Abstract
Purpose
Interphotoreceptor retinol-binding protein (IRBP), composed of two or four homologous modules in tandem, plays an important role in retinoid trafficking between the retinal pigmented epithelium, photoreceptors, and Müller cells. The exact nature of this role is not yet clear. Attempts to purify the full-length retinal IRBP to homogeneity for crystallization purposes have largely been unsuccessful, owing primarily to instability and denaturation of the protein at high concentrations in aqueous media.
Methods
A bacterial expression system was used for the production of the recombinant full-length four modules-containing Xenopus IRBP (xIRBP; 1197 amino acids; 131 kDa). An optimized purification strategy and the presence of molar excesses of a thiol-based reducing agent yielded highly pure xIRBP in a soluble, stable and active form, free of its fusion partner. Binding of all-trans retinol was characterized by fluorescence spectroscopy monitoring ligand-fluorescence enhancement, quenching of endogenous protein fluorescence, and energy transfer.
Results
We grew the first diffraction-quality crystal of xIRBP. We have gathered diffraction data from these crystals to 2.46 Å resolution, sufficient to yield an atomic model of the tertiary structure of IRBP. Retinol-binding results determined by fluorescence spectroscopy show roughly one retinol-binding site per polypeptide chain.
Conclusions
The binding stoichiometry taken together with modeling data suggest that not all modules are functionally equivalent. Determination of the full-length IRBP structure will be a significant breakthrough in understanding the functional roles of IRBP in the visual cycle. The advances presented here will not only lead to the structure of the full-length IRBP, but will allow us to understand how the modules interact in the function of IRBP. Furthermore, these studies will allow characterization of the ligand-binding site(s) with bound ligand(s).
Introduction
The functional significance of the module structure of interphotoreceptor retinoid-binding protein (IRBP) remains unknown. IRBP, roughly a 135 kDa (~1200 amino acids) glycoprotein, is composed of four homologous “modules.” Each are approximately 300 amino acids in length. Rods and cones secrete IRBP into subretinal space, where it is the major soluble protein component of the interphotoreceptor matrix (IPM). In the IPM, IRBP is rapidly turned over by a combination of retinal pigmented epithelium (RPE) and photoreceptor endocytosis [1–3]. Although insight into its structure, cellular physiology, and function has been accumulating [4–8], progress is severely hampered by lack of information about its complete three-dimensional structure.
IRBP is positioned to mediate interactions between cells bordering the IPM. IRBP carries visual cycle retinoids in a light-dependent manner, and was originally thought to only solubilize retinoids in the IPM. Its role is rather complex as it functions in four steps of the visual cycle: promoting (1) the removal of all-trans retinol from bleached rod outer segments [9,10], (2) the delivery of all-trans retinol to the RPE [11], (3) the release of 11-cis retinal from the RPE [12–14], and (4) the delivery of 11-cis retinal to the outer segments [14]. The role of IRBP in these steps, although sufficient, may not be strictly necessary, as IRBP-deficient mice (transgenic “knockout” mice) do not show a depressed rate of retinoid exchange between the retina and RPE [15]. The rate of recovery is more rapid than that in controls [16,17]. This could be explained by a buffering activity, a property that has been observed in studies monitoring the exchange of retinoids between lipid vesicles [18]. The importance of IRBP as a buffer may be related to its biochemical activity in preventing oxidative and isomeric degradation of visual cycle retinoids [19]. The mechanism of IRBP’s antioxidant activity is not known. Finally, it should be mentioned that IRBP has been implicated in functions outside of the visual cycle including retinal development (reviewed in [5,8]), and fatty acid binding [20,21]. The emerging picture is that IRBP has multiple functions in the visual cycle, and probably has roles outside the cycle. Understanding the mechanisms involved will require a clearer picture of its structure.
Xenopus provides a particularly useful experimental system to study the trafficking, and function of IRBP in the visual cycle and in development. We have, thus, continued to focus on Xenopus IRBP (xIRBP; 131 kDa; 1197 amino acids without the signal peptide) as an alternative model for human IRBP, which is 60% identical to xIRBP in amino acid sequence. Furthermore, xIRBP is composed of four modules that are homologous to that of mammalian and avian IRBPs [22,23]. In the current study, we have overcome the problems related to generating IRBP crystals. Here, we extend our previous study [24] by preparing a full-length xIRBP in a soluble form free of its thioredoxin (Trx) fusion partner, establish conditions that allow for concentrating the protein without precipitation, and identify conditions that permit routine growth of diffraction quality crystals. These advances will not only lead to the structure of the full-length IRBP, but will also allow characterization of ligand-binding domain(s) with bound ligand.
