized on glutathione–agarose beads, and then used for affinity chromatography. (B) Affinity chromatography. Buffer or total detergent soluble extracts of placental microvilli were mixed with GST–agarose beads or GST–EBP50–agarose beads containing the first PDZ domain (P1 or P1+), the second PDZ domain (P2), or both in tandem (P1 + P2). The beads were then washed extensively in buffer at 0.3 M NaCl, and bound proteins were eluted and resolved on a 6–20% silver-stained gradient SDS gel. (C) One-fourth the amount of the samples shown in B were resolved by SDS-PAGE, transferred to PVDF, and then probed with biotinylated EBP50. Arrowheads and brackets indicate specific 120-kD and 64/65-kD binding candidates, respectively. The mobilities of molecular mass standards in kD are indicated at left. DF, dye front. (D) The heterogeneity of the 64/65-kD candidate is due to phosphorylation. A PDZ domain affinity bead eluate from B that was enriched in the 64/65-kD binding candidate was incubated in the presence (+AP) or absence (−AP) of calf intestinal alkaline phosphatase and then resolved by SDS-PAGE on a 10% gel, transferred to PVDF, and overlaid with biotinyl EBP50 probe as in C.
Proteins retained on the GST–PDZ domain fusion protein beads may bind directly or indirectly. To identify those that bound directly and as a further test of specificity, biotin-labeled purified recombinant EBP50 was used as a soluble probe in a blot overlay assay on duplicates of the samples shown in Fig. 1 B (Fig. 1 C). A strong signal was seen for the candidates at 64 and 65 kD (Fig. 1 C, lanes 7–10), and a specific signal, although weaker on a relative scale, was seen for the 120-kD candidate (Fig. 1 C, lanes 8 and 10; and on longer exposure, lanes 7 and 9; data not shown). The EBP50 probe also recognized these candidates in the starting total microvillar extracts (data not shown). These results indicate that the interaction between the PDZ domains of EBP50 and the 120-, 65-, and 64-kD polypeptides is direct.
The 64/65-kD Polypeptides Contain a TBC/rabGAP Domain, and the 120-kD Polypeptide Is an Isoform of Nadrin, a rhoGAP Domain–containing Protein
To determine the identity of the proteins retained on the P1+ beads, a larger scale purification was performed, and the 64-, 65-, and 120-kD PDZ-binding polypeptides were subjected to mass spectrometry-based sequence analysis. A correlative search of the GenBank/EMBL/DDBJ and EST databases with the masses of peptides derived from the 64- and 65-kD candidates identified the same set of human ESTs coding for a novel human protein, indicating that their difference in migration was likely due to some form of posttranslational modification. Therefore, to examine the heterogeneity of the 64/65 kD doublet, an enriched sample from placental microvilli was treated with calf intestinal alkaline phosphatase. As shown in Fig. 1 D, this resulted in the collapse of the 65-kD band into the 64-kD species. Since immunoblots with antiphosphotyrosine antibodies did not detect either of these species (data not shown), the resulting modification is presumably due to either serine and/or threonine phosphorylation. We refer to these two polypeptides as EPI64.
The EST cDNA clones encoding peptides present in EPI64 were obtained and sequenced. One of these, from a human prostate library, contained an ∼1.97-kb insert with an open reading frame possessing all of the EPI64 derived peptides (Fig. 2 A). In addition, it also contained a portion of the 5′ untranslated region as well as the full 3′ untranslated region, and a poly A tail. The reading frame encodes a 508–amino acid protein with no predicted transmembrane domains and a calculated molecular mass of 57.1 kD. Interestingly, the deduced protein sequence of EPI64 ends in the amino acids DTYL, a motif that matches the optimal peptide binding consensus sequence, D-(S/T)-(R/Y)-L, determined for the first PDZ domain of EBP50 (Hall et al. 1998a,Hall et al. 1998b; Wang et al. 1998). Thus, EPI64 appears to be a novel cytoplasmic protein that probably associates with the PDZ domains of EBP50 via a typical PDZ to COOH-terminal interaction.
Figure 2.

cDNA sequence and genomic structure of the EPI64 gene. (A) Nucleotide and derived protein sequences of human EPI64 cDNA. The TBC/rabGAP domain is underlined. Intron/exon boundaries are indicated by arrowheads, and the COOH-terminal PDZ–binding motif is boxed. These sequence data are available from GenBank/EMBL/DDBJ under accession number AF331038. (B) Schematic diagram of the intron and exon segments comprising the EPI64 gene. Boxes indicate exons and are numbered sequentially from 5′ to 3′.
Database searches revealed that the human EPI64 gene resides at chromosome locus 22q12.1-qter. A comparison of the cDNA and genomic sequences revealed that the EPI64 gene consists of eight introns and nine exons, with the first intron being exceptionally large, and spanning over 22 kb (Fig. 2 B). Database searches also identified a 200–amino acid region in EPI64, between residues G111 and L311 (Fig. 2 A), that shows significant sequence identity to a broad range of proteins from diverse species. This domain is generally referred to as the TBC/rabGAP domain (Richardson and Zon 1995; Neuwald 1997; Albert et al. 1999), and it is conserved from humans to plants. Fig. 3 A shows an alignment of the EPI64 TBC/rabGAP domain with those in proteins from a selection of various species. A key feature of a rabGAP domain is a well-conserved arginine, located at residue 160 in the EPI64 sequence, that has been shown to be critical for the rab GTPase activity of the yeast proteins Gyp1p and Gyp7p (Albert et al. 1999). Thus, EPI64 has a TBC/rabGAP domain.
Figure 3.
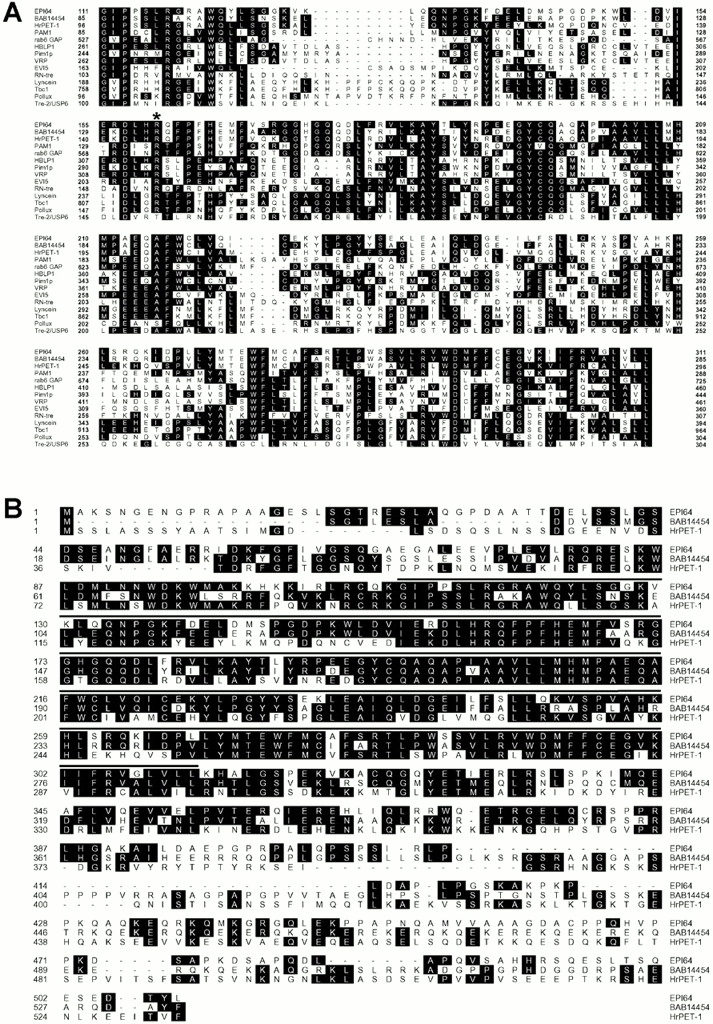
Alignment of EPI64 with related protein sequences. (A) EPI64 has a TBC/rabGAP domain. TBC/rabGAP domains of proteins from diverse species were aligned with EPI64 sequences using the CLUSTAL algorithm of the MEGALIGN software program (DNASTAR). The complete sequences of these proteins are available at the following GenBank/EMBL/DDBJ accession numbers: Homo sapiens BAB14454 (BAB14454), Halocynthia roretzi HrPET-1 (BAA81906), Arabidopsis thaliana PAM1 (AAC33763), H. sapiens rab6 GAP (NP_036329), Mus musculus HBLP1 (NP_061245), Saccharomyces cerevisiae Pim1p (AAB01977), H. sapiens VRP (NP_008994), H. sapiens EVI5 (NP_005656), Drosophila melanogaster RN-tre (AAC48286), Bos taurus Lyncein (CAA76943), M. musculus Tbc1 (AAA85223), D. melanogaster Pollux (AAB02200), and H. sapiens Tre-2 USP6 (NP_004496). Residues matching the consensus sequence are shaded. A well-conserved arginine residue, critical for yeast Gyp1p and Gyp7p GAP activity, is indicated (*). (B) EPI64 demonstrates significant homology to HrPET-1 and is closely related to the unknown human protein BAB14454. An alignment of the full protein sequences of EPI64, HrPET-1, and BAB14454 is shown. The TBC/rabGAP domain is overlined. Sequence identities matching the consensus are shaded.
