Abstract
In the salivary glands of the dipteran Chironomus tentans, a specific messenger ribonucleoprotein (mRNP) particle, the Balbiani ring (BR) granule, can be visualized during its assembly on the gene and during its nucleocytoplasmic transport. We now show with immunoelectron microscopy that actin becomes associated with the BR particle concomitantly with transcription and is present in the particle in the nucleoplasm. DNase I affinity chromatography experiments with extracts from tissue culture cells indicate that both nuclear and cytoplasmic actin are bound to the heterogeneous RNP (hnRNP) protein hrp36, but not to the hnRNP proteins hrp23 and hrp45. The interaction is likely to be direct as purified actin binds to recombinant hrp36 in vitro. Furthermore, it is demonstrated by cross linking that nuclear as well as cytoplasmic actin are bound to hrp36 in vivo. It is known that hrp36 is added cotranscriptionally along the BR mRNA molecule and accompanies the RNA through the nuclear pores and into polysomes. We conclude that actin is likely to be bound to the BR transcript via hrp36 during the transfer of the mRNA from the gene all the way into polysomes.
Keywords: actin, heterogeneous nuclear ribonucleoprotein, premessenger ribonucleoprotein particles, Balbiani rings, nucleocytoplasmic transport
Introduction
Immediately upon transcription, premessenger RNA molecules become associated with proteins to form ribonucleoprotein (RNP) complexes, usually referred to as pre-mRNP or hnRNP (heterogeneous nuclear RNP) particles. The proteins, designated hnRNP proteins, have been extensively characterized (Dreyfuss et al. 1993). In human, for example, there are ∼30 major components (designated A-U) and a large number of minor ones (Krecic and Swanson 1999). The binding of hnRNP proteins to pre-mRNA is nonrandom, and each transcript seems to carry a specific subset of proteins (Matunis et al. 1993; Wurtz et al. 1996). It is likely that the hnRNP proteins package and present the RNA in a manner that affects the fate of the mRNA both in the nucleus and the cytoplasm. It is known that hnRNP proteins regulate splicing and 3′-end processing, retain hnRNA in the nucleus, or mediate RNA transport. Furthermore, some of the proteins leave the nucleus and are involved in processes such as translation, mRNA degradation, and transport of mRNA in the cytoplasm (for recent reviews, see Krecic and Swanson 1999; Nakielny and Dreyfuss 1999).
When further exploring how hnRNP proteins participate in nuclear as well as cytoplasmic events, it is an advantage to be able to follow the fate of individual hnRNP proteins in a specific mRNP particle from the site of synthesis of the mRNA to the site of translation. This has proven feasible in the Balbiani ring experimental system in the salivary glands in Chironomus tentans (Daneholt 1997). The Balbiani rings are exceptionally large, puffed regions of polytene chromosomes and contain genes, 35–40 kb in size, encoding large-sized secretory proteins. The transcription rate is exceptionally high, and the densely spaced transcription products can be visualized while being formed on the gene. Furthermore, the completed products can be readily recognized as Balbiani ring (BR) granules, 50 nm in diameter, in the nucleoplasm and as rod-like structures when passing through the nuclear pores (Mehlin et al. 1992). On the cytoplasmic side, the BR mRNA is immediately engaged in polysome formation and becomes loaded onto the tubular endoplasmic reticulum. Using immunoelectron microscopy, we have studied the flow patterns of several hnRNP proteins (Daneholt 1997). One of the proteins, hrp36, was shown to be added along the BR transcript concomitantly with transcription, to accompany the RNA through the nuclear pore and to be present along the mRNA molecule during translation (Visa et al. 1996). It was proposed that hrp36, like its mammalian homologue hnRNP A1 (Michael et al. 1995), acts as a transport mediator.
In our efforts to identify an export receptor bound to the hnRNP A1-like hrp36, we made the unexpected observation that actin is associated with hrp36. This result prompted us to carry out a systematic study of actin in relation to the assembly and transport of the BR particle.
Materials and Methods
Antibodies
We raised the polyclonal antiactin antibody 9771-4 against complexes of recombinant human cofilin and rabbit skeletal muscle actin. The complexes were prepared from F-actin mixed (∼1:1) with cofilin at pH 8.0 and centrifuged to remove residual F-actin. The antibody was affinity purified on Sepharose-actin.
The affinity-purified, polyclonal, anti–SCP3 antibody was raised against the bacterially expressed rat synaptonemal complex protein SCP3. The monoclonal antisera against hrp23, hrp36, and hrp45 were produced in our laboratory as described (Wurtz et al. 1996; see also Sun et al. 1998).