Methods
Expression of the recombinant full-length Xenopus interphotoreceptor retinol-binding protein and purification by Ni-affinity chromatography
Preparation of the clone expressing xIRBP as a thioredoxin fusion protein that contains a His-patch has been previously described [24]. The fusion protein DNA was sub-cloned into pThiohisA vector (Invitrogen, Carlsbad, CA); the bacterial cell used for protein expression was Top10 (Invitrogen). Cell lysate in a binding buffer of 25 mM Tris-HCl, pH 7.5 at 4 °C, containing 200 mM NaCl and 1mM Dithiothreitol (DTT; Inalco, San Luis Obispo, CA) was applied to a pre-equilibrated 5ml HisTrap HP column (GE Healthcare, Piscataway, NJ). All purification columns were run on an Äkta FPLC (GE Healthcare). Protein was eluted over a 20-column volume linear gradient from 0 to 300 mM imidazole in an elution buffer composed of 25 mM Tris-HCl, pH 7.5 at 4 °C, 50 mM NaCl, and 0.5 mM DTT. A major protein peak eluted at 50 mM imidazole. The eluted fractions were pooled, and the protein concentration was determined by measuring the optical density at 280 nm (OD280) and by sodium dodecyl sulfate-PAGE (SDS–PAGE) analysis performed with 12% Tris-HCl pre-cast gels (Bio-Rad Laboratories, Hercules, CA). Pooled fractions were diluted such that the imidazole concentration was 20 mM or less. To remove the N-terminal Trx fragment, we added EnterokinaseMax (EK; Invitrogen) in a ratio of 2 U EK to 1 mg protein. After incubation for ~36 h at 4 °C, EK was removed using EK-Away resin (Invitrogen) according to the manufacturer’s direction.
The cleaved xIRBP was then applied to a second 5 ml HisTrap HP column (GE Healthcare) pre-equilibrated with the binding buffer. The same chromatography method was used on a Äkta FPLC (GE Healthcare). Protein was eluted over a 20-column volume linear gradient from 0 to 300 mM imidazole with the following elution buffer: 25 mM Tris-HCl, pH 7.5 at 4 °C, 50 mM NaCl, and 0.5 mM DTT. While a part of the cleaved xIRBP flowed through the column as expected, some bound the column, presumably through the endogenous histidine side chains, and eluted as a major peak at an imidazole concentration of 60 mM. The bound fractions were pooled, and the protein concentration was determined by SDS–PAGE analysis, and absorbance spectroscopy using extinction coefficients calculated from the primary sequence [24]. Proteolysis was prevented by adding 0.4 mM Pefabloc SC (Roche, Indianapolis, IN) to the pooled fractions.
Gel filtration
The DTT concentration of the protein solution was then adjusted to 1 mM, and resulting solution was concentrated to a final volume of about 6 ml by ultrafiltration using a YM-50 centricon (Millipore, Billerica, MA). The protein was then loaded onto a previously equilibrated Sephacryl S-300HR (S300) column (GE Healthcare), which measured 2.6 × 100 cm. The gel filtration procedure was run on a Äkta FPLC with one column volume isocratic elution using the following binding buffer: 25 mM Tris-HCl, pH 7.5 at 4 °C, 50 mM NaCl, and 1 mM DTT. xIRBP eluted as dimeric as well as monomeric peaks, each of which were pooled and analyzed by SDS–PAGE. The purest xIRBP fractions eluted as a dimer between elution volumes 212 and 250 ml. The procedure yielded 15 to 20 mg of ~97% pure xIRBP from 1.25 L of cell culture.
Protein assay
Final IRBP-containing fractions were pooled, and protein concentration was determined by OD280 and 12% SDS–PAGE analysis.
N-terminal sequence
The xIRBP band from the SDS–PAGE analysis was blotted onto a PVDF membrane. The band was cut out, dried, and sent to Proseq (Boxford, MA) for sequencing by the Edman degradation method.
