Abstract
We examined the mobilities of nucleolar components that act at various steps of the ribosome biogenesis pathway. Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) analyses demonstrate that factors involved in rRNA transcription (upstream-binding factor [UBF]), processing (nucleolin, fibrillarin, and RNase MRP subunits, Rpp29), and ribosome assembly (B23) exchange rapidly between the nucleoplasm and nucleolus. In contrast, the mobilities of ribosomal subunit proteins (S5, L9) are much slower. Selective inhibition of RNA polymerase I transcription does not prevent the exchanges but influences the rates of exchange differentially for different nucleolar components. These findings suggest that the rapid exchange of nucleolar components between the nucleolus and nucleoplasm may represent a new level of regulation for rRNA synthesis. The different dynamic properties of proteins involved in different steps of ribosome biogenesis imply that the nucleolar association of these proteins is due to their specific functional roles rather than simply their specific nucleolar-targeting events.
Keywords: nucleolar proteins, ribosome biogenesis, dynamics, living cells
Introduction
The nucleolus is a prominent nonmembrane-bound nuclear substructure that organizes around chromosome segments containing nucleolar-organizing regions (NORs). It is the center of rDNA transcription and ribosome biogenesis (for reviews see Busch and Smetana 1970; Scheer and Hock 1999; Carmo-Fonseca et al. 2000; Olson et al. 2000). More recently, additional functions have been attributed to the nucleolus, including cell cycle regulation, telomerase activity, signal recognition particle biogenesis, p53 metabolism, small RNA processing, and mRNA transport (for reviews see Pederson 1998; Garcia and Pillus 1999; Olson et al. 2000).
Ribosome biogenesis involves rRNA synthesis, maturation, and assembly of rRNA and ribosomal proteins into the large and small ribosome subunits. This process is regulated throughout the cell cycle, primarily at the level of rRNA synthesis (for review see Hannan et al. 1998). rDNA transcription peaks during the S and G2 phases, stops as cells enter mitosis (for review see Grummt 1999), and then reactivates as cells exit from mitosis. The transcription–initiation complex consists of an upstream-binding factor (UBF)1, SL1 factors containing the TATA-binding protein (TBP), and RNA polymerase (pol) I. It remains to be clarified whether the initiation complex is assembled onto the DNA template in a stepwise fashion or as a preassembled complex (for reviews see Sollner-Webb and Tower 1986; Reeder 1989; Sollner-Webb and Mougey 1991; Moss and Stefanovsky 1995; Grummt 1999). The pol I transcription machinery is associated with the NORs at all times, including the period of transcription silencing during mitosis (Scheer and Rose 1984; Roussel et al. 1996; Gebrane-Younes et al. 1997). Newly synthesized pre-rRNAs undergo a complex series of modifications including 3′-external transcribed spacer cleavage, 2′-O-ribose methylation, and pseudouridylation, followed by 5′-external transcribed spacer and internal transcribed spacer elimination (for review see Venema and Tollervey 1999). The steps and mechanisms involved in the processing of pre-rRNA have been investigated extensively in many organisms both in vivo and in vitro, especially in yeast (Venema and Tollervey 1999). Many factors have been shown to participate in various steps of the processing. These extensive modifications generate 18S, 5.8S, and 28S rRNA. The mature rRNAs are subsequently assembled with ribosomal proteins into preribosomal particles in the nucleolus.
In spite of extensive and detailed analyses in the steps and mechanisms regarding the syntheses and assembly of the ribosome, little is known as to the spatial and temporal dynamics of these processes in living cells. For example, it is not clear whether specific proteins are stably associated with the nucleolus or are exchanged with the nucleoplasm. The mechanisms that control their entry and exit from the nucleolus are also unknown, as well as those that help engage or disengage them from their tasks. To begin addressing these problems, we have compared the mobilities of factors involved in various steps of ribosome biogenesis in living cell nucleoli. FRAP and fluorescence loss in photobleaching (FLIP) were used to determine the movement of green fluorescent protein (GFP)-tagged proteins. The nucleolar components investigated in this report include UBF1, nucleolin, fibrillarin, Rpp29, B23, and ribosomal proteins S5 and L9. UBF1 is involved in rDNA transcription (Grummt 1999). Nucleolin, fibrillarin, and an RNase MRP subunit, Rpp29, have been shown to participate in various steps of pre-rRNA processing (Venema and Tollervey 1999). Nucleolin and B23 are involved in ribosome assembly and other functions (Ginisty et al. 1999; Szebeni and Olson 1999; Philpott et al. 2000). S5 and L9 are ribosomal proteins that are parts of the small and large subunits, respectively.
Comparisons of the mobilities among the examined GFP fusion proteins reveal that proteins involved in different steps of ribosome biogenesis have different dynamics in living cells. UBF1, nucleolin, B23, fibrillarin, and Rpp29 rapidly exchange between the nucleolus and nucleoplasm, whereas ribosomal proteins move relatively slower. Selective inhibition of pol I transcription activity does not prevent the movement of any of the examined proteins but does differentially influence the rate of movement for various factors. These findings provide new insights into the temporal and spatial dynamics of proteins involved in ribosome biogenesis.