Two proteins in particular were found to be quite closely related to EPI64: a 533–amino acid unnamed human protein (sequence data available from GenBank/EMBL/DDBJ under accession number BAB14454), and a 532–amino acid uncharacterized ascidian protein, HrPET-1 (sequence data available from GenBank/EMBL/DDBJ under accession number BAA81906); an alignment of these proteins with EPI64 is shown in Fig. 3 B. The BAB14454 protein and HrPET-1 show ∼67 and ∼40% overall sequence identity to EPI64, and ∼82 and ∼54% identity with the TBC/rabGAP domain of EPI64, respectively. Information submitted to GenBank/EMBL/DDBJ for HrPET-1 suggests that it may play some role in a localization pathway of maternal RNAs at the posterior–vegetal cytoplasm in early ascidian embryos. No additional information is available for the BAB14454 database entry. Although these two proteins do not share identical COOH-terminal sequences with EPI64, they do show conservation of sequence with EPI64 outside the TBC/rabGAP domain. These two proteins also demonstrate ∼40% sequence identity to each other, and as such, it is difficult to discern whether HrPET-1 is the homologue of EPI64 or of the human BAB14454 protein. Based on total residue length, it is perhaps more likely that BAB14454 and HrPET-1 are homologues. Regardless, both BAB14454 and HrPET-1 are close relatives of EPI64.
Several cDNA clones containing peptides identical to those found in the 120-kD PDZ domain binding candidate were also obtained from the human EST database. Complete nucleotide sequencing of two clones from placental and nasal epithelial libraries revealed them to be incomplete, but showed that the derived COOH-terminal protein sequences terminated with the amino acids STAL. This COOH-terminal sequence is predicted to bind EBP50 PDZ-1 and fits the more general consensus sequence X-(S/T)-X-(L/V/I) for a broad spectrum of PDZ domain–binding proteins (Saras and Heldin 1996; Songyang et al. 1997). This terminus is followed by the same 3′ untranslated and poly A regions in these clones. Therefore, it seems likely that the 120-kD placental protein binds, like EPI64, via its COOH terminus to the PDZ domains of EBP50.
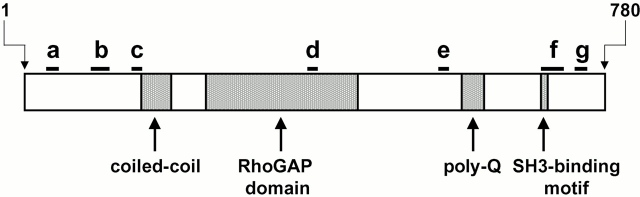
BLAST searches of GenBank/EMBL/DDBJ with either the nucleotide or deduced protein sequences from these clones identified two protein sequences that had been entered into the database during the course of this work. One of these was a partial sequence of a hypothetical human protein (sequence data available from GenBank/EMBL/DDBJ under accession number BAA91532), and the other is a complete sequence coding for rat brain nadrin, a 780–amino acid protein described as a novel neuron-specific rho GTPase-activating protein involved in regulated exocytosis (Harada et al. 2000). The hypothetical human protein, which was isolated as a clone from a neuronal precursor cell library (sequence data available from GenBank/EMBL/DDBJ under accession number BAB14454), is almost certainly a human isoform of rat brain nadrin based on protein sequence alignments exhibiting very few, minor, and conservative residue differences. (The deposited nucleotide sequence appears to have a four-base deletion after nucleotide 616; the downstream homology inferred here assumes a correction of this presumed error.) Peptides derived from the 120-kD placental protein align with rat brain nadrin throughout its sequence (Fig. 4). However, the human EST clones described above have regions of perfect alignment with rat brain nadrin punctuated by at least two regions where the sequence has no similarity: we interpret this as suggesting that nadrin is alternatively spliced and that we have identified the human placental isoform. One of the peptides derived from the 120-kD placental protein lies in the rhoGAP domain of nadrin, indicating that the 120-kD protein has this domain.
Figure 4.
The 120-kD human placental protein is an isoform of rat brain nadrin. A schematic diagram showing the domain structure of rat brain nadrin is shown with peptides derived from the sequencing of the human 120-kD protein mapped onto it. Peptides are lettered a–g and have the following sequences: a, TEVLSEDLLQIER; b, MLETCGDAENQLALELSQHEVFVEK; c, LVLDWDSVR; d, LAQTSDVNK; e, KPAPAPPK; f, NRPSVPPPPQPPGVHSAGDSSLTNTAPTASK; g, SIFPEMHSDSASK.
Expression of Recombinant EPI64 and Mapping of the EBP50 Association Site to the Extreme COOH Terminus of EPI64
EPI64 was expressed as a recombinant protein in E. coli to better characterize it and its association with EBP50. Both untagged and GST fusions of full-length EPI64 were generated. Fig. 5 A shows that, when untagged EPI64 is purified from bacterial extracts on EBP50 PDZ-1+ affinity beads, it comigrates with the 64-kD species from human placenta. These results suggest that the cDNA contains the full-length coding sequence of EPI64. In addition, they indicate that EPI64 migrates somewhat anomalously in SDS-PAGE relative to its calculated molecular mass of 57.1 kD.
Figure 5.