Crystallization
The buffer components of purified xIRBP were adjusted to 50 mM Tris-HCl, pH7.5 at 4 °C, 100 mM NaCl, and 4 mM DTT. The protein solution was then concentrated to12 mg/ml of xIRBP. At the final concentration, 0.5 mM oleic acid was added to the solution, and the mixture was incubated. Crystal screening experiments were conducted with freshly purified xIRBP using the MbClass II Suite commercial screen (Qiagen/Nextal, Montreal, Canada) cocktail solutions. The screening was performed at 23 °C in 24-well sitting drop plates. Crystals initially grew at 23 °C from 0.1M MgCl2, 0.1M NaCl, 0.1 M Na HEPES pH 7.5, 12% polyethylene glycol (PEG) 4000, as well as from 0.1M MgCl2, 0.1 M NaCl, 0.1M 3-(Cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (CAPSO); pH 9.5, 12% PEG 4000 when the protein and the reservoir (cocktail) solutions were mixed in 1:1 ratio and the resulting droplets were allowed to undergo vapor diffusion against reservoirs in sealed wells. Following optimization of the growth conditions, best diffraction-quality crystals were routinely grown at 23 °C from 0.1M MgCl2, 0.1M NaCl, 0.1M Tris-HCl pH 8.0, 14%–22% PEG 4000 in a 3:1 protein:cocktail ratio.
X-ray diffraction experiment
Initial X-ray diffraction experiments were performed on the in-house R-AXIS IVC image plate and rotating anode X-ray system, with the crystals maintained both at ambient and cryogenic temperatures. X-ray diffraction data collection was performed at the A-1 station of the Cornell High Energy Synchrotron Source (CHESS), Ithaca, New York. The crystals were cooled at cryogenic temperature by plunging them into liquid nitrogen and maintaining them in a stream of liquid nitrogen during data collection. Glycerol was added to the crystallization droplets to adjust the final glycerol concentration to 40%–50%. This liquid mixture served as the cryoprotectant for the crystals. The diffraction images were recorded on an ADSC Quantum-210 Charge Coupled Device (CCD) detector. The diffraction data were processed with HKL2000 [25] and MOSFLM [26] routines.
Ligand-binding assays by fluorescence spectroscopy
The binding of all-trans retinol to Xenopus IRBP was characterized in titrations using a DM 45 scanning spectrofluorimeter (Online Instrument Systems, Inc., Bogart, Georgia) [27]. Titrations monitoring enhancement of retinol fluorescence, quenching of the intrinsic protein fluorescence, and energy transfer were performed as previously described [28]. Enhancement of retinol fluorescence was followed by monitoring the increase in retinol fluorescence (excitation: 330 nm; emission: 480 nm) compared to a fluorescence-matched solution of N-acetyl-L-tryptophanamide [29]. The concentration of the N-acetyl-L-tryptophanamide solution was adjusted to match the fluorescence of the apo-IRBP solution. The fluorescence of all-trans retinol in the presence of N-acetyl-L-tryptophanamide was subtracted from that in the presence of xIRBP. Assays following quenching of protein fluorescence used an excitation wavelength of 280 nm and an emission wavelength of 340 nm. In these experiments the inner filter effect was accounted for by graphical correction [30]. Energy transfer assays monitored retinol fluorescence at 480 nm (excitation: 280 nm). The dissociation constant (Kd) and number of binding sites (N) was determined by nonlinear least square fit to a binding equation that assumed a single type of noninteracting site(s) [31].
Results
Stability and integrity of purified Xenopus interphotoreceptor retinol-binding protein
The expession and purification procedure described in the previous section yielded 15 to 20 mg of ~97% pure xIRBP from 1.25 L of cell culture. A gel electrophoresis of the final product is shown in Figure 1. In contrast to previous reports of xIRBP instability at high concentrations [32], the purified xIRBP, free of its fusion partner Trx, maintained its integrity, high solubility, and ligand-binding ability for weeks at 4 °C at a concentration of ~10 mg/ml. The enhanced protein stability and sustained solubility were partially attributable to the presence of molar excess of DTT in solution. Furthermore, the absence of n-octyl-β-D-glucopyranoside or any other detergent was probably a stabilizing factor as well, since for soluble proteins, detergents could add to instability promoting gradual unfolding. Thus, under the reducing conditions of the purification experiments, highly pure xIRBP maintained its naturally folded stable tertiary structure, and, therefore, was fully soluble and functionally active for a prolonged period of time.
Figure 1.