Materials and Methods
Cell Culture and Transfection
HeLa cells were maintained in DME supplemented with 10% FBS at 37°C and 5% CO2. Cells were diluted twice a week. Expression constructs were transiently transfected into HeLa cells by electroporation (Sambrook et al. 1989). In brief, subconfluent cells in a 100-mm culture dish were collected by trypsinization and mixed with 20 μg of DNA, including 4 μg target DNA and 16 μg sheared salmon sperm DNA. A 280-μl mixture of cells in DME containing 10% FBS and DNA was electroporated in a Bio-Rad Laboratories electroporator at 250 V and 950 μF. Cells were subsequently seeded onto glass coverslips that were mounted on the bottom of 35-mm petri dishes with an opening in the center (Mactek) and grown for 48 h. Cells were incubated in 0.04 μg/ml actinomycin D (ActD) for 2 h to selectively inhibit pol I transcription before observation.
Construction of GFP Fusion Proteins
GFP-Rpp29 was provided by Dr. S. Altman (Yale University, New Haven, CT) (Jarrous et al. 1999). All other GFP fusion proteins were constructed using PCR cloning into pEGFP-C1 (CLONTECH Laboratories, Inc.). In all cases, GFP was fused to the NH2 terminus of the proteins. The primers that amplified UBF1 are GGGGTACCATGAACGGAGAAGCCGACTGC for the NH2 terminus and CGGGATCCCGTCAGTTGGAGTCAGAGTCTGAGGA for the COOH terminus. Those for nucleolin are GGGGTACCATGGTGAAGCTCGCGAAGGCA for the NH2 terminus and CGGGATCCCGCTATTCAAACTTCGTCTTCTTTCC for the COOH terminus. Fibrillarin primers are GGGGTACCATGAAGCCAGGATTCAGTCCC for the NH2 terminus and CGGGATCCTCAGTTCTTCACCTTGGGGGG for the COOH terminus. B23 primers are GGGGTACCATGGAAGATTCGATGGACATG for the NH2 terminus and CGGGATCCTTAAAGAGACTTCCTCCACTG for the COOH terminus. Ribosomal protein S5 primers are GGGGTACCATGACCGAGTGGGAGACAGCA for the NH2 terminus and CGGGATCCTCAGCGGTTGGACTTGGCCAC for the COOH terminus. Ribosomal protein L9 primers are GGGGTACCATGAAGACTATTCTCAGCAAT for the NH2 terminus and CGGGATCCTTATTCATCAGCCTGCTGAAC for the COOH terminus. All fusion constructs were sequenced and shown to be faithful copies of the corresponding genes.
Immunolabeling after Transfection
48 h after transfection, cells were fixed with 2% paraformaldehyde for 15 min and permeabilized with 0.3% Triton X-100 for 5 min. Antibodies specifically recognizing UBF (Chan et al. 1991), nucleolin (Pinol-Roma 1999), fibrillarin (Sigma-Aldrich), B23 (Santa Cruz Biotechnology, Inc.), and ribosomal S6 (Chan and Wool 1988) were incubated with cells for 1 h at room temperature. The immunolabeling signals were subsequently detected by incubating cells with Texas red–conjugated secondary antibodies.
Photobleaching and Live Cell Imaging
48 h after transfection, cells were maintained in DME supplemented with 30 mM Hepes, pH 7.1, to stabilize the pH of the medium during imaging. The 35-mm dishes with coverslip bottoms were directly mounted onto a ZEISS 510 confocal laser scanning microscope equipped with an argon-krypton laser (ZEISS). The medium was kept at 37°C using an ASI 400 Air Stream incubator (Nevtek). The 488-nm laser and a 63× plan Apo lens with a 1.4 NA were used in bleaching and imaging experiments. A laser power of 1.1% of 3.75 mW was used in image acquisitions, and 100% of 3.75 mW was used in photobleaching. The time for each image acquisition is 3.9 s, which did not significantly influence the fluorescent intensity through multiple acquisitions. An area of 2 μm2 was bleached with an iteration of 60. In FRAP analyses, images were collected before, immediately after, and at 9-s intervals after bleaching for the nucleolar FRAP and 1-s intervals for the nucleoplasmic FRAP. For FLIP analyses, an image was collected before and then after every 20 s of bleaching. At least 10 data sets were analyzed for each result. Photobleach analyses of GFP-tagged molecules in living cells or organisms raise concerns since the results could be influenced by phototoxicity. Several studies investigating this problem have shown that photobleaching using a low laser power does not significantly damage the examined cells (White and Stelzer 1999; Kruhlak et al. 2000; Phair and Misteli 2000). In addition, we have monitored cells over 24 h after receiving similar and higher doses of laser irradiation that we used in these studies. The results showed that cells survived well and that some underwent mitosis during this period of time.