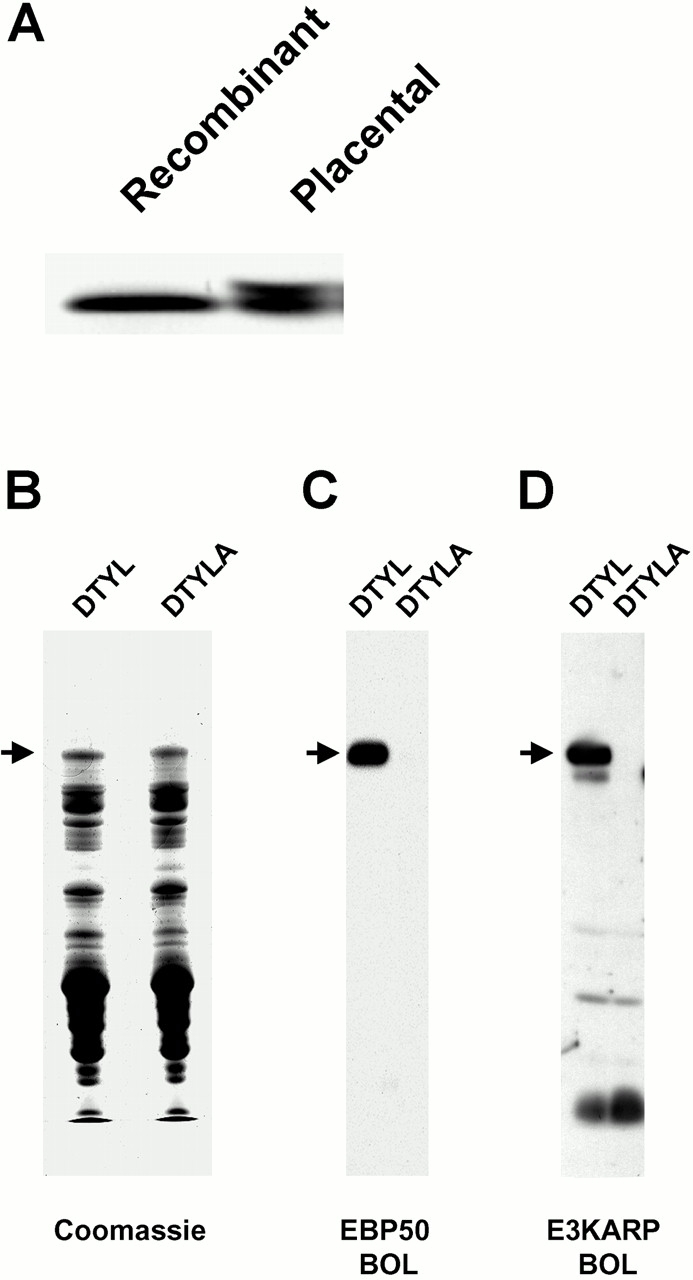
Expression of recombinant EPI64 and mapping of the EBP50 and E3KARP association sites to the extreme COOH terminus of EPI64. (A) Recombinant EPI64 comigates with the 64-kD species from placental microvilli. Recombinant EPI64 purified from bacterial extracts was resolved by SDS-PAGE adjacent to a sample of EPI64 purified from placental microvilli. The proteins were transferred to PVDF and then detected via an EBP50 PDZ domain blot overlay. (B) Coomassie-stained gel of wild-type (DTYL) and COOH-terminal mutant (DTYLA) GST–EPI64 after purification on glutathione–agarose. A fraction of the samples from B were resolved by SDS-PAGE, transferred to PVDF membrane, and probed with biotinyl EBP50 (C), or biotinyl E3KARP (D). Arrows indicate the migration of the full-length GST–EPI64 fusion proteins. Additional signal for lower molecular mass bands in D result from background seen in control blots.
The presence of the COOH-terminal -DTYL sequence in EPI64 suggests that it might bind to a PDZ domain of EBP50 through this motif. To test this, GST–EPI64 fusion proteins possessing either the wild-type COOH terminus, -DTYL, or a mutated COOH terminus with an additional alanine, -DTYLA, were generated. Based on previous work (Hall et al. 1998a; Short et al. 1998; Cao et al. 1999), we surmised that the addition of a single alanine residue might be sufficient to disrupt the predicted PDZ–COOH terminus interaction and chose to use a blot overlay assay to assess this. The GST–EPI64–DTYL and -DTYLA proteins were purified on glutathione–agarose and eluted, and a sample of each was resolved by SDS-PAGE. These proteins were then either stained in the gel with Coomassie blue to evaluate their status after purification (Fig. 5 B), or were transferred to a membrane and incubated with biotin-labeled EBP50 probe (Fig. 5 C). Although considerable degradation of the fusion proteins occurred during these manipulations, the EBP50 probe clearly bound to the construct containing the wild-type COOH terminus but not to the construct in which the COOH terminus was mutated, indicating that the association of EPI64 with the PDZ domains of EBP50 requires its extreme COOH terminus.
E3KARP is a human protein closely related to EBP50 that shares ∼55% overall sequence identity, the predominance of which is found over the two PDZ domains (Reczek et al. 1997; Yun et al. 1997). Like EBP50, E3KARP has also been found to bind members of the ERM family, although it does not appear to bind merlin (Murthy et al. 1998; Reczek and Bretscher 1998; Yun et al. 1998; Nguyen et al. 2001). Based on these similarities, we explored the possibility that EPI64 might also bind the PDZ domains of E3KARP. A blot overlay assay using a biotinylated E3KARP probe was performed on the same samples described above. Like EBP50, E3KARP also bound tightly to wild-type EPI64 but not to the mutant form, indicating that it can also associate with EPI64, and it can do so in a manner requiring the extreme COOH terminus (Fig. 5 D). It is of interest to note that the numerous degradation products of GST–EPI64 present after purification (Fig. 5 B) that contain the NH2 terminally fused GST moiety, as confirmed by immunoblot analysis, are not recognized, as they lack the EPI64 COOH-terminal sequences (Fig. 5C and Fig. D). This further substantiates that the interaction with the PDZ domains is COOH terminally mediated.
Tissue and Cell Distribution and Localization of EPI64
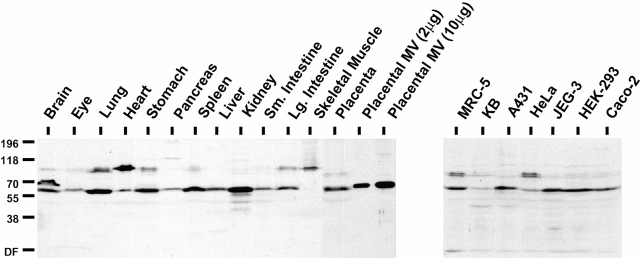
Recombinant GST–EPI64 was used to elicit antibodies in rabbits that were then affinity purified from the resulting antiserum and used to examine the tissue and cell distribution of endogenous EPI64. Immunoblots of normalized total SDS-soluble lysates of murine and human tissues indicated that EPI64 is present to some extent in most of the tissues examined, with the exception of skeletal muscle (Fig. 6, left). It is most abundant in kidney and is also found in lung, stomach, spleen, and placenta. As such, it shows a striking correspondence with the distribution of EBP50 in these tissues (compare with Figure 8 in Reczek et al. 1997). Database searches reveal that cDNAs for EPI64 have been isolated from neurons, germ cell tumors, infant brain, breast, prostate, and uterus. Moreover, immunoblots show that EPI64 is significantly enriched in microvillar proteins over total placental proteins (Fig. 6, left). We conclude that EPI64 is widely distributed in tissues and is enriched in purified placental microvilli.
Figure 6.
Tissue and cell distribution of EPI64. 100 μg of total proteins from murine tissues and human placenta and 2 and 10 μg of total proteins from isolated human placental microvilli (left), or 400 μg of total proteins from the indicated cultured human cell lines (right) were resolved by SDS-PAGE, transferred to PVDF membrane, and probed with affinity-purified antibodies to EPI64. The mobilities of molecular mass standards in kD are indicated at left. DF, dye front.
Immunoblotting of total SDS-soluble lysates from different human cell cultures shows that EPI64 is seen to varying extents in all the lines examined (Fig. 6, right), similarly to EBP50 (data not shown). A doublet band at ∼93–95 kD is also recognized by the antibodies in MRC-5 and HeLa cell lysates and may represent one or more potential alternatively spliced EPI64 isoforms, or a closely related protein(s). Interestingly, similar polypeptide bands are seen in some of the lysates of murine tissues, particularly those containing cells of muscle origin, and may be indicative of a similar scenario (Fig. 6, left). The presence of a major ∼70-kD immunoreactive form in brain (Fig. 6) may also be a related protein, or result from alternative splicing of the gene.