Preparation of Actin, hrp36, and hrp23
Nonmuscle Actin.
Nonmuscle actin was prepared from calf thymus cells as described by Rozycki et al. 1991.
hrp23.
The hrp23 protein was expressed from a randomly primed λgt11 phage C. tentans salivary gland cDNA library according to the Promega instruction manual and affinity purified on an anti–hrp23 antibody NHS-Sepharose column (Amersham Pharmacia Biotech).
hrp36.
Recombinant hrp36 was expressed in Escherichia coli BL21 (DE3) transformed with a plasmid, p36-1, consisting of hrp36 cDNA inserted into a pET21d vector. In brief, the transformed cells were induced with 1 mM IPTG for 3 h, and subsequently sonicated. The bacterial lysate was clarified by centrifugation, loaded onto a Q-Sepharose column on an FPLC (Amersham Pharmacia Biotech), and fractionated using a 0–100% 2 M NaCl linear gradient. Fractions containing hrp36 were then pooled, dialyzed, and loaded onto an SP-Sepharose column (Amersham Pharmacia Biotech). The hrp36 protein was eluted and dialyzed.
Culturing Conditions
C. tentans was reared as described by Case and Daneholt 1978, and salivary glands were isolated from fourth instar larvae. C. tentans tissue culture cells were grown as described by Wyss 1982.
Preparation of Extracts from C. tentans Tissue Culture Cells
The nuclear and cytoplasmic extracts were prepared as described by Wurtz et al. 1996.
SDS-PAGE and Western Blot Analysis
Proteins were fractionated in 10% polyacrylamide gels containing 0.1% SDS, and analyzed by Western blot according to Sun et al. 1998.
Isolation and Immunostaining of Polytene Chromosomes
The procedures have been previously described (Sun et al. 1998).
Immunoelectron Microscopy on Ultrathin Cryosections
Immunoelectron microscopy was carried out according to Visa et al. 1996. The density of gold particles in a region was determined by analysis of 15 randomly chosen unit areas, each being 1.9 μm2.
DNase I-binding Assay
DNase I (Sigma-Aldrich) was covalently linked to CNBr-activated Sepharose beads (Sigma-Aldrich) at a concentration of 1 mg/ml according to the manufacturer's protocol. BSA-Sepharose beads, which served as control, were prepared in the same way. Coupling efficiency, as calculated by UV spectrophotometry, was >95%.
The DNase I binding experiments were performed in either one or two steps. In the single step procedure, nuclear and cytosolic extracts, prepared from 500 ml of C. tentans tissue culture cells, were incubated with 100 μl of DNase I beads for 30–40 min at 4°C. The beads were washed batchwise six times with 1 ml PBS, containing 1% NP-40, 0.1% deoxycholate, 1 mM DTT, and 1 mM PMSF, and heat denatured in Laemmli buffer. The solubilized proteins were resolved by SDS-PAGE and stained with Coomassie blue. The two-step binding experiments were carried out with purified proteins. Samples of actin were individually incubated with either hrp23 or hrp36 in equimolar amounts in G buffer (5 mM Tris, pH 7.6, 0.5 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT) for 30–40 min at 4°C, and subsequently incubated with 100 μl of DNase I beads; purified actin, hrp23, and hrp36 were also individually incubated with the same amount of DNase I beads. After washing of the beads, bound proteins were analyzed with SDS-PAGE as described above. Control experiments were performed in parallel with BSA-Sepharose.
In Vivo Cross Linking
Proteins in C. tentans tissue culture cells were cross linked for 10 min at room temperature with 0.5 mM dithiobis-succinimidylpropionate (DSP; Sigma-Aldrich) in the standard cultivation medium. Nuclear and cytosolic extracts were prepared as described above and where appropriate incubated briefly with 8 M urea. The extracts were diluted 10× with PBS, containing 0.2% NP-40 and 1 mM PMSF, and immediately incubated with DNase I beads as described above.