Quantitation of Relative Fluorescence Intensity
Fluorescence intensity was measured using Metamorph (Universal Imaging Corp.) imaging software. The average intensities of the areas of interest in images, including before, immediately after, and a series of time points after bleaching, were measured under the same condition for each data set. The fluorescence intensity of a nonbleached nucleolus in the same nucleus was also measured. The relative fluorescence intensity (RFI) in the FRAP analyses was calculated as RFI = (Net/N11)/(Ne0/N10). Net is the average intensity of the bleached nucleolus at various time points after bleaching. N1t is the average intensity of the control nonbleached nucleolus at the corresponding time points. Ne0 is the average intensity of the bleached nucleolus before bleaching. N10 is the average intensity of a control nonbleached nucleolus in the same nucleus before bleaching. When Ne0/N10 equals Net/N1t, namely when RFI is 1, fluorescence recovery of the bleached nucleolus reaches 100%. The Net of the images acquired immediately after bleaching were either set at 0 or used as they were. Both calculations show the same type of dynamics during the fluorescence recovery with small differences in their rates at the beginning of the recovery. The Net of the images acquired immediately after bleaching equal 0 and are used to represent the raw data in this report. Using this equation, we have taken into consideration the overall fluorescence change if any during subsequent image acquisitions. For FLIP analyses, images were taken at 20-s intervals of each bleaching. The RFI in FLIP analyses was calculated as RFI = Net/Ne0. The effective diffusion coefficients (D) were calculated as described by Endow and Piston 1998 and Yguerabide et al. 1982. In brief, two equations were used: F I/F 0 = K −1(1−e− K), where F I represents the fluorescence intensity immediately after bleaching, and F 0 represents the fluorescence intensity before bleaching; and D = βw 2/4 t 1/2, where the β value is derived from K and w represents the width of the bleaching area (Yguerabide et al. 1982).
Results
GFP Fusion Proteins Behave Similarly to Endogenous Proteins
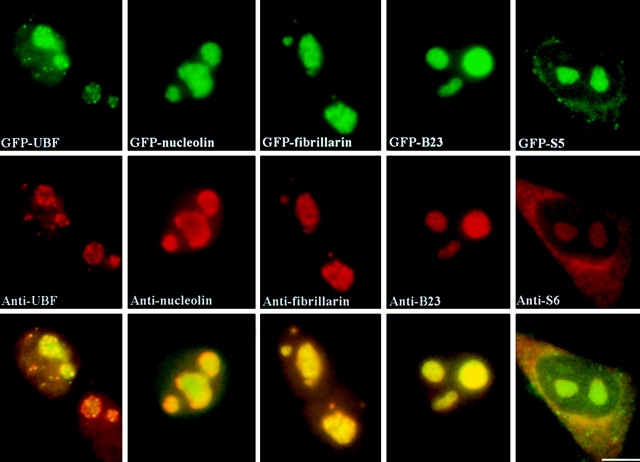
The GFP-tagged proteins used in these studies included GFP-UBF1, -nucleolin, -fibrillarin, -Rpp29 (Jarrous et al. 1999), -B23, and the ribosomal subunit proteins -S5 and -L9. The expression vectors were transiently transfected into HeLa cells, and the localizations of GFP fusion proteins were compared with the endogenous proteins. Cells were examined 48 h after transfection since cells overexpressing fusion proteins at a cytotoxic level underwent cell death by this time, whereas cells that successfully progressed through at least one cell division could be easily identified as sister cells were often found near one another. Therefore, the level of GFP fusion protein expressed in these surviving cells was within a physiologically tolerable concentration. All GFP fusion proteins are localized to the same subcellular regions as their endogenous counterparts, as shown previously and also as determined by immunostaining using the respective antibodies (Fig. 1). Due to the lack of antibodies to ribosomal proteins S5 and L9, an anti-S6 antibody was used to evaluate the localization of these ribosome proteins. GFP-UBF1, -fibrillarin, -nucleolin, -Rpp29, and -B23 were localized in the nucleolus and were also diffusely distributed throughout the nucleoplasm. GFP-S5 and -L9 were concentrated both in the nucleolus and in the cytoplasm (Fig. 1). Western blot analyses using anti-GFP antibody (CLONTECH Laboratories, Inc.) showed the expected size of fusion proteins in transfected cells, demonstrating that the fusion proteins were expressed as full-length proteins (data not shown). Furthermore, the GFP fusion proteins exhibited biochemical extraction profiles similar to their endogenous counterparts. Extractions with 100 mM KCl and 0.1% Triton X-100 did not significantly alter the localization of either endogenous proteins or GFP fusion proteins. Extractions with 400 mM KCl and 0.1% Triton X-100 removed most of the nucleoplasmic UBF1, fibrillarin, nucleolin, Rpp29, and B23. However, substantial portions of these proteins remained associated with nucleoli (data not shown). A large proportion of GFP-S5 and -L9 were observed in both the nucleolus and the cytoplasm following the 400-mM KCl extraction as was the case for the endogenous proteins (data not shown). In addition, GFP-S5 and -L9 were also detected in the ribosome fraction using subcellular fractionation assays (data not shown). These results demonstrate that the GFP fusion proteins are similar to their endogenous counterparts with regard to their localization and biochemical behavior. To best represent the native proteins, cells with the minimal expression level of the GFP fusion proteins were generally chosen for observations throughout this study.
Figure 1.
GFP fusion proteins and their corresponding endogenous proteins localize to the same subcellular regions. Top panels show the localization of GFP fusion proteins as indicated. The middle panels show the localization of the endogenous proteins immunolabeled with specific antibodies. Lower panels show the mergers of the corresponding red and green panels. Bar, 10 μm.