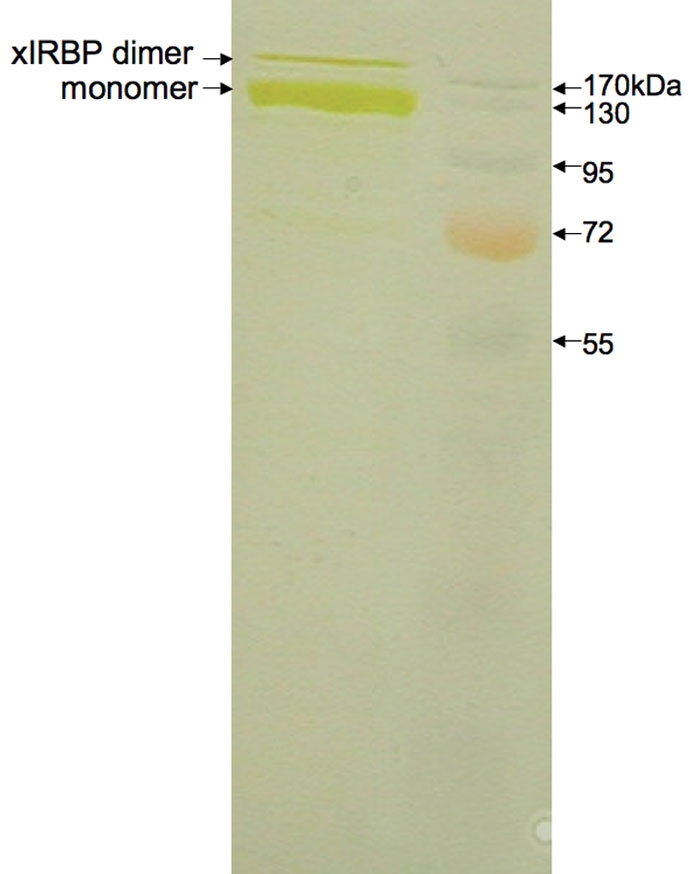
Gel electrophoresis (SDS–PAGE; silver stained) analysis of the purified four-module containing Xenopus interphotoreceptor retinol-binding protein. The major band corresponds to monomeric Xenopus interphotoreceptor retinol-binding protein (xIRBP) molecular weight of 131 kDa. The minor higher molecular weight band represents dimeric form of the protein, which may or may not be an artifact of the analysis itself.
N-terminus sequence
The N-terminal sequence for the full-length recombinant xIRBP was unambiguously determined to be G-L-G-D-P-F-Q-P-S-L. The actual N-terminal sequence of xIRBP is F-Q-P-S-L… The first part of the determined sequence, namely G-L-G-D-P, belonged to the vector pThioHisA (Invitrogen) that was included in the cloning sequence of xIRBP. This result, along with the molecular weight of the protein band (Figure 1), confirmed the identity of the purified protein to be the full-length recombinant xIRBP, free of its fusion partner Trx.
Binding assays with all-trans retinol
Binding curves for fluorescence studies for binding of all-trans retinol are shown in Figure 2. For the increase in retinol fluorescence measurements (excitation: 330 nm; emission: 480 nm), the number of binding sites per molecule of protein (n) was 1.30±0.04 with Kd=0.15±0.03 μM. Titration monitoring quenching of tryptophan fluorescence by bound retinol (excitation: 280 nm; emission: 340 nm) yielded binding parameters n=1.32±0.06 and Kd=0.16 ± 0.04 μM. Finally, the calculated binding parameters were n=1.05 ± 0.03 and Kd=0.06± 0.01 μM for titration monitoring of energy transfer.
Figure 2.
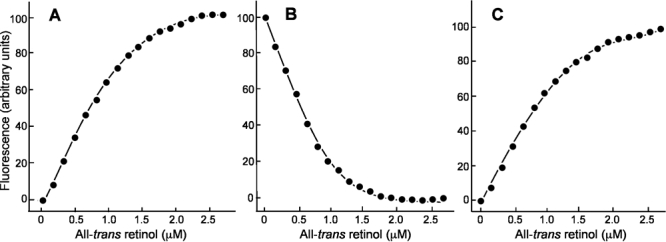
Fluorescence titrations are shown of Xenopus interphotoreceptor retinol-binding protein (xIRBP) binding all-trans retinol. The concentration of xIRBP was 0.89 μM in each panel. A: The titration of IRBP with all-trans retinol in this panel is followed by monitoring the increase in retinol fluorescence compared to a fluorescence matched solution of N-acetyl-L-tryptophanamide (excitation: 330 nm; emission: 480 nm). B: The titration shown follows the quenching of intrinsic protein fluorescence by bound retinol (excitation: 280 nm; emission: 340 nm). C: The titration follows ligand binding by monitoring energy transfer (excitation: 280 nm; emission: 480 nm).
The retinol-binding data compare well with the data from groups purifying the protein in the presence of DTT, albeit at concentrations lower than the present work of Chen et al. [33]. Older studies, which apparently did not include a reducing agent in the purification of xIRBP, tended to obtain values for N and Kd of roughly one site and 1 μM, respectively [11,34].