Results and Discussion
Nuclear Actin Is Located in Transcriptional Puffs on Polytene Chromosomes
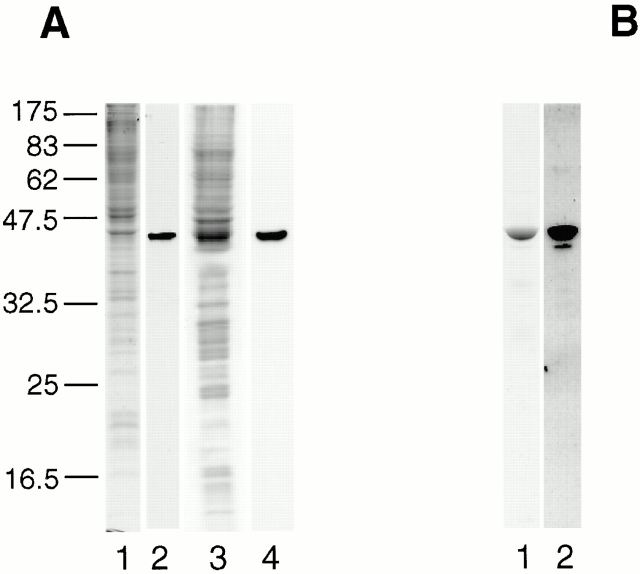
The distribution of actin in C. tentans cells was investigated using a polyclonal, affinity-purified antiactin antibody, 9771-4. In a Western-blot experiment, the antiactin antibody detected specifically a single protein of 42,000 Mr in both nuclear and cytoplasmic extracts of cultured C. tentans cells (Fig. 1 A, lanes 2 and 4, respectively). The antibody recognized C. tentans actin, which had been affinity purified on a DNase I column from a C. tentans nuclear extract (Fig. 1 B); mass spectrometric analysis confirmed that the protein was actin (see Fig. 5). Thus, the affinity-purified, antiactin antibody cross reacts with C. tentans actin and shows high immunospecificity.
Figure 1.
Western blot analysis of nuclear and cytoplasmic actin in C. tentans. (A) Nuclear and cytoplasmic extracts from C. tentans tissue culture cells were analyzed by SDS-PAGE (1 and 3, respectively) or further by Western blotting using an antiactin antibody (2 and 4, respectively). (B) For comparison, actin was purified from cultured cells by DNase I affinity chromatography and analyzed as above by SDS-PAGE (1) or further by Western blotting (2). A molecular mass calibration is shown (kD) on the left.
Figure 5.
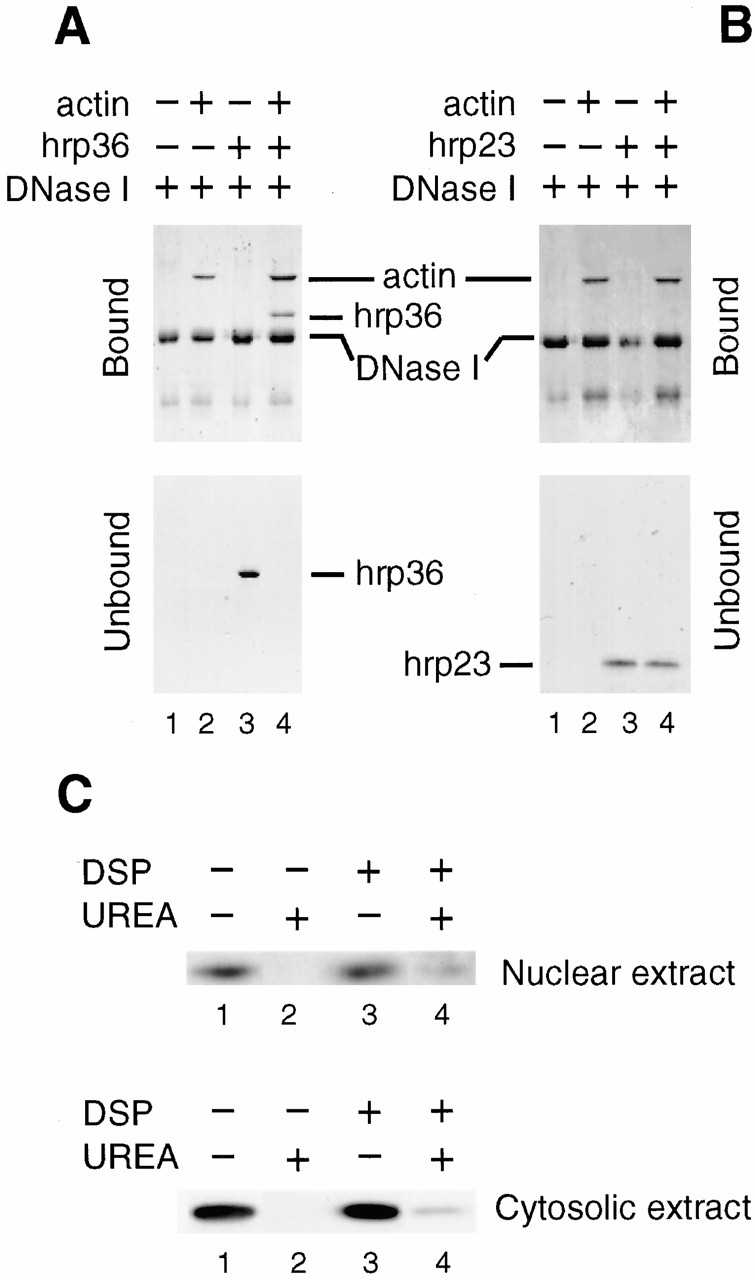
In vitro and in vivo binding of actin to hrp36. (A) Purified G-actin and recombinant hrp36 were preincubated in equimolar amounts. The proteins were added to DNase I-Sepharose beads, and both bound and unbound fractions separated by SDS-PAGE and visualized by Coomassie blue staining. No additions (1), plus actin (2), plus hrp36 (3), or with actin and hrp36 (4). (B) The experiment in A was repeated with the sole difference that recombinant hrp23 was used in place of hrp36. (C) Cross linking of actin to hrp36 in C. tentans tissue culture cells. Cultured cells were treated with DSP and control cells were incubated in parallel without DSP. Nuclear and cytosolic extracts were prepared, and each extract was split into two equal portions, one being treated with urea. All eight samples were subjected to DNase I affinity chromatography and bound proteins were analyzed by SDS-PAGE and Western blotting using an anti–hrp36 antibody.