Nucleolar Components of Ribosome Biogenesis Move Rapidly
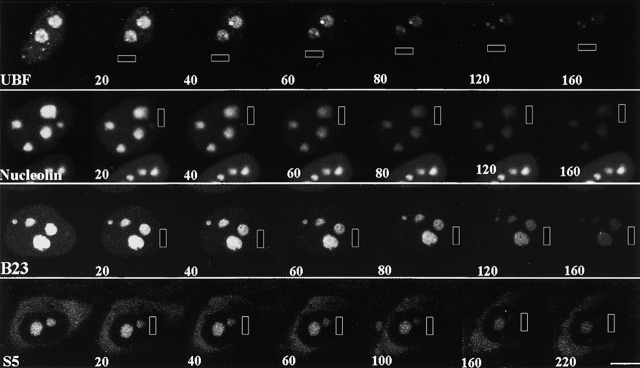
The mobility of factors that are involved in different steps of rRNA transcription, processing, and ribosomal particle assembly were evaluated using FRAP analyses. HeLa cells were transfected with GFP-UBF1, -nucleolin, -fibrillarin, -Rpp29, -B23, -S5, or -L9. 48 h after transfection, FRAP analyses were performed using a ZEISS 510 laser scanning microscope. Selected regions of the nucleolus, either a portion of or an entire nucleolus, were bleached using the 488-nm laser line. Images were obtained immediately after bleaching and subsequently every 9 s for ≤5 min (Fig. 2). Changes in fluorescence intensity within bleached areas were quantitatively measured at each time point (see Materials and Methods). The FRAP rates of GFP-UBF1, -nucleolin, -fibrillarin, -Rpp29, and -B23 are rapid (Fig. 2). Similar recovery rates for GFP-fibrillarin have been recently described by Phair and Misteli 2000. The half times (t 1/2) for the fluorescence recovery of these fusion proteins are within 20 s (Fig. 3 and Table ). The fluorescence recovery in the bleached nucleolus is unlikely to be due to spontaneous recovery of the bleached GFP protein since the fluorescence did not recover in formaldehyde-fixed cells (data not shown). In addition, previous studies have also shown that GFP fusion proteins of less mobile molecules, such as histone H2B or nuclear lamin B, do not recover bleached fluorescence for long periods of time (Moir et al. 2000; Phair and Misteli 2000). Thus, the fluorescence recovery of the bleached area resulted chiefly from the influx of the unbleached GFP fusion proteins. Photobleaching of an entire nucleus prevented detectable fluorescence recovery within 5 min, suggesting that the vast majority of the GFP fusion proteins entering the bleached nucleolus were derived from preexisting nuclear GFP fusion proteins (data not shown). This notion is further supported by the observation that inhibition of protein synthesis by cycloheximide treatment for 2 h did not significantly affect the fluorescence recovery (data not shown). These findings demonstrate that there is a rapid exchange of the GFP fusion proteins between the nucleolus and nucleoplasm.
Figure 2.
FRAP analyses demonstrate that the examined nucleolar components exchange rapidly between the nucleolus and nucleoplasm. Arrowheads indicate the sites of bleaching and numbers represent the time (s) after photobleaching. BL is the first image obtained immediately after photobleaching. Bar, 10 μm.
Figure 3.
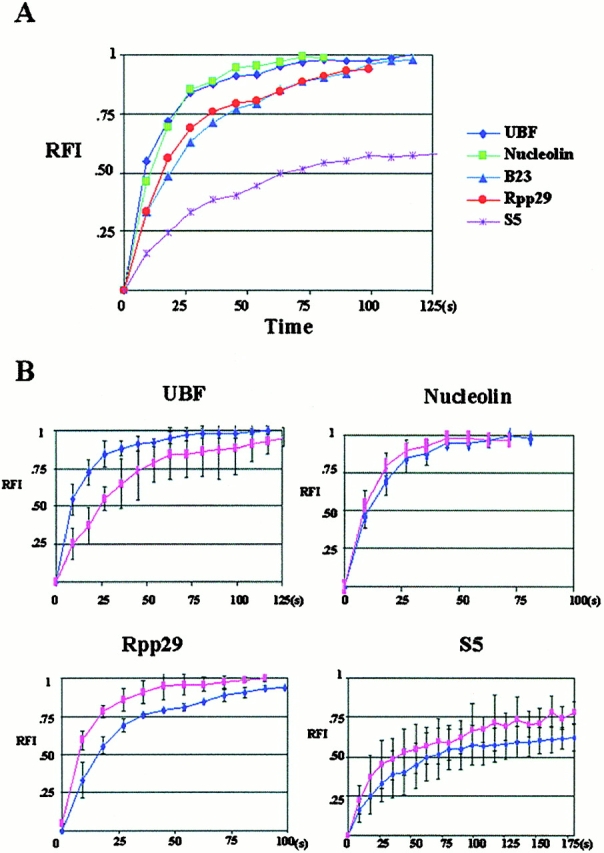
Quantitative analyses of FRAP demonstrate that the FRAP rate of GFP ribosomal proteins is significantly slower than UBF and some of the factors involved in pre-rRNA processing. x, time after the photobleaching (s); y, relative fluorescence intensity in the testing nucleolus. (A) A summary of FRAP rates of various nucleolar components examined in this study. (B) Comparisons of the FRAP rates of some of the examined factors in cells treated or not treated with a low concentration of ActD that selectively inhibits pol I transcription. The dark blue line indicates the FRAP rate in untreated cells, and the pink line indicates the FRAP rate in treated cells.