The information obtained from our protein sequence analysis and phosphatase treatment of placental EPI64 (Fig. 1 D) indicated that it exists as both a 64-kD species and a phosphorylated 65-kD species. Both of these are recognized by our antiserum; however, they could only be resolved upon shorter exposure of the immunoblot (data not shown). The resolution of EPI64 in the other total lysate samples was insufficient to detect such multiple species.
Using the same antibodies, the localization of EPI64 was compared with that of EBP50 and ezrin in human placenta. Freshly cut cryosections were double labeled for either EPI64 and actin, EBP50 and actin, or ezrin and actin (Fig. 7). EPI64 appeared to be enriched in the region of the syncytiotrophoblast containing abundant actin-rich microvilli. Diffuse and less intense staining for EPI64 was also seen in some of the cell layers beneath this region (Fig. 7 A). This staining pattern is very similar to that seen for EBP50 and ezrin (Fig. 7D and Fig. G).
Figure 7.
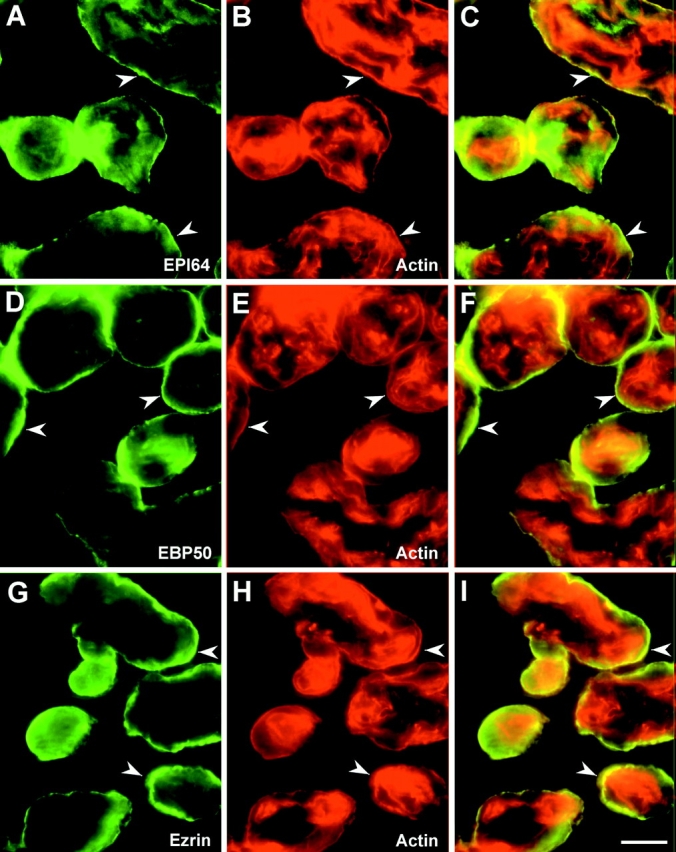
Localization of EPI64 in human placenta. Cryosections of placenta were stained with affinity-purified antibodies to EPI64 (A), EBP50 (D), and ezrin (G) and double-labeled for actin using rhodamine phalloidin (B, E, and H). (C, F, and I) Merged images. EPI64 colocalizes with actin in the microvilli-rich region of the syncytiotrophoblast (arrowheads) in a pattern that is very similar to that seen for EBP50 and ezrin. Bar, 40 μm.
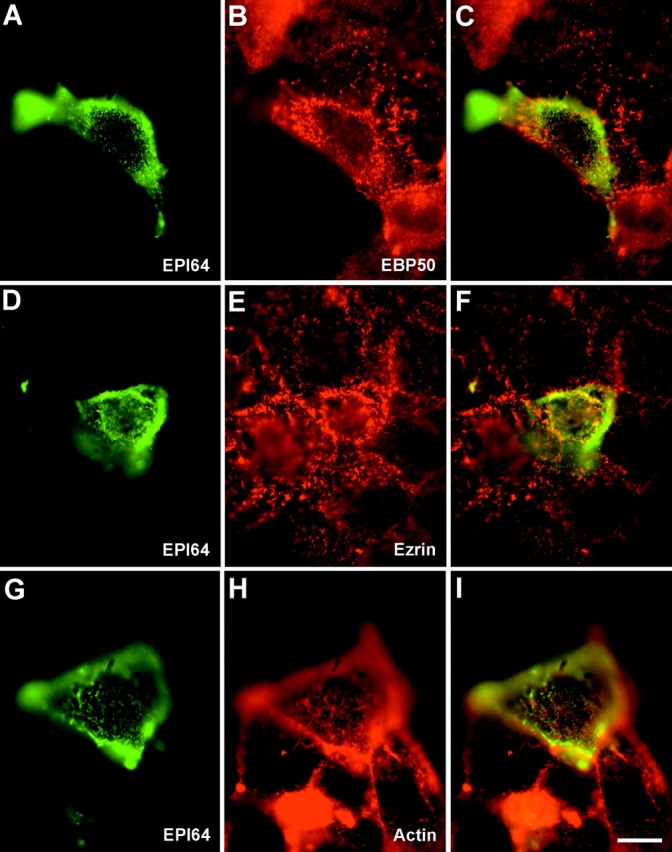
The localization of EPI64 in cultured human cells using the affinity-purified EPI64 antibodies was unreliable, as they did not yield appreciable specific staining. This was somewhat surprising as this reagent worked well for the immunohistochemical staining of placenta. We suspect that the poor signal is due to the low levels of EPI64 in cultured cells (Fig. 6, right). Therefore, human JEG-3 cultured cells were transiently transfected to express epitope-tagged EPI64. In transfected cells, EPI64 specifically localizes to the numerous surface microvilli (Fig. 8A, Fig. D, and Fig. G). The presence of the epitope tag permitted double labeling to compare the distribution of EPI64 with EBP50, ezrin, or actin. The staining in double-labeled cells revealed that EPI64 colocalizes precisely with EBP50 in microvilli and shows significant overlapping staining with ezrin and cortical F-actin in these structures (Fig. 8, D–I). Expression of EPI64 also appeared to enhance the level of EBP50 in microvilli (Fig. 8 B).
Figure 8.
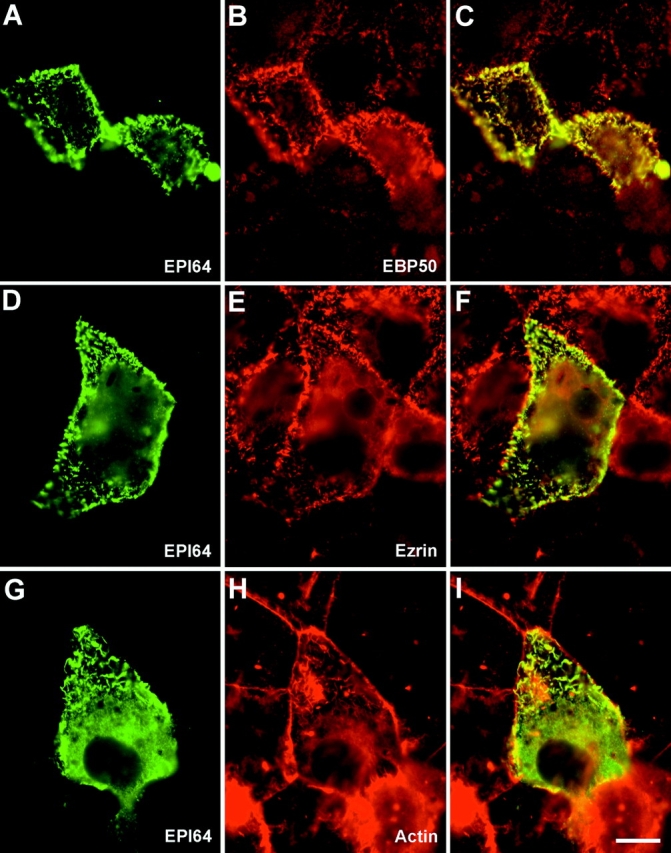
Localization of EPI64 in JEG-3 cells. Human JEG-3 cells transiently transfected with Xpress epitope-tagged EPI64 were fixed and double labeled to detect Xpress-tagged EPI64 (A, D, and G), and EBP50 (B), or ezrin (E), or F-actin (H). (C, F, and I) Merged images. Bar, 10 μm.