Crystallization and X-ray diffraction
Single crystals of xIRBP appeared in 5–10 days at 23 °C and continued to grow for weeks. The crystals had two different morphologies: plate-like and parallelepiped-shaped. The largest and best-formed crystals were typically about 0.2 mm in the longest dimension. Figure 3 is a photomicrograph of crystals of xIRBP that grew from a buffer containing 0.1 M MgCl2, 0.1 M NaCl, 0.1 M Tris-HCl (pH8.0) with 20%–22% PEG 4000 as the precipitant, at 2:1 and 3:1 protein to reservoir ratios. A typical diffraction image recorded with synchrotron X-rays at the CHESS is shown in Figure 4. Crystals of both morphologies had the same space group and showed similar diffraction characteristics, typically to a resolution of about 2.45 Å. The space group was P21 and the cell parameters were a=93.53 Å, b=134.04 Å, c=121.29 Å, β=109.8°. There were two 131 kDa polypeptide chains in the asymmetric unit, giving rise to a specific volume of 2.65 Å3/Da, which was within the range for protein crystals [35]. From one crystal a total of 180 diffraction images, each of 1° oscillation and 15 s exposure, were recorded on a CCD detector yielding 318,878 total observations for 99,902 unique reflections. The data was 99.3% complete to 2.46 Å resolution, with an Rmerge of 0.099. The overall intensity to standard deviation ratio is 16.9 and that in the highest resolution shell (2.55–2.46 Å) is 3.6.
Figure 3.
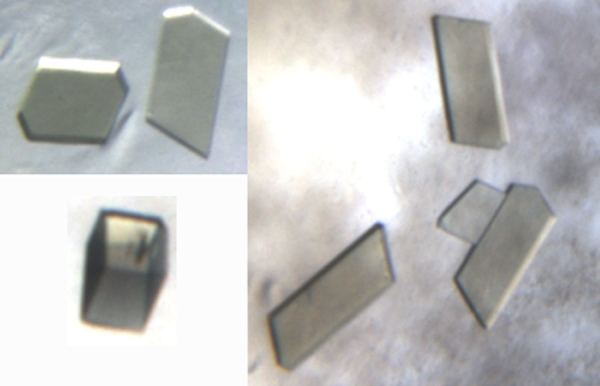
Photomicrographs of Xenopus interphotoreceptor retinol-binding protein crystals grown in the presence of oleic acid. The crystals were typically about 0.2 mm in the longest dimension.
Figure 4.
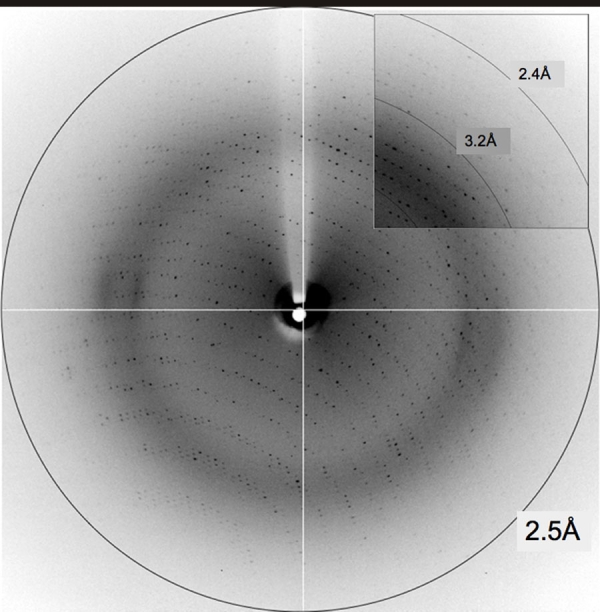
An X-ray diffraction pattern from a Xenopus interphotoreceptor retinol-binding protein crystal recorded at the A-1 station of the Cornell High Energy Synchrotron Source. The detector was at a distance 250 mm; the wavelength, the oscillation angle and the exposure time were 0.978 Å, 1°, and 15 s, respectively. The right upper corner segment is shown in higher contrast to demonstrate the limiting resolution of diffraction of ~2.45 Å.
Discussion
IRBP is the only known retinoid-binding protein that possesses a multiple domain structure. However, the exact nature of its role has remained a mystery, owing partially to the lack of structural insights into the nature of binding pockets, and correlation of the functional properties with structural details. Biochemical and homology modeling studies suggest that the ligand-binding domains in each module are not equivalent, and that the modules interact with each other [24]. Thus, understanding IRBP’s complex function in the visual cycle will require a detailed knowledge of the structural and functional relationships of its modules. The structure of the xIRBP module 2 [36] has provided some clues to that effect. Nonetheless, since a functional full-length Xenopus and mammalian IRBP molecule contains four homologous modules, these structures in the ligand-bound form are likely to yield more comprehensive understanding of the structure-function relationship. Our effort to investigate the structure of full-length xIRBP in complex with fatty acids, and retinoids is a step toward achieving that goal.