Table 1.
Summary of the Diffusion Coefficient (D) of Examined GFP-nucleolar Components
| Proteins | Nucleolus | Nucleoplasm | |||
|---|---|---|---|---|---|
| t 1/2 recovery | D | t 1/2 recovery | D | ||
| s | μm2/s | s | μm2/s | ||
| GFP-UBF | Non | 9 | 0.13667 | 2.8 | 0.56786 |
| ActD low | 25 | 0.05684 | 2.8 | 0.51786 | |
| GFP-nucleolin | Non | 9 | 0.14444 | 1.3 | 1.15385 |
| ActD low | 8 | 0.152941 | 1 | 1.34657 | |
| GFP-Rpp29 | Non | 18 | 0.075 | 1 | 1.5436 |
| ActD low | 8 | 0.164706 | 1.4 | 1.25833 | |
| GFP-B23 | Non | 20 | 0.0785 | 1 | 1.51 |
| ActD low | 17 | 0.085294 | 1 | 1.56748 | |
| S5 | Non | 72 | 0.019028 | 1.2 | 1.258333 |
| ActD low | 32 | 0.042813 | 1 | 1.59 | |
Summary of the t 1/2 recovery and D of examined GFP fusion proteins in the nucleolus and nucleoplasm in cells treated or not treated (Non) with a low concentration of ActD. The calculation of D is detailed in Materials and Methods.
The FRAP rate does not appear to be dependent on the size of the bleached region. However, bleaching a larger region (an entire large nucleolus ranging from 3 to 5 μm in diameter) reduced the level of fluorescence intensity at the maximum recovery in the bleached and nonbleached nucleoli in the same nucleus (for example, UBF1 in Fig. 1). This could be explained by the loss of a substantial portion of the nuclear pool of the emission-competent GFP fusion protein. The loss of fluorescence intensity in the unbleached nucleolus also demonstrates a rapid movement of the protein between multiple nucleoli and the nucleoplasm. Thus, the RFI used in quantifying the fluorescence intensity included the information of changes in fluorescence intensity in the nonbleached nucleolus (see Materials and Methods). The quantitative analyses show that the influx of the nucleolar components follows a growth with a constant rate of decay model (Fig. 3), suggesting a rapid influx of GFP fusion proteins at the early stage of the recovery, subsequently reaching equilibrium where the rate of influx matches the rate of efflux. In addition, the rapid fluorescence recovery of these factors appears to be complete (Fig. 3 A), suggesting that the majority of the proteins are exchangeable and that they are only associated with the nucleolus for a short period of time. Moreover, the nucleoplasmic populations of these components recover much faster (Fig. 4 A) than the nucleolar fractions, suggesting that the two fractions are involved in different activities or are associated with different nuclear factors. Even though the fluorescence intensity, size of the cells, bleaching laser power, and bleaching area were kept similar between the examined cells, there were variations in the FRAP rate from cell to cell that expressed a particular construct (Fig. 3 B and Fig. 4 B).
Figure 4.

Quantitative analyses of FRAP demonstrate that the FRAP rate of all nucleolar GFP fusion proteins are similarly rapid in the nucleoplasm and are not affected during pol I transcription inhibition. x, time after the photobleaching (s); y, relative fluorescence intensity in the testing region. (A) A summary of FRAP rates of various nucleolar components examined in this study. (B) Comparisons of the FRAP rates of some of the examined factors in cells treated or not treated with a low concentration of ActD that selectively inhibits pol I transcription. The dark blue line indicates the FRAP rate in untreated cells, and the pink line indicates the FRAP rate in treated cells.
In comparison to factors involved in rRNA transcription and processing, ribosomal proteins S5 and L9 show relatively slower FRAP rates with a t 1/2 of ∼72 s (FRAP of GFP-S5 is shown in Fig. 3 A and Table ). However, the FRAP rate of these proteins is much more rapid in the nucleoplasm (Fig. 4 A and Table ). S5 and L9 are components of the large and small ribosomal subunits, respectively, and the fluorescence recovery rates of S5 and L9 in the nucleolus probably represent the influx of proteins to be assembled into preribosomal particles. The slower FRAP rate of the ribosomal proteins is consistent with the notion that the assembly of ribosomal subunits is a slower process compared with transcription and rRNA processing.