To determine if the localization of EPI64 was dependent on its COOH-terminal -DTYL sequence that binds EBP50, the localization of epitope-tagged EPI64 with the COOH-terminal alanine mutation was examined after transient transfection (Fig. 9). Introduction of this construct into JEG-3 cells showed that the addition of a single alanine residue to the COOH terminus was sufficient to perturb the localization to microvilli and resulted in a grainy and diffuse staining pattern (Fig. 9A, Fig. D, and Fig. G). Furthermore, mutant EPI64 did not colocalize with EBP50, ezrin, or cortical F-actin (Fig. 9C, Fig. F, and Fig. I). These in vivo results strongly support the in vitro data demonstrating an association between EPI64 and EBP50 and suggests that this interaction is needed for the microvillar localization of EPI64.
Figure 9.
Localization of the EPI64 COOH-terminal mutant in JEG-3 cells. Human JEG-3 cells transiently transfected with Xpress epitope-tagged EPI64 COOH-terminal mutant were fixed and double labeled to detect Xpress-tagged mutant EPI64 (A, D, and G), and EBP50 (B), or ezrin (E), or F-actin (H). Bar, 10 μm.
Isolation of Protein Complexes on the Isolated Domains of EBP50 and Ezrin
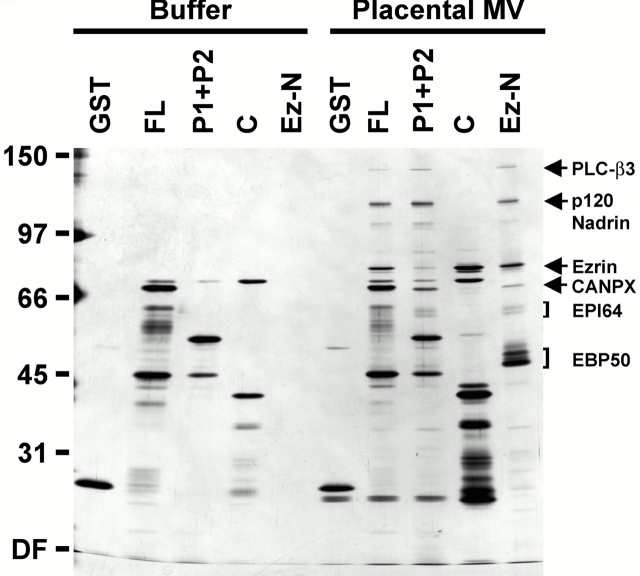
The data presented here, in conjunction with our previous biochemical studies (Reczek et al. 1997; Reczek and Bretscher 1998), suggested that it should be possible to assemble complexes of ezrin, EBP50, and EPI64, as well as other potentially relevant interacting proteins, from extracts of human placental microvilli. To test this hypothesis, agarose beads with immobilized full-length EBP50 (FL), its two PDZ domains (P1 + P2), its COOH-terminal fragment containing the ERM binding site (C) (Fig. 1 A), or the ezrin NH2-terminal domain (Ez-N) were generated. Total detergent soluble extracts of microvillar proteins were incubated with the beads, unbound proteins were removed by washing, and retained polypeptides were eluted and analyzed by SDS-PAGE. (Fig. 10). As before (Fig. 1), it is important to subtract the polypeptides contributed by the beads in buffer alone to identify polypeptides derived from the microvillar extract. The picture that emerges from the retained polypeptide profile is remarkably consistent with the documented protein–protein interactions. Thus, the NH2-terminal domain of ezrin (lane labeled Ez-N) retained EBP50, EPI64 and p120 nadrin (both presumably through EBP50), and ezrin, as well as some other polypeptides. The polypeptides retained by the COOH-terminal domain of EBP50 (lane C), together with those retained by the two PDZ domains of EBP50 (lane labled P1 + P2), recapitulate the sum of the polypeptides retained on the ezrin NH2-terminal domain, except of course for EBP50 itself. The polypeptides retained on full-length EBP50 (lane FL) are the sum of the polypeptides retained on the two halves of EBP50. The self-consistency of this analysis provides considerable support for the protein–protein interactions documented here and elsewhere.
Figure 10.
Assembly of protein complexes on the isolated domains of EBP50 and ezrin. Buffer or total detergent-soluble extracts of placental microvilli (MV) were mixed with either GST–agarose beads, agarose beads with GST fused to full-length EBP50 (FL), its two PDZ domains (P1 + P2), or its COOH-terminal half containing the ERM binding site (C), or agarose beads with the ezrin NH2-terminal domain (Ez-N). The beads were then washed, bound proteins eluted and resolved by SDS-PAGE, and the resulting gel was silver stained as described in the legend to Fig. 1 B. The mobilities of molecular mass standards are indicated in kD. The migration of specifically bound polypeptides of determined identity are indicated at right. DF, dye front.
A recent study has suggested that EBP50 can self-associate through PDZ–PDZ domain interactions (Fouassier et al. 2000). In this study, we found no indication of the ability of the immobilized PDZ domains to retain intact EBP50 (Fig. 1 B and 10; P1 + P2); at present we do not know the reason for this apparent discrepancy.
This analysis also allowed us to identify additional components that bind to the PDZ domains of EBP50. These include ∼140-, ∼107-, ∼68-, and ∼35-kD polypeptides seen in eluates in the FL, P1 + P2 and Ez-N lanes. Scaling up this binding assay, sufficient quantities of the 140- and 68-kD bands were recovered for protein sequence analysis. This revealed that the 140-kD band is the β3 isoform of phospholipase-C (PLC-β3), and the 68-kD band is a novel 641–amino acid protein described as an X-linked calpain-like protease according to unpublished information submitted to GenBank/EMBL/DDBJ (under accession number CAA04051). Interestingly, both PLC-β3 and the putative protease end in amino acid sequences predicted to bind PDZ domains NTQL and LTEL, respectively. The specific 81-kD band seen in the FL, C, and Ez-N eluates is ezrin, as determined by immunoblot analysis (data not shown). An additional specifically bound polypeptide of ∼43 kD was seen only in the COOH bead eluate, but has not yet been identified. These data demonstrate that it is possible to recover in vivo protein complexes (consisting of ezrin, EBP50, EPI64, placental nadrin, PLC-β3, etc.) over the isolated protein domains of some of their constituents. Whether all of these represent interactions that occur in microvilli in vivo will have to await future localization studies.
Discussion
In this paper, we extend our analysis of the molecular structure of the ezrin-based cortical cytoskeleton. Previously, we had identified EBP50 as a microvillar protein isolated from placental extracts that bound to the NH2-terminal domain of ezrin (Reczek et al. 1997), and we subsequently demonstrated that EBP50 only binds to activated ezrin in which its binding site is unmasked (Reczek and Bretscher 1998). Using a similar approach to find potentially relevant microvillar proteins that bind EBP50, we have identified several polypeptides that have COOH-terminal sequences characteristic of proteins that bind directly to the PDZ domains of EBP50.
We refer to one of these EBP50 PDZ–binding polypeptides as EPI64. Compared with the other candidates, it demonstrated the most robust and reproducible association with the PDZ domains of EBP50 and, like EBP50 and ezrin, it is highly enriched in isolated placental microvilli. Our analysis of in vitro and in vivo PDZ binding, cell and tissue distribution, and localization data all indicate that EPI64 is a physiologically relevant binding partner of EBP50.