Determining the structure of IRBP has proven difficult. First, IRBP is large (~131 kDa), and undergoes ligand-dependent conformational changes [37]. This together with its glycosylation properties [38] could contribute to molecular and dynamical heterogeneities. Purification of tens of milligrams of xIRBP to high homogeneity and in pristine form as well as successful growth of the high diffraction-quality crystals demonstrate that by using the recombinant protein, a bacterial expression system, and optimized purification strategies, we have been able to eliminate the heterogeneities and instabilities of the past. The use of a thiol-based reducing agent in prevention of denaturation is significant since IRBPs contain several cysteine residues [39]. Homology modeling of modules 1, 3, and 4 of xIRBP after the module 2 structure suggests that many, if not all, of its seven cysteines do not participate in disulfide formation and remain as free –SH groups exposed to the solvent. Interestingly, of the 10 cysteines in bovine IRBP, eight were experimentally determined to have free –SH groups [39]. Our data for bovine IRBP indicate that molecules aggregate, unfold, and denature irreversibly by cross-linking through free sulfhydryl groups (unpublished results), which may result in previously observed limited solubility [32]. Although some enhancement of solubility can be achieved by the addition of detergents, such as n-octyl-β-D-glucopyranoside, slow precipitation through denaturation of the protein cannot be prevented [32]. Although the function of IRBP’s free –SH groups are unknown, it is plausible that they may have a role in IRBP’s possible antioxidant activities for the protection of reduced retinol molecules in the visual cycle [19].
The all-trans retinol binding results derived with the structurally pristine xIRBP from three separate experiments, namely (1) enhancement in retinol fluorescence, (2) quenching of intrinsic protein fluorescence, and (3) titration monitoring of energy transfer, are internally consistent and suggest roughly one retinol binding site per four-module full-length molecule with a dissociation constant of about 0.1μM. This data can be interpreted as the retinol-binding site being restricted to one of the four modules, or to an intramodule site defined by multiple modules, consistent with the notion that not all four modules are functionally equivalent as the homology modeling results indicated. Interestingly, homology modeling identified the presence of a highly hydrophobic cavity, the so-called site I, suitable for retinol binding in modules 1 and 2, and predicted that this site in modules 3 and 4 may not exist [24]. Some differences between the two retinol-binding sites in modules 1 and 2 were identified. It is likely that only one of these site I sites actually binds retinol, and the other is unavailable owing to the quaternary association of the four modules. The second type of sites in each module, the so-called site II, was more open, hydrophilic, and predicted to be lower affinity retinol or possibly fatty acid binding sites. Interestingly, the current xIRBP crystals were grown in the presence of 0.5 mM oleic acid, prompting us to speculate that binding of ligands to the specific sites may provide additional dynamical stability necessary for the growth of diffraction-quality crystals. Efforts to grow xIRBP crystals in the presence of retinol and other ligands are now in progress. Elucidation of the nature and number of the ligand-binding sites and their implications in overall IRBP function must await the determination of the crystal structures of some of these complexes.
Acknowledgements
This work was supported by a National Institutes of Health grant EY09412 (F.G-F., D.G.), a Challenge Grant from Research to Prevent Blindness to the Department of Ophthalmology, and a Merit Review Award, Veterans Affairs Research Service (F.G.-F.).