Nucleolar Components Rapidly Exit the Nucleolus
The FLIP approach was employed to evaluate the nucleolar residence time of various components involved in the ribosome biogenesis pathway. A defined area of the nucleoplasm distant from the nucleolus or nuclear region of measurement was bleached continuously until the entire nucleus was nearly depleted of emission-competent GFP fusion proteins (see Materials and Methods). Images were obtained at 20-s intervals, and the depletion of emission-competent GFP-UBF1, -nucleolin, -fibrillarin, -Rpp29, -B23, -S5, and -L9 from nucleoli and the nucleoplasm was quantified by measuring the RFI at the corresponding locus. If a protein is highly mobile throughout the nucleus, a rapid depletion of fluorescence is expected when one particular nuclear region is bleached repeatedly. The FLIP rates of all GFP fusion proteins were obtained under similar conditions, including cell size, expression level, and the area of bleaching (Fig. 5). The loss of fluorescence at the regions of measurement is not due to the nonspecific bleaching outside of the designated area by the scattered laser light, since cells immediately adjacent to the targeted cell maintained similar fluorescence intensity throughout the period of the bleaching process (for example, Fig. 5, Nucleolin panels). Quantitative analyses demonstrate that the majority of the GFP-UBF1, -nucleolin, -B23, -fibrillarin, and -Rpp29 show a rapid rate of depletion from the nucleolus (Fig. 6) with the t 1/2 ranging from 50 to 80 s. These dynamics follow the exponential decay model, and the fluorescence can be reduced to an undetectable level in all nucleoli of the tested cell. In comparison, the FLIP rates of ribosomal proteins S5 and L9 are slower with t 1/2 ∼ 140 s (Fig. 6). The slower FLIP rate implies a longer residence time of ribosomal proteins in the nucleolus. These FLIP observations are in agreement with the FRAP analyses that nucleolar proteins exchange rapidly between the nucleolus and the nucleoplasm.
Figure 5.
FLIP analyses demonstrate that the examined nucleolar components exit the interphase nucleolus rapidly. The rectangles indicate the areas of bleaching, and the numbers represent the cumulative bleaching duration (s). Bar, 10 μm.
Figure 6.
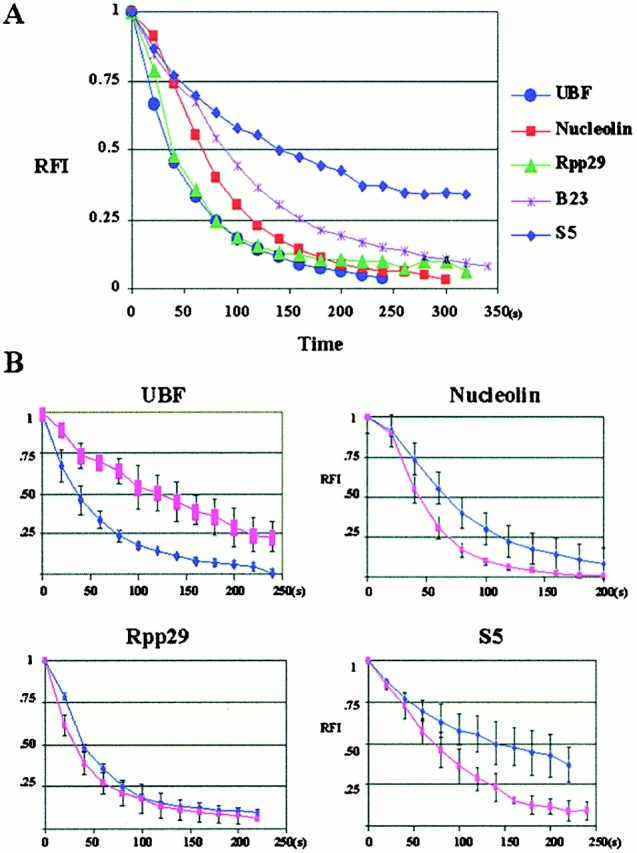
Quantitative analyses of FLIP demonstrate that the GFP ribosomal proteins exit the nucleolus slower than UBF and some of the factors involved in pre-rRNA processing. x, cumulative duration of photobleaching (s); y, relative fluorescence intensity in the testing nucleolus. (A) A summary of FLIP rates of various nucleolar components examined in this study. (B) Comparisons of the FLIP rates of some of the examined factors in cells treated or not treated with a low concentration of ActD that selectively inhibits pol I transcription. The dark blue line indicates the FLIP rate in untreated cells, and the pink line indicates the FLIP rate in treated cells.
Selective Pol I Transcription Inhibition Does Not Prevent the Exchange between the Nucleoplasm and the Nucleolus
To examine the mobility of the nucleolar components when pol I transcription is inhibited, cells were treated with 0.04 μg/ml ActD, which selectively inhibits pol I activity. The FRAP rates of GFP-UBF1, -nucleolin, -B23, -fibrillarin, -Rpp29, -S5, and -L9 were compared with those of untreated cells (Fig. 3 B, Fig. 4 B, and Table ). Inhibition of pol I transcription does not prevent the movement of any of the examined proteins. However, quantitative analyses demonstrate that FRAP (Fig. 3 B and Table ) and FLIP rates (Fig. 5 B) change differentially for different nucleolar components. GFP-UBF1 recovers and loses fluorescence slower, whereas GFP-nucleolin, -fibrillarin, -B23, and -Rpp29 recover and lose fluorescence faster. In comparison, the FRAP and FLIP rates of GFP-S5 and -L9 increase significantly (Fig. 3 B and Fig. 5 B). These results demonstrate that the nucleolar dynamics of factors involved in different steps of ribosome biogenesis respond differently to pol I transcriptional inhibition. In contrast, there is very little change in the mobility of these proteins in the nucleoplasm when pol I transcription is inhibited (Fig. 4 and Table ).