EPI64 interacts directly with the PDZ domains of EBP50, and this interaction requires a canonical PDZ association with the COOH terminus of EPI64 that ends in DTYL. Since this sequence matches an optimal EBP50 PDZ-1 binding consensus (Hall et al. 1998a; Wang et al. 1998), EPI64 is predicted to have preference for the first PDZ domain in vivo. This prediction is further substantiated by the finding that soluble PDZ-1 fusion protein is much more effective than PDZ-2 at competing the binding of full-length EBP50 probe to endogenous EPI64 in ligand overlay assays (data not shown). Interestingly, however, immobilized fusion proteins containing either PDZ-1 or PDZ-2 were seen to retain EPI64 equally well by affinity chromatography, indicating that in this assay there did not seem to be an apparent preference. (Fig. 1B and Fig. C). Yet, neither PDZ domain on its own bound EPI64 as well as the fusion proteins containing PDZ-1 followed by ∼40 flanking amino acids (P1+, residues 1–138) or the two PDZ domains in tandem (P1 + P2, residues 1–248) (Fig. 1B and Fig. C). This suggests that additional EBP50 sequences flanking PDZ-1 may serve to fully stabilize the association with EPI64, either by allowing better stabilization or accessibility of the PDZ domain itself, or by making other contacts with EPI64.
The ability of E3KARP to bind EPI64 in vitro (Fig. 5) is likely to rely on a similar mode of association given the high level of overall sequence identity, particularly within the PDZ domains, that it shares with EBP50 (Yun et al. 1997). Using the atomic structure for the third PDZ domain of the synaptic protein PSD-95 as a model (Doyle et al. 1996), it is possible to identify and compare the critical residues in the PDZ domains of EBP50 and E3KARP that are predicted to make contacts with a bound peptide. These residues in EBP50 (K19, Y24, F26, H27, L28, G30, R40, H72, and I79 in PDZ-1 and K159; Y164, F166, N167, L168, S170, R180, H212, and I129 in PDZ-2) are identical or well conserved compared with those in E3KARP, suggesting that the peptide binding preferences of the EBP50 and E3KARP PDZ domains are likely to be extremely similar, if not identical. Thus, a careful study comparing the respective distribution and localization of EBP50 and E3KARP with those PDZ ligands that have the potential to bind both proteins will be needed to establish their biological relevance.
EPI64 is a 508–amino acid protein with no predicted transmembrane domains, signal sequences, or acylation motifs, suggesting it is a cytoplasmic protein. The most salient feature of EPI64 is the presence of the ∼200–amino acid TBC/rabGAP domain between residues 111 and 311. These domains are well conserved across species and have been found in a wide range of different proteins from plant adhesion molecules to mammalian oncogenes (Zhang et al. 1996; Neuwald 1997). The name TBC derives from the name of the murine protein, Tbc1, in which this domain was first identified based on its similarity to sequences in the tre-2 oncogene, and the yeast regulators of mitosis, BUB2 and cdc16 (Richardson and Zon 1995). The connection of this domain with rab GTPase activation stems from subsequent in-depth sequence analyses and alignments (Neuwald 1997) and recent work demonstrating that it appears to contain the catalytic activities of the yeast rabGAPs, Gyp1p, and Gyp7p (Albert et al. 1999). The yeast study has far provided the best evidence that these domains function as rabGAPs. In addition to mapping the catalytic activity to this region, Albert et al. 1999 identified a critical arginine residue within this domain that is necessary for the full GAP activities. The recent crystal structure of the TBC/rabGAP domain of Gyp1p supports the idea that this residue functions as a catalytic arginine finger analogous to that seen in ras- and cdc42-GAPs (Rak et al. 2000). Although the overall sequence identity that Gyp1p and Gyp7p share with EPI64 is rather low (∼10%), it is interesting to note that this identity is clustered across motifs within the TBC/rabGAP domain that are best conserved with other TBC/rabGAP–containing proteins (Neuwald 1997; Albert and Gallwitz 1999). Furthermore, the critical arginine in these yeast GAPs is conserved within the TBC/rabGAP domain of EPI64 (Fig. 3 A).
The lack of obvious effects of expressing wild-type or mutant EPI64 in cultured cells on cell morphology or the localization of EBP50, ezrin, or cortical F-actin (Fig. 8 and Fig. 9) suggests that EPI64 has functions independent of this linkage. With its distinct localization to microvilli, it would be uniquely positioned to regulate rab-mediated membrane fusion events, such as those occurring during the regulatory endocytic or exocytic trafficking of vesicles carrying the transmembrane ion channels or receptors to which EBP50 has also been shown to bind. As a rabGAP, EPI64 would be expected to enhance the intrinsic rate of hydrolysis of GTP on the active rab on an incoming vesicle, perhaps providing a signal that it has reached an appropriate target site. With respect to the potential regulation of EPI64 itself, it is interesting to note that EPI64 is differentially phosphorylated, which does not seem to affect its association with EBP50 (Fig. 1 and Fig. 10), but may have a role in directly regulating GAP domain activity or perhaps in modulating the associations EPI64 makes with other ligands.
The assembly of microvillar protein complexes on the different domains of ezrin and EBP50 revealed that EPI64 is a component of a multiprotein complex in these structures. In addition to EPI64 and human p120 nadrin, we also identified the β3 isoform of PLC, and the novel putative calpain-like protease, CANPX, in eluates from the EBP50 PDZ domain beads (Fig. 10). PLC-β3 and CANPX both end in COOH-terminal sequences predicted to bind the PDZ domains of EBP50; thus, it is very likely that they are binding directly to EBP50 in these complexes like EPI64 and nadrin. The evidence presented in Fig. 1 B suggests that CANPX even appears to have a distinct preference for PDZ-1. A recent study by Hwang et al. 2000 demonstrating the binding of PLC-β3 to the second PDZ domain of E3KARP indicates that the interaction of this protein with the PDZ domains of EBP50 is direct. Moreover, Tang et al. 2000 have shown that the β1 and β2 isoforms of PLC can bind the first PDZ domain of EBP50. Interestingly, the recovery of PLC-β3 in our affinity bead eluates was not consistently robust (compare Fig. 1 B and 10) and may reflect a respectively lower affinity for the EBP50 PDZ domains than the other ligands we identified.
The plethora of proteins that have been suggested to interact with the PDZ domains of EBP50 and/or E3KARP begs the critical question of whether all, or just some, might be physiologically important. In Table , the experimentally determined optimal EBP50 PDZ–binding peptide consensus sequences are compared with identified ligands and their COOH-terminal sequences. Many of the identified ligands fit the optimal binding sequences well, but this is a biased set, as many were suggested based on their COOH-terminal sequences alone. Aside from the proteins identified in this study, only the β2AR was identified as a potential ligand biochemically and without regard to sequence. An independent criterion might be whether the identified ligand is colocalized in cells and subcellular structures with EBP50 and/or E3KARP. For example, in cultured cells, EBP50 colocalizes very closely with ezrin (Reczek et al. 1997), and EPI64 colocalizes precisely with EBP50. However, such an analysis has not yet been achieved for most of the other candidates. Is it possible that many different proteins interact with the PDZ domains of EBP50? Ezrin and EBP50 are both major components of isolated placental microvilli (Berryman et al. 1995; Reczek et al. 1997), yet the candidates that bind the PDZ domains of EBP50 appear to be less abundant. This apparent difference in stoichiometry would suggest that many different proteins can bind EBP50 even in one structure, and the finding that many different EBP50 binding candidates can be retained with EBP50 from extracts of placental microvilli argues for this heterogeneity. If this is indeed the case, it would offer one explanation as to how the presence of EPI64 in such complexes might influence the trafficking of other EBP50-associated proteins such as the β2AR, CFTR, or NHE3. It might also allow a protease like CANPX to locally remodel the cytoskeleton to facilitate such events. It will therefore be very important in the future to determine binding hierarchies and how the binding of one protein to a PDZ domain might affect the binding of other proteins to that or the adjacent PDZ domain in EBP50.