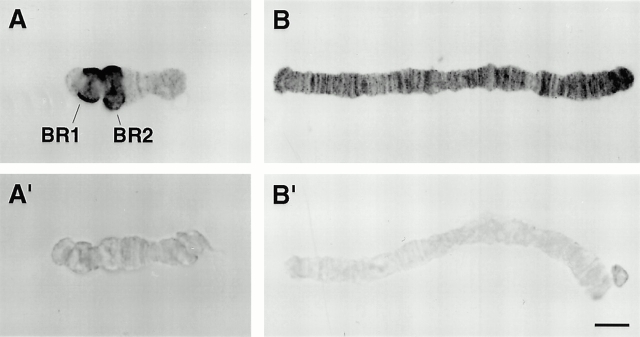
Immunocytology studies were performed on isolated polytene chromosomes using the antiactin antibody to test whether actin is associated with any of the chromosomal puffs active in transcription. All four chromosomes from C. tentans were isolated from fixed salivary glands with their morphology well preserved, as shown for chromosomes I and IV in Fig. 2. The most conspicuous result was the strong immunolabeling of the most giant puffs, the Balbiani rings on chromosome IV, in particular Balbiani rings 1 and 2. (Fig. 2 A). A large number of smaller puffs were also decorated on all four chromosomes (Fig. 2A and Fig. B). No labeling of the polytene chromosomes was observed in controls using anti–SCP3 antibody instead of the antiactin antibody (data not shown). To decide whether actin is bound to chromatin and/or the transcription products, isolated polytene chromosomes were digested with RNase A before the immunolabeling. After the RNase treatment, the chromosomes displayed weak or no immunolabeling (Fig. 2A′ and B′). We conclude that actin is present in the BRs and many other chromosomal puffs and is likely to be associated with the transcription products, the pre-mRNP particles.
Figure 2.
Immunocytochemical localization of actin on isolated C. tentans polytene chromosomes. (A and A′) Chromosome IV; (B and B′) chromosome I. (A and B) No RNase; (A′ and B′) RNase treated. The expanded, heavily stained regions on chromosome IV (A) represent the large Balbiani rings. Bar, 10 μm.
Actin Resides in BR pre-mRNP Particles In Vivo
The Balbiani rings exhibited a particularly strong labeling with the antiactin antibody. It seemed, therefore, feasible to make use of the BR experimental system to study the association of actin with a specific pre-mRNP particle during the assembly and intranuclear transport of the particle (Fig. 3 H). We adopted the immunoelectron microscopic approach earlier used (Visa et al. 1996). Ultrathin cryosections were immunostained with the antiactin antibody and subsequently with a secondary anti–rabbit antibody coupled to colloidal gold. An anti–SCP3 antibody replaced the antiactin antibody in the control samples.
Figure 3.