Discussion
Nucleolar Components Rapidly Exchange between the Nucleolus and Nucleoplasm
Our observations demonstrate that components involved in various steps of ribosome biogenesis are not stationary in the nucleolus during interphase but move rapidly between the nucleolus and the nucleoplasm. Our findings of GFP-fibrillarin are similar to the recent study of GFP-fibrillarin in living cells (Phair and Misteli 2000; Snaar et al. 2000). Since the resolution of FRAP and FLIP analyses does not reach the molecular level, our results represent the sum of movement of molecules in all of their possible forms: bound, free, activated, inactivated, or bound to different partners. Nevertheless, these analyses provide basic concepts and allow comparisons between the dynamics of different proteins evaluated under the same conditions. The exchange between the nucleoplasm and nucleolus on a scale of seconds suggests that the examined nucleolar factors (stable proteins) cycle between the two compartments more than once during their lifetime, possibly after each functional act (that is, one round of transcription, pre-rRNA processing, or ribosome assembly). These findings are consistent with several explanations. (a) Proteins may disengage from their sites of function and exchange with the nucleoplasmic pool. The nucleolar association may be the result of assembly into an active complex. This idea is further supported by the observation that the fluorescence recovery of the GFP-nucleolar proteins is nearly 100%, demonstrating that most if not all of the nucleolar population are replaced within a short period of time. (b) Proteins could be inactivated after each round of functional act when the active complex dissociates. The inactivated proteins may exchange with the nucleoplasmic pool before becoming reactivated. Reactivated proteins may then cycle back into the nucleolus and incorporate into active complexes and such a switch could be provided by phosphorylation. In fact, many of the proteins involved in ribosomal biogenesis are regulated by phosphorylation and dephosphorylation (Grummt 1999). For example, cdc2 kinase phosphorylates UBF1, thus inactivating the protein during mitosis (Heix et al. 1998; Kuhn et al. 1998; Klein and Grummt 1999). Alternatively, (c) proteins could cycle through the nucleoplasm as protein complexes without dissociating into single proteins after each functional act. The complex could be recharged or modified before reengaging in activities in the nucleolus. These possibilities are not mutually exclusive. Future studies will attempt to distinguish between these possibilities.
We also found variations between the mobility of nucleolar components involved at different steps of ribosome biogenesis. The simplest explanation is that each step of ribosome biogenesis may require a certain length of time that leads to a defined retention period for the corresponding participants in the nucleolus as reflected in their differential mobilities. Indeed, observations that ribosomal proteins move significantly slower in the nucleolus than UBF and pre-rRNA processing factors are consistent with a slower rate of ribosome assembly as compared with rRNA transcription and processing. The movement of ribosomal proteins may represent the influx of protein subunits and the export of assembled ribosome subunits to the cytoplasm, as opposed to the cyclic movement of the transcription and processing factors between the nucleolus and nucleoplasm. In addition, all of these proteins move more rapidly in the nucleoplasm, while they are not actively engaged in ribosome biogenesis. However, the nucleoplasmic mobility is still significantly lower than GFP alone, suggesting potential large protein complex or enzymatic activities. These findings imply that the nucleolar localization of proteins may be attributed to their specific molecular activities rather than to specific organelle-targeting events such as active nuclear import. This implication is consistent with the notion that no single consensus sequence or motif is responsible for the import of various nucleolar proteins (Scheer and Weisenberger 1994). On the other hand, when pol I transcription is inactivated, nucleolar components continue to be highly concentrated in the structurally segregated nucleolus in spite of the absence of substrates for pre-rRNA processing or ribosome assembly. We speculate that the continuous association with the nucleolus could be explained by a separation of binding to form active complexes and executing specific functions. Proteins may still be capable of forming functionally viable complexes during transcription inhibition. The lack of continuing output of substrates (pre-rRNA) prevents functional acts (pre-rRNA processing and ribosome assembly) from being performed, thus leading to a more rapid dissociation of these active complexes. Our findings that pre-rRNA processing factors and ribosomal proteins move faster through the nucleolus during pol I transcriptional inhibition support this hypothesis. In contrast, GFP-UBF moves slower during transcription inhibition. UBF is involved in the preinitiation of pol I transcription (for review see Grummt 1999). The structural disruption of DNA by ActD could significantly alter the dynamics of the preinitiation complex formation and dissociation leading to the alteration of UBF mobility.