Table 1.
COOH-terminal Consensus Sequences and Ligands for the PDZ Domains of EBP50 and E3KARP
| Sequence position | PDZ preference | Location | References | ||||
|---|---|---|---|---|---|---|---|
| −3 | −2 | −1 | 0 | ||||
| Peptide consensus | |||||||
| D | S/T | R/F/Y | L | PDZ-1 | — | Wang et al. 1998 | |
| D | S/T | W | L | PDZ-2 | — | Wang et al. 1998 | |
| D | S/T | A/R/F/L | L | PDZ-1 | — | Hall et al. 1998a | |
| Protein | |||||||
| EPI64 | D | T | Y | L | PDZ-1 | Microvillar | This study |
| CFTR | D | T | R | L | PDZ-1 | Apical | Hall et al. 1998a; Short et al. 1998; Wang et al. 1998 |
| PLC-β1 | D | T | P | L | PDZ-1 | — | Tang et al. 2000 |
| H+ATPase β1 | D | T | A | L | PDZ-1 | — | Breton et al. 2000 |
| P2YR | D | T | S | L | PDZ-1 | — | Hall et al. 1998a |
| β2AR | D | S | S | L | PDZ-1 | — | Hall et al. 1998b |
| PDGFR | D | S | F | L | PDZ-1 | — | Maudsley et al. 2000 |
| Trp4 | T | T | R | L | PDZ-1 | — | Tang et al. 2000 |
| Trp5 | T | T | R | L | PDZ-1 | — | Tang et al. 2000 |
| PLC-β2 | E | S | R | L | PDZ-1 | — | Tang et al. 2000 |
| Nadrin | S | T | A | L | PDZ-1 | — | This study |
| CANPX | L | T | E | L | PDZ-1 | — | This study |
| PLC-β3 | N | T | Q | L | PDZ-1/-2 | — | This study; Hwang et al. 2000 |
| GRK6A | P | T | R | L | PDZ-1 | — | Hall et al. 1999 |
| YAP65 | L | T | W | L | PDZ-2 | Apical | Mohler et al. 1999 |
Heterogeneous scaffolding complexes are becoming a common feature of eukaryotic cell organization, with the best example being those of the PSD in the dendritic spines of neurons (Sheng and Pak 2000). In these complexes, there is also precedence for an EBP50–EPI64-like interaction where the PDZ domain–containing protein PSD-95 associates with the rasGAP, SynGAP (Chen et al. 1998; Kim et al. 1998). Another particularly relevant nonneuronal example is the association of the fourth PDZ domain of the FERM domain–containing protein PTPL1 with the rhoGAP, PARG1 (Saras et al. 1997). The functional significance of each of these GAP proteins in their respective cellular milieus has yet to be elucidated. Likewise, determination of the functional implications of the EPI64–EBP50 interaction in the context of the ezrin-based cytoskeleton will be an important challenge for future studies.
Acknowledgments
We are grateful to Dr. C. Yun for the E3KARP cDNA constructs. D. Reczek would like to especially thank Isabel Angeline Comella for providing useful materials for this research.
This work was supported by grant GM36652 from the National Institutes of Health.
Footnotes
Abbreviations used in this paper: β2AR, β2-adrenergic receptor; CFTR, cystic fibrosis transmembrane conductance regulator; E3KARP, NHE3 kinase A regulatory protein; EBP50, ERM-binding phosphoprotein 50; EPI64; EBP50–PDZ interactor of 64; ERM, ezrin/radixin/moesin; ERMAD, ERM association domain; FERM, 4.1 ERM; GRK6A, G protein–coupled receptor kinase 6A; GST, glutathione S-transferase; PLC-β3, phospholipase-C-β3; PSD, postsynaptic density; TBC, Tre/Bub2/Cdc16; PDZ, PSD-95/DlgA/ZO-1–like.
References
- Albert S., Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Albert S., Will E., Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret C., Roy C., Montcourrier P., Mangeat P., Niggli V. Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP2) binding site in the NH2-terminal domain of ezrin correlates with its altered cellular distribution. J. Cell Biol. 2000;151:1067–1080. doi: 10.1083/jcb.151.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M., Bretscher A. Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol. Biol. Cell. 2000;11:1509–1521. doi: 10.1091/mbc.11.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M., Franck Z., Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J. Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- Berryman M., Gary R., Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvillia proposal for their involvement in cortical morphogenesis. J. Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S., Wiederhold T., Marshansky V., Nsumu N.N., Ramesh V., Brown D. The B1 subunit of the H+ ATPase is a PDZ domain-binding protein. Colocalization with NHE-RF in renal B-intercalated cells. J. Biol. Chem. 2000;275:18219–18224. doi: 10.1074/jbc.M909857199. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J. Cell Biol. 1983;97:425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A., Chamber C., Nguyen R., Reczek D. ERM-merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- Cao T.T., Deacon H.W., Reczek D., Bretscher A., von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- Chen H.J., Rojas-Soto M., Oguni A., Kennedy M.B. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- Chishti A.H., Kim A.C., Marfatia S.M., Lutchman M., Hanspal M., Jindal H., Liu S.C., Low P.S., Rouleau G.A., Mohandas N. The FERM domaina unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein- binding domainmolecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Fouassier L., Yun C.C., Fitz J.G., Doctor R.B. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J. Biol. Chem. 2000;275:25039–25045. doi: 10.1074/jbc.C000092200. [DOI] [PubMed] [Google Scholar]
- Franck Z., Gary R., Bretscher A. Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J. Cell Sci. 1993;105:219–231. doi: 10.1242/jcs.105.1.219. [DOI] [PubMed] [Google Scholar]
- Gary R., Bretscher A. Heterotypic and homotypic associations between ezrin and moesin, two putative membrane-cytoskeletal linking proteins. Proc. Natl. Acad. Sci. USA. 1993;90:10846–10850. doi: 10.1073/pnas.90.22.10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R., Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Ostedgaard L.S., Premont R.T., Blitzer J.T., Rahman N., Welsh M.J., Lefkowitz R.J. A C-terminal motif found in the beta2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins Proc. Natl. Acad. Sci. USA 95 1998. 8496 8501a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Premont R.T., Chow C.W., Blitzer J.T., Pitcher J.A., Claing A., Stoffel R.H., Barak L.S., Shenolikar S., Weinman E.J. The beta2-adrenergic receptor interacts with the Na+/H+ exchanger regulatory factor to control Na+/H+ exchange Nature 392 1998. 626 630b [DOI] [PubMed] [Google Scholar]
- Hall R.A., Spurney R.F., Premont R.T., Rahman N., Blitzer J.T., Pitcher J.A., Lefkowitz R.J. G protein-coupled receptor kinase 6A phosphorylates the Na+/H+ exchanger regulatory factor via a PDZ domain-mediated interaction. J. Biol. Chem. 1999;274:24328–24334. doi: 10.1074/jbc.274.34.24328. [DOI] [PubMed] [Google Scholar]
- Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Furuta B., Takeuchi K., Itakura M., Takahashi M., Umeda M. Nadrin, a novel neuron-specific GTPase-activating protein involved in regulated exocytosis. J. Biol. Chem. 2000;275:36885–36891. doi: 10.1074/jbc.M004069200. [DOI] [PubMed] [Google Scholar]
- Heiska L., Alfthan K., Gronholm M., Vilja P., Vaheri A., Carpen O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5- bisphosphate. J. Biol. Chem. 1998;273:21893–21900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- Helander T.S., Carpen O., Turunen O., Kovanen P.E., Vaheri A., Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- Hirao M., Sato N., Kondo T., Yonemura S., Monden M., Sasaki T., Takai Y., Tsukita S., Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane associationpossible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Wong T.Y., Lin R.C., Furthmayr H. Replacement of threonine 558, a critical site of phosphorylation of moesin in vivo, with aspartate activates F-actin binding of moesin. Regulation by conformational change. J. Biol. Chem. 1999;274:12803–12810. doi: 10.1074/jbc.274.18.12803. [DOI] [PubMed] [Google Scholar]