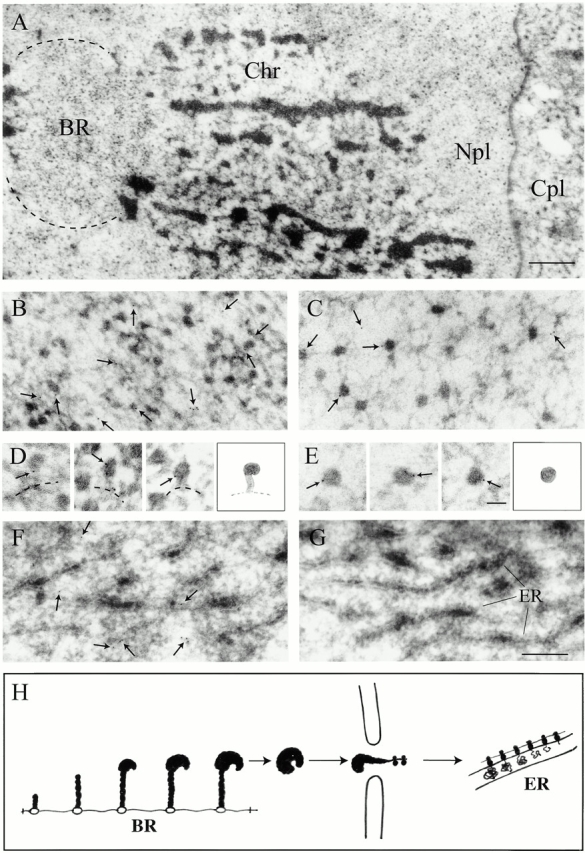
Immunoelectron microscopic localization of actin in C. tentans salivary gland cells. (A) Nucleus in a salivary gland cell. Chr, polytene chromosome; Npl, nucleoplasm; Cpl, cytoplasm. (B) Balbiani ring. (C) Nucleoplasm. (D) Growing BR particles and a schematic drawing. Broken line, putative position of chromatin axis. (E) Nucleoplasmic BR particles and a schematic drawing. (F and G) Cytoplasm. (H) Schematic presentation of the assembly and transport of BR particles. (B–F) Antiactin antibody; (G) control antibody. Gold particles have been marked with small arrows. Bar, 1 μm (A), 200 nm (B, C, F, and G), and 50 nm (D and E).
A portion of a salivary gland cell is shown at low magnification in Fig. 3 A. At higher magnification, a considerable number of gold particles can be observed in the BRs (Fig. 3 B) as well as in the nucleoplasmic (C) and cytoplasmic (F) regions. A cytoplasmic control specimen is also shown (Fig. 3 G). As there are very few gold particles in the control specimens (<3% of the number in the antiactin antibody sample), essentially all the gold particles in the experimental samples are significant. The average density of particles in the BRs and nucleoplasm is almost twice that in the cytoplasm (8.0, 9.1, and 4.8 particles/1.9 μm2).
Within the BR, a growing BR particle is initially present as a slightly coiled RNP fibril, which later on during transcription is being packed into a dense globular structure (Fig. 3 H). As seen in Fig. 3 B, some gold particles are associated with dense RNP particles, constituting almost completed transcription products. Such late transcription products, often described as stalked granules, are presented at higher magnification in Fig. 3 D. Thus, we conclude that actin is associated with a growing BR particle and becomes bound to the particle during its assembly concomitantly with transcription.
Also in the nucleoplasm, gold particles appear bound to the dense BR RNP granules (Fig. 3 C). Examples of gold-labeled nucleoplasmic BR particles are shown at higher magnification in Fig. 3 E. Thus, actin remains associated with the completed BR RNP particles released into the nucleoplasm.
BR particles can be seen at a relatively low frequency either bound to the nuclear basket of the nuclear pore complex or in an extended conformation passing through the nuclear pores (Fig. 3 E; see e.g., Mehlin et al. 1992). Due to the fact that BR particles in transit are so sparsely distributed, it has not been possible to demonstrate whether or not actin is associated with BR particles moving through the nuclear pores. However, gold particles can be seen associated with the nuclear envelope, also within the nuclear pore complexes themselves (data not shown), but, as hnRNP complexes other than BR granules are difficult to recognize, we cannot conclude that actin is bound to translocating hnRNP particles. Finally, we noted that gold particles are also observed in the cytoplasm (Fig. 3 F).
We conclude from the immunoelectron microscopy that actin becomes associated with the BR RNP particles during their assembly on the gene concomitant with transcription, and actin remains associated with the completed and released BR particles present in the nucleoplasm. Whether or not actin also accompanies the BR RNP particle through the nuclear pores and remains associated with BR mRNA during protein synthesis cannot be determined from these data, but the results are compatible with such a hypothesis.