References
- 1.Cunningham LL, Gonzalez-Fernandez F. Coordination between production and turnover of interphotoreceptor retinoid-binding protein in zebrafish. Invest Ophthal Vis Sci. 2000;41:3590–9. [PubMed] [Google Scholar]
- 2.Cunningham LL, Yang L, Gonzalez-Fernandez F. Interphotoreceptor retinoid-binding protein (IRBP) is rapidly cleared from the Xenopus interphotoreceptor matrix. Exp Eye Res. 1999;68:399–410. doi: 10.1006/exer.1998.0633. [DOI] [PubMed] [Google Scholar]
- 3.Hollyfield JG, Varner HH, Rayborn ME, Liou GI, Bridges CD. Endocytosis and degradation of interstitial retinol-binding protein: differential capabilities of cells that border the interphotoreceptor matrix. J Cell Biol. 1985;100:1676–81. doi: 10.1083/jcb.100.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Fernandez F, Ghosh D. Focus on Molecules: Interphotoreceptor retinoid-binding protein (IRBP). Exp Eye Res. 2007 doi: 10.1016/j.exer.2006.09.003. In press. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Fernandez F. Interphotoreceptor retinoid-binding protein–an old gene for new eyes. Vision Res. 2003;43:3021–36. doi: 10.1016/j.visres.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Pepperberg DR, Okajima TL, Wiggert B, Ripps H, Crouch RK, Chader GJ. Interphotoreceptor retinoid-binding protein (IRBP). Molecular biology and physiological role in the visual cycle of rhodopsin. Mol Neurobiol. 1993;7:61–85. doi: 10.1007/BF02780609. [DOI] [PubMed] [Google Scholar]
- 7.Liou GI, Geng L, Baehr W. Interphotoreceptor retinoid-binding protein: biochemistry and molecular biology. Prog Clin Biol Res. 1991;362:115–37. [PubMed] [Google Scholar]
- 8.Gonzalez-Fernandez F. Evolution of the visual cycle: the role of retinoid-binding proteins. J Endocrinol. 2002;175:75–88. doi: 10.1677/joe.0.1750075. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Blakeley LR, Cornwall MC, Crouch RK, Wiggert BN, Koutalos Y. Interphotoreceptor retinoid-binding protein is the physiologically relevant carrier that removes retinol from rod photoreceptor outer segments. Biochemistry. 2007;46:8669–79. doi: 10.1021/bi7004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qtaishat NM, Wiggert B, Pepperberg DR. Interphotoreceptor retinoid-binding protein (IRBP) promotes the release of all-trans retinol from the isolated retina following rhodopsin bleaching illumination. Exp Eye Res. 2005;81:455–63. doi: 10.1016/j.exer.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid-binding protein: role in delivery of retinol to the pigment epithelium. Exp Eye Res. 1989;49:629–44. doi: 10.1016/s0014-4835(89)80059-4. [DOI] [PubMed] [Google Scholar]
- 12.Carlson A, Bok D. Promotion of the release of 11-cis-retinal from cultured retinal pigment epithelium by interphotoreceptor retinoid-binding protein. Biochemistry. 1992;31:9056–62. doi: 10.1021/bi00152a049. [DOI] [PubMed] [Google Scholar]
- 13.Okajima TI, Wiggert B, Chader GJ, Pepperberg DR. Retinoid processing in retinal pigment epithelium of toad (Bufo marinus). J Biol Chem. 1994;269:21983–9. [PubMed] [Google Scholar]
- 14.Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid-binding protein promotes rhodopsin regeneration in toad photoreceptors. Proc Natl Acad Sci USA. 1990;87:6907–11. doi: 10.1073/pnas.87.17.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC. Kinetics of visual pigment regeneration in excised mouse eyes and in mice with a targeted disruption of the gene encoding interphotoreceptor retinoid-binding protein or arrestin. Biochemistry. 1999;38:12012–9. doi: 10.1021/bi990504d. [DOI] [PubMed] [Google Scholar]
- 16.Ripps H, Peachey NS, Xu X, Nozell SE, Smith SB, Liou GI. The rhodopsin cycle is preserved in IRBP “knockout” mice despite abnormalities in retinal structure and function. Vis Neurosci. 2000;17:97–105. doi: 10.1017/s095252380017110x. [DOI] [PubMed] [Google Scholar]
- 17.Ripps H. The rhodopsin cycle: a twist in the tale. Prog Brain Res. 2001;131:335–50. doi: 10.1016/s0079-6123(01)31027-0. [DOI] [PubMed] [Google Scholar]
- 18.Ho MT, Massey JB, Pownall HJ, Anderson RE, Hollyfield JG. Mechanism of vitamin A movement between rod outer segments, interphotoreceptor retinoid-binding protein, and liposomes. J Biol Chem. 1989;264:928–35. [PubMed] [Google Scholar]
- 19.Crouch RK, Hazard ES, Lind T, Wiggert B, Chader G, Corson DW. Interphotoreceptor retinoid-binding protein and alpha-tocopherol preserve the isomeric and oxidation state of retinol. Photochem Photobiol. 1992;56:251–5. doi: 10.1111/j.1751-1097.1992.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 20.Bazan NG, Reddy TS, Redmond TM, Wiggert B, Chader GJ. Endogenous fatty acids are covalently and noncovalently bound to interphotoreceptor retinoid-binding protein in the monkey retina. J Biol Chem. 1985;260:13677–80. [PubMed] [Google Scholar]