Although our observations are consistent with the possibility that nucleolar factors involved the ribosome biogenesis cycle between the nucleolus and nucleoplasm, it remains to be determined whether these proteins cycle individually or in complexes. We found that several proteins involved in rRNA metabolism, including nucleolin, B23, fibrillarin, and Rpp29, share similar mobility when pol I transcription is either active or inactive. This similarity suggests that factors involved in rRNA processing may be part of a common active complex in the nucleolus, and the complex may not dissociate while cycling through the nucleoplasm. This speculation is consistent with previous studies demonstrating that a specific antibody recognizing nucleolin coimmunoprecipitates both B23 and fibrillarin and that the association is sensitive to RNase treatment (Pinol-Roma 1999). Thus, these factors, at least transiently, are bound to the same complex with their common target, pre-rRNA. More recently, studies from two groups and our laboratory showed that partially processed pre-rRNA complexes reenter newly formed daughter cell nucleoli at the beginning of the cell cycle (Dundr and Olson 1998; Dousset et al. 2000; Dundr et al. 2000). These findings demonstrate that complexes containing pre-rRNA and its processing factors could be present outside of the nucleolus and are able to reenter the nucleolus. These observations open a possibility that some of these components could cycle through the nucleolus in a complex.
In summary, we have demonstrated that components participating in various steps of ribosome biogenesis rapidly cycle between the nucleolus and nucleoplasm, probably at intervals relating to each functional act of rRNA transcription, processing, and ribosome assembly. Proteins involved at different steps of the biogenesis demonstrate different dynamics, suggesting that their nucleolar association may be due to their specific functional activities. However, it remains to be determined whether the cycling may also involve reactivation and modification of specific functional components.
Acknowledgments
We would like to thank Drs. S. Altman, E.D.K. Chan, S. Pinol-Roma, D.L. Spector, and R. Tjian for their generosity in sharing their constructs and antibodies. We are also grateful to D. Leary and Drs. S. Adam, R. Goldman, R. Kamath, R. Moir, T. Spann, D.L. Spector, S. Pendergrast, and B. Feldman for their critical comments in the preparation of the manuscript.
This study was supported by grants from the National Cancer Institute, National Institutes of Health to S. Huang (1 R01 CA 77560-01A1 and 5 K01 CA74988-03).
Footnotes
Abbreviations used in this paper: ActD, actinomycin D; FLIP, fluorescence loss in photobleaching; GFP, green fluorescent protein; NOR, nucleolar-organizing region; pol, RNA polymerase; RFI, relative fluorescence intensity; TBP, TATA-binding protein; UBF, upstream-binding factor.
References
- Busch H., Smetana K. Nucleoli of tumor cells. In: Busch H., editor. The Nucleolus. Academic Press; New York: 1970. [Google Scholar]
- Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Chan E.K.L., Imai H., Hamel J.C., Tan E.M. Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J. Exp. Med. 1991;174:1239–1244. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.L., Wool I.G. The primary structure of rat ribosomal protein S6. J. Biol. Chem. 1988;263:2891–2896. [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Olson M.O.J. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli T., Olson M.O. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., Piston D.W. Methods and protocols. In: Chalfie M., Kain S., editors. GFP Green Fluorescent Protein. Properties, Applications, and Protocols. Wiley-Liss; New York: 1998. pp. 271–360. [Google Scholar]
- Garcia S.N., Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- Gebrane-Younes J., Fomproix N., Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- Hannan K.M., Hannan R.D., Rothblum L.I. Transcription by RNA polymerase I. Front. Biosci. 1998;3:d376–d398. doi: 10.2741/a282. [DOI] [PubMed] [Google Scholar]
- Heix J., Vente A., Voit R., Budde A., Michaelidis T.M., Grummt I. Mitotic silencing of human rRNA synthesisinactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N., Wolenski J.S., Wesolowski D., Lee C., Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J. Cell Biol. 1999;146:559–572. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcriptionthe nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M.J., Lever M.A., Fischle W., Verdin E., Bazett-Jones D.P., Hendzel M.J. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 2000;150:41–51. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Vente A., Doree M., Grummt I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 1998;284:1–5. doi: 10.1006/jmbi.1998.2164. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., Goldman R.D. Nuclear lamins A and B1different pathways of assembly during nuclear envelop formation in living cells. J. Cell Biol. 2000;151:1–15. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Stefanovsky V.Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- Olson M.O., Dundr M., Szebeni A. The nucleolusan old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Philpott A., Krude T., Laskey R.A. Nuclear chaperones. Semin. Cell Dev. Biol. 2000;11:7–14. doi: 10.1006/scdb.1999.0346. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H. Regulatory elements of the generic ribosomal gene. Curr. Opin. Cell Biol. 1989;1:466–474. doi: 10.1016/0955-0674(89)90007-0. [DOI] [PubMed] [Google Scholar]
- Roussel P., Andre C., Comai L., Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs. J. Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning 1989. Cold Spring Harbor Press; Cold Spring Harbor, New York: pp. 1659 [Google Scholar]
- Scheer U., Hock R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Scheer U., Rose K.M. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc. Natl. Acad. Sci. USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Weisenberger D. The nucleolus. Curr. Opin. Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Snaar S., Wiesmeijer K., Jochemsen A.G., Tanke H.J., Dirks R.W. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 2000;151:653–662. doi: 10.1083/jcb.151.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Mougey E.B. News from the nucleolusrRNA gene expression. Trends Biochem. Sci. 1991;16:58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu. Rev. Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Szebeni A., Olson M.O. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae . Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- White J., Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Yguerabide J., Schmidt J.A., Yguerabide E.E. Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Phys. J. 1982;40:69–75. doi: 10.1016/S0006-3495(82)84459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]