Our finding that actin is associated with BR pre-mRNP particles and other pre-mRNP particles in the nuclei of salivary gland cells in C. tentans is in agreement with the growing amount of evidence that actin is present in nuclei of somatic cells as well as oocytes (for recent review, see Rando et al. 2000). At the ultrastructural level, it has been noted that nuclear actin appears in pre-mRNA containing fibrogranular material in somatic cells (Nakayasu and Ueda 1985), but our study seems to be the first one to demonstrate that actin is present in well-defined pre-mRNP particles.
Actin Binds Specifically to the hnRNP Protein hrp36
The interaction between actin and BR particles cannot be studied directly as it is not feasible to prepare BR particles in large quantities. Instead, we investigated the interaction between actin and the whole population of pre-mRNP particles isolated from C. tentans tissue culture cells. Since actin binds tightly to DNase I, we used DNase I affinity chromatography to identify proteins binding to actin. DNase I-Sepharose beads were incubated with either nuclear or cytoplasmic extracts from C. tentans tissue culture cells. The DNase I bound proteins (B) were analyzed by SDS-PAGE and Western blotting; unfractionated nuclear (N) or cytoplasmic (C) extracts were studied in parallel. The analysis of the nuclear extract is presented in Fig. 4 A. The Coomassie blue–stained SDS gel showed that the DNase I-bound proteins (lane 2) represent only a small subset of the large number of proteins in the nuclear extract (lane 1). The two predominant DNase I-binding proteins comigrated with actin and hrp36, and their identity was confirmed by Western blotting (Fig. 4 A, lanes 3 and 4, and 7 and 8, respectively). Often the hrp36 band runs as a doublet corresponding to the two known isoforms of hrp36 (Wurtz et al. 1996; our unpublished sequence information). In contrast to hrp36, the two BR particle proteins hrp23 and hrp45 also present in the nuclear extract were not pulled down with the DNase I beads (Fig. 4 A, lanes 5 and 6, and 9 and 10, respectively). The analysis of the cytoplasmic extract is shown in Fig. 4 B. A subset of the proteins was bound to DNase I-Sepharose (lane 2 vs. 1), and actin and hrp36 again constituted the predominant proteins (lane 2; for identification, see lanes 4 and 8, respectively). Neither hrp23 nor hrp45 proteins were present in the cytoplasm, in agreement with earlier studies (hrp23: Sun et al. 1998; hrp45: Alzhanova-Ericsson et al. 1996). We conclude that actin is associated with hrp36 in both the nucleus and cytoplasm, but not with hrp23 or hrp45.
Figure 4.
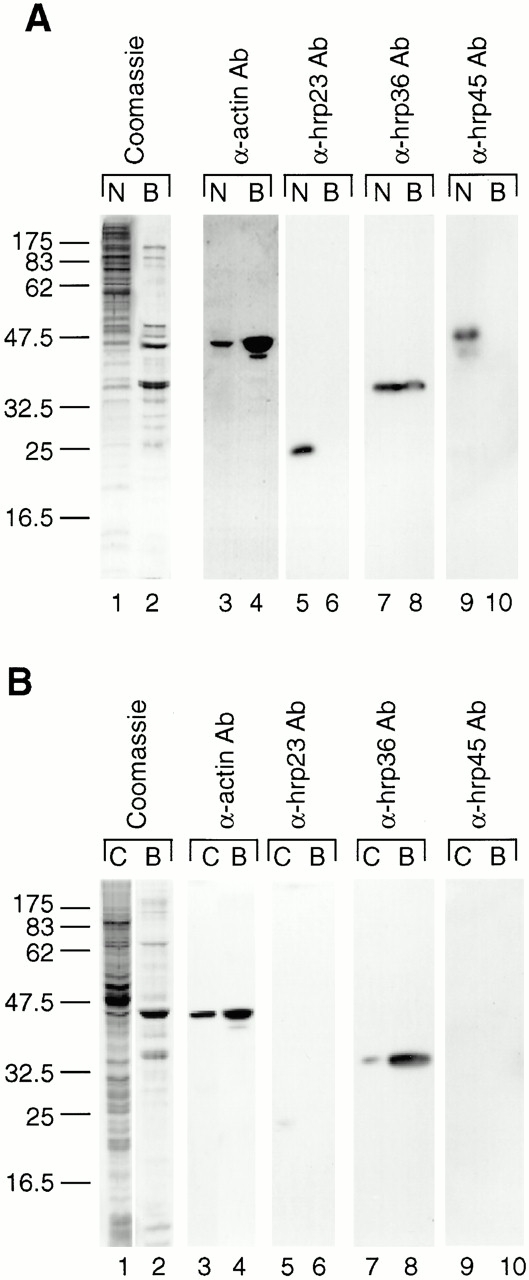
Nuclear and cytoplasmic actin-associated proteins studied by DNase I affinity chromatography. (A) Nuclear extracts were prepared from C. tentans tissue culture cells and split into two portions, one was directly fractionated by SDS-PAGE (N), while the other one was mixed with DNase I-Sepharose beads, and the bound proteins were eluted and separated by SDS-PAGE (B). Gels were Coomassie blue stained (1 and 2) or analyzed by Western blotting using an antiactin antibody (3 and 4) or monoclonal antibodies against three C. tentans hnRNP proteins: hrp23 (5 and 6, respectively), hrp36 (7 and 8, respectively), and hrp45 (9 and 10, respectively). The bound proteins were also transferred onto PVDF membrane, and the band corresponding to actin was excised, proteolytically digested, and analyzed by mass spectrometry. Three peptides were derived (SYELPDGQVITIGNER, VAPEEHPVLLTEAPLNPK, DLYANTVLSGGTTBYPGIADR), all of them consistent with the protein being actin as established by database searching. (B) Parallel experiments with cytoplasmic extracts. C, proteins in extract; B, proteins bound to DNase I-Sepharose. α-actin Ab, antiactin antibody.