- Hwang J.I., Heo K., Shin K.J., Kim E., Yun C., Ryu S.H., Shin H.S., Suh P.G. Regulation of phospholipase C-β 3 activity by Na+/H+ exchanger regulatory factor 2. J. Biol. Chem. 2000;275:16632–16637. doi: 10.1074/jbc.M001410200. [DOI] [PubMed] [Google Scholar]
- Keller P., Simons K. Post-Golgi biosynthetic trafficking. J. Cell Sci. 1997;110:3001–3009. doi: 10.1242/jcs.110.24.3001. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Liao D., Lau L.F., Huganir R.L. SynGAPa synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- Kornau H.C., Seeburg P.H., Kennedy M.B. Interaction of ion channels and receptors with PDZ domain proteins. Curr. Opin. Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legg J.W., Isacke C.M. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr. Biol. 1998;8:705–708. doi: 10.1016/s0960-9822(98)70277-5. [DOI] [PubMed] [Google Scholar]
- Magendantz M., Henry M.D., Lander A., Solomon F. Interdomain interactions of radixin in vitro. J. Biol. Chem. 1995;270:25324–25327. doi: 10.1074/jbc.270.43.25324. [DOI] [PubMed] [Google Scholar]
- Mangeat P., Roy C., Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S., Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley S., Zamah A.M., Rahman N., Blitzer J.T., Luttrell L.M., Lefkowitz R.J., Hall R.A. Platelet-derived growth factor receptor association with Na+/H+ exchanger regulatory factor potentiates receptor activity. Mol. Cell. Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler P.J., Kreda S.M., Boucher R.C., Sudol M., Stutts M.J., Milgram S.L. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J. Cell Biol. 1999;147:879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A., Gonzalez-Agosti C., Cordero E., Pinney D., Candia C., Solomon F., Gusella J., Ramesh V. NHE-RF, a regulatory cofactor for Na+/H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J. Biol. Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- Nakamura F., Amieva M.R., Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J. Biol. Chem. 1995;270:31377–31385. doi: 10.1074/jbc.270.52.31377. [DOI] [PubMed] [Google Scholar]
- Nakamura F., Huang L., Pestonjamasp K., Luna E.J., Furthmayr H. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell. 1999;10:2669–2685. doi: 10.1091/mbc.10.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem. Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- Nguyen R., Reczek D., Bretscher A. Hierarchy of merlin and ezrin N- and C-terminal domain interactions in homo- and heterotypic associations and their relationship to binding of scaffolding proteins EBP50 and E3KARP. J. Biol. Chem. 2001;276:7621–7629. doi: 10.1074/jbc.M006708200. [DOI] [PubMed] [Google Scholar]
- Niggli V., Andreoli C., Roy C., Mangeat P. Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 1995;376:172–176. doi: 10.1016/0014-5793(95)01270-1. [DOI] [PubMed] [Google Scholar]
- Oakley B.R., Kirsch D.R., Morris N.R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Pearson M., Reczek D., Bretscher A., Karplus P. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp K., Amieva M.R., Strassel C.P., Nauseef W.M., Furthmayr H., Luna E.J. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol. Biol. Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietromonaco S.F., Simons P.C., Altman A., Elias L. Protein kinase C-theta phosphorylation of moesin in the actin-binding sequence. J. Biol. Chem. 1998;273:7594–7603. doi: 10.1074/jbc.273.13.7594. [DOI] [PubMed] [Google Scholar]
- Rak A., Fedorov R., Alexandrov K., Albert S., Goody R.S., Gallwitz D., Scheidig A.J. Crystal structure of the GAP domain of gyp1pfirst insights into interaction with Ypt/Rab proteins. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:5105–5113. doi: 10.1093/emboj/19.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D., Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino- terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Reczek D., Berryman M., Bretscher A. Identification of EBP50a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.M., Zon L.I. Molecular cloning of a cDNA with a novel domain present in the tre-2 oncogene and the yeast cell cycle regulators BUB2 and cdc16. Oncogene. 1995;11:1139–1148. [PubMed] [Google Scholar]
- Rouleau G.A., Merel P., Lutchman M., Sanson M., Zucman J., Marineau C., Hoang-Xuan K., Demczuk S., Desmaze C., Plougastel B. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Saras J., Heldin C.H. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem. Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- Saras J., Franzen P., Aspenstrom P., Hellman U., Gonez L.J., Heldin C.H. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J. Biol. Chem. 1997;272:24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- Serrador J.M., Alonso-Lebrero J.L., del Pozo M.A., Furthmayr H., Schwartz-Albiez R., Calvo J., Lozano F., Sanchez-Madrid F. Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 1997;138:1409–1423. doi: 10.1083/jcb.138.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador J.M., Nieto M., Alonso-Lebrero J.L., del Pozo M.A., Calvo J., Furthmayr H., Schwartz-Albiez R., Lozano F., Gonzalez-Amaro R., Sanchez-Mateos P., Sanchez-Madrid F. CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood. 1998;91:4632–4644. [PubMed] [Google Scholar]
- Sheng M., Pak D.T. Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu. Rev. Physiol. 2000;62:755–778. doi: 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- Short D.B., Trotter K.W., Reczek D., Kreda S.M., Bretscher A., Boucher R.C., Stutts M.J., Milgram S.L. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Tang Y., Tang J., Chen Z., Trost C., Flockerzi V., Li M., Ramesh V., Zhu M.X. Association of mammalian Trp4 and phospholipase C isozymes with a PDZ domain-containing protein, NHERF. J. Biol. Chem. 2000;275:37559–37564. doi: 10.1074/jbc.M006635200. [DOI] [PubMed] [Google Scholar]
- Trofatter J.A., MacCollin M.M., Rutter J.L., Murrell J.R., Duyao M.P., Parry D.M., Eldridge R., Kley N., Menon A.G., Pulaski K. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;75:826. doi: 10.1016/0092-8674(93)90501-g. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen O., Wahlstrom T., Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Raab R.W., Schatz P.J., Guggino W.B., Li M. Peptide binding consensus of the NHE-RF-PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR) FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- Weinman E.J., Minkoff C., Shenolikar S. Signal complex regulation of renal transport proteinsNHERF and regulation of NHE3 by PKA. Am. J. Physiol. Renal. Physiol. 2000;279:F393–F399. doi: 10.1152/ajprenal.2000.279.3.F393. [DOI] [PubMed] [Google Scholar]
- Yang Q., Tonks N.K. Isolation of a cDNA clone encoding a human protein-tyrosine phosphatase with homology to the cytoskeletal-associated proteins band 4.1, ezrin, and talin. Proc. Natl. Acad. Sci. USA. 1991;88:5949–5953. doi: 10.1073/pnas.88.14.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S., Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998;140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.H., Oh S., Zizak M., Steplock D., Tsao S., Tse C.M., Weinman E.J., Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.H., Lamprecht G., Forster D.V., Sidor A. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J. Biol. Chem. 1998;273:25856–25863. doi: 10.1074/jbc.273.40.25856. [DOI] [PubMed] [Google Scholar]
- Zhang S.D., Kassis J., Olde B., Mellerick D.M., Oderwald W.F. Pollux, a novel Drosophila adhesion molecule, belongs to a family of proteins expressed in plants, yeast, nematodes, and man. Genes Dev. 1996;10:1108–1119. doi: 10.1101/gad.10.9.1108. [DOI] [PubMed] [Google Scholar]