We performed reconstitution experiments with purified recombinant hrp36 and native monomeric β/γ-actin to ascertain whether actin and hrp36 associate directly, using the DNase I pull-down assay. To keep actin in the monomeric G form throughout the experiment, binding studies were performed in low-salt G buffer. After stringent washes of the DNase I-Sepharose beads, bound and unbound proteins were resolved by SDS-PAGE. In control experiments, actin and hrp36 were separately incubated with DNase I-Sepharose beads, confirming that actin binds to DNase I, whereas hrp36 does not (Fig. 5 A, lanes 2 and 3, respectively). Some noncovalently bound DNase I was released from the column by SDS (Fig. 5 A, lane 1). When hrp36 was incubated with equimolar amounts of actin, both hrp36 and actin were present in the bound fraction (Fig. 5 A, lane 4). Since only actin binds to DNase I, this demonstrates a complex between hrp36 and actin. Parallel experiments were carried out with purified recombinant hrp23, which confirmed that hrp23 does not bind to actin (Fig. 5 B). Experiments using BSA-Sepharose or uncoupled Sepharose confirmed that the material retained was not due to nonspecific binding. In no instance did we detect binding of hrp36 to these control resins (data not shown). Therefore, binding to DNase I-Sepharose represents a specific interaction. We conclude that hrp36, but not hrp23, forms a specific complex with G actin.
Actin Can Be Cross Linked In Vivo to hrp36
To establish whether actin and hrp36 are associated in living cells, we treated C. tentans tissue culture cells with the membrane-permeable, cleavable cross-linking reagent, DSP. Cross-linked nuclear and cytoplasmic extracts were treated with urea and assayed by DNase I pull-down experiments. Non–urea-treated samples were analyzed in parallel. The bound fractions were studied by SDS-PAGE followed by Western blotting using an anti–hrp36 antibody (Fig. 5 C). After treatment with DSP and urea, hrp36 bound to the DNase I-Sepharose column in both the nuclear and cytoplasmic extracts. In the absence of DSP but in the presence of urea, no hrp36 bound to the column. When urea treatment was omitted in both the DSP-treated and untreated cells, hrp36 bound to the column. Coomassie staining of the gels suggested that binding of actin to the DNase I-Sepharose column was largely unaffected under the experimental conditions used (data not shown). We conclude that actin and hrp36 are part of the same complex in vivo in both the nucleus and the cytoplasm.
Actin-hrp36 Complexes Associated with BR RNA
We have demonstrated that actin becomes associated with the BR particles concomitant with transcription and is present in nucleoplasmic particles released from the genes. Furthermore, we have shown that actin is associated with hrp36 in both nuclear and cytoplasmic extracts, that the interaction is likely to be direct, and that the actin–hrp36 complexes are formed in vivo both in the nucleus and cytoplasm. As hrp36 is known to be bound to BR RNA both in the nucleus and the cytoplasm (Visa et al. 1996), we conclude that actin becomes coupled cotranscriptionally to BR RNA via hrp36 and is associated with the transcript during its transport from the nucleus into the cytoplasm and further into polysomes.
A central issue is whether the hrp36-bound actin is in a monomeric or polymeric form. We have found no evidence for actin filaments in the fixed salivary gland nuclei. However, as microfilaments could be extremely sensitive to fixation, we cannot exclude the possibility that they could have disassembled. In fact, early microdissection experiments with C. tentans salivary gland cells showed that the polytene chromosomes are embedded in a labile gel (D'Angelo 1946), which has properties like the actin gel in amphibian oocyte nuclei (Clark and Merriam 1977). Thus, presently, the issue of the state of actin in the nuclear actin–hrp36 complex has to be left open. The state of actin in the cytoplasmic actin-hrp36 complex is also unclear.
Possible Roles of BR RNP-associated Actin
It can be speculated that the hrp36–actin complex is important for packing the RNA into a BR RNP fibril and further into well-defined higher-order structures. Other possibilities would be that actin promotes interaction of the BR particle with a fibrous network in the nucleoplasm, allows binding to export receptors, or is involved in the dramatic conformational change of the particle upon translocation through the nuclear pore. As actin remains bound to hrp36 in the cytoplasm, it is finally important to recall that hnRNP proteins have been found to affect translation efficiency, mRNA stability, and RNA location within cytoplasm (Krecic and Swanson 1999). Evidently, a wide range of functional options have to be considered for the actin–hrp36 complex.
Acknowledgments
We are grateful to Birgitta Björkroth and Lise-Marie Fjelkestam for technical assistance and Sergej Masich for preparing the electron microscopic figures. The p36-1 plasmid was a gift from Alla Alzhanova-Ericsson and the polyclonal anti–SCP3 antibody from Christer Höög. The Protein Analysis Center at Karolinska Institutet carried out the mass spectrometry.
This work was supported by the Swedish Natural Science Research Council and the Human Frontier Science Program Organization.
Footnotes
Abbreviations used in this paper: BR, Balbiani ring; DSP, dithiobis-succinimidylpropionate; RNP, ribonucleoprotein.
References
- Alzhanova-Ericsson A., Sun X., Visa N., Kiseleva E., Wurtz T., Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- Case S.T., Daneholt B. The site of the transcription unit in Balbiani ring 2 of Chironomus tentans as derived from analysis of the primary transcript and 75S RNA. J. Mol. Biol. 1978;124:223–241. doi: 10.1016/0022-2836(78)90157-2. [DOI] [PubMed] [Google Scholar]
- Clark T.G., Merriam R.W. Diffusible and bound actin in nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–891. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- D'Angelo E.G. Micrurgical studies on Chironomus salivary gland chromosomes. Biol. Bull. 1946;90:71–87. [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Byrd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Krecic A.M., Swanson M.S. HnRNP complexescomposition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Matunis E.L., Matunis M.J., Dreyfuss G. Association of individual hnRNP proteins with nascent transcripts. J. Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin H., Daneholt B., Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Choi M., Dreyfuss G. A nuclear export signal in hnRNP A1a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Nakayasu H., Ueda K. Association of rapidly-labelled RNAs with actin in nuclear matrix from mouse L5178Y cells. Exp. Cell Res. 1985;160:319–330. doi: 10.1016/0014-4827(85)90179-x. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Rando O.J., Zhao K., Crabtree G.R. Searching for a function for nuclear actin. Trends Cell Biol. 2000;10:92–97. doi: 10.1016/s0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- Rozycki M., Schutt C.E., Lindberg U. Affinity chromatography-based purification of profilin:actin. Methods Enzymol. 1991;196:100–118. doi: 10.1016/0076-6879(91)96012-g. [DOI] [PubMed] [Google Scholar]
- Sun X., Alzhanova-Ericsson A.T., Visa N., Aissouni Y., Zhao J., Daneholt B. The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J. Cell Biol. 1998;142:1181–1193. doi: 10.1083/jcb.142.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson A.T., Sun X., Kiseleva E., Björkroth B., Wurtz T., Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Wurtz T., Kiseleva E., Nacheva G., Alzhanova-Ericsson A.T., Rosén A., Daneholt B. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1996;16:1425–1435. doi: 10.1128/mcb.16.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteriods, juvenile hormone, insulin and heat shock. Exp. Cell Res. 1982;139:309–319. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]