- 21.Semenova EM, Converse CA. Comparison between oleic acid and docosahexaenoic acid binding to interphotoreceptor retinoid-binding protein. Vision Res. 2003;43:3063–7. doi: 10.1016/j.visres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Fernandez F, Van Niel E, Edmonds C, Beaver H, Nickerson JM, Garcia-Fernandez JM, Campochiaro PA, Foster RG. Differential expression of interphotoreceptor retinoid-binding protein, opsin, cellular retinaldehyde-binding protein, and basic fibroblastic growth factor. Exp Eye Res. 1993;56:411–27. doi: 10.1006/exer.1993.1055. [DOI] [PubMed] [Google Scholar]
- 23.Stenkamp DL, Calderwood JL, Van Niel EE, Daniels LM, Gonzalez-Fernandez F. The interphotoreceptor retinoid-binding protein (IRBP) of the chicken (Gallus gallus domesticus). Mol Vis. 2005;11:833–45. [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Fernandez F, Baer CA, Ghosh D. Module structure of interphotoreceptor retinoid-binding protein (IRBP) may provide bases for its complex role in the visual cycle - structure / function study of Xenopus IRBP. BMC Biochem. 2007;8:15. doi: 10.1186/1471-2091-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. The HKL Program Suit. In HKL Manual. New Haven, CT.: Yale University Press; 1995. [Google Scholar]
- 26.Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr D Biol Crystallogr. 1999;55:1690–5. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- 27.Ward LD. Measurement of ligand binding to protein by fluorescence spectroscopy. In Methods Enzymol. Edited by Hirs CHW, Timasheff SN. New York: Academic press; 1985: 117: 400 −414. [DOI] [PubMed] [Google Scholar]
- 28.Baer CA, Retief JD, Van Niel E, Braiman MS, Gonzalez-Fernandez F. Soluble expression in E. coli of a functional interphotoreceptor retinoid-binding protein module fused to thioredoxin: correlation of vitamin A binding regions with conserved domains of C-terminal processing proteases. Exp Eye Res. 1998;66:249–62. doi: 10.1006/exer.1997.0418. [DOI] [PubMed] [Google Scholar]
- 29.Cogan U, Kopelman M, Mokady S, Shinitzky M. Binding affinities of retinol and related compounds to retinol binding proteins. Eur J Biochem. 1976;65:71–8. doi: 10.1111/j.1432-1033.1976.tb10390.x. [DOI] [PubMed] [Google Scholar]
- 30.Mertens ML, Kagi JH. A graphical correction procedure for inner filter effect in fluorescence quenching titrations. Anal Biochem. 1979;96:448–55. doi: 10.1016/0003-2697(79)90605-5. [DOI] [PubMed] [Google Scholar]
- 31.Baer CA, Kittredge KL, Klinger AL, Briercheck DM, Braiman MS, Gonzalez-Fernandez F. Expression and characterization of the fourth repeat of Xenopus interphotoreceptor retinoid-binding protein in E. coli. Curr Eye Res. 1994;13:391–400. doi: 10.3109/02713689408999866. [DOI] [PubMed] [Google Scholar]
- 32.Fong SL, Liou GI, Landers RA, Alvarez RA, Bridges CD. Purification and characterization of a retinol-binding glycoprotein synthesized and secreted by bovine neural retina. J Biol Chem. 1984;259:6534–42. [PubMed] [Google Scholar]
- 33.Chen Y, Saari JC, Noy N. Interactions of all-trans-retinol and long-chain fatty acids with interphotoreceptor retinoid-binding protein. Biochemistry. 1993;32:11311–8. doi: 10.1021/bi00093a007. [DOI] [PubMed] [Google Scholar]
- 34.Adler AJ, Evans CD, Stafford WF., III Molecular properties of bovine interphotoreceptor retinol-binding protein. J Biol Chem. 1985;260:4850–5. [PubMed] [Google Scholar]
- 35.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–7. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 36.Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10:43–9. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 37.Adler AJ, Stafford WF, III, Slayter HS. Size and shape of bovine interphotoreceptor retinoid-binding protein by electron microscopy and hydrodynamic analysis. J Biol Chem. 1987;262:13198–203. [PubMed] [Google Scholar]
- 38.Taniguchi T, Adler AJ, Mizuochi T, Kochibe N, Kobata A. The structures of the asparagine-linked sugar chains of bovine interphotoreceptor retinol-binding protein. Occurrence of fucosylated hybrid-type oligosaccharides. J Biol Chem. 1986;261:1730–6. [PubMed] [Google Scholar]
- 39.Fedorovich IB, Semenova EM, Grant K, Converse CA, Ostrovsky MA. Photosensitized light-induced damage of IRBP (interphotoreceptor retinoid-binding protein): effects on binding properties. Curr Eye Res. 2000;21:975–80. doi: 10.1076/ceyr.21.6.975.6984. [DOI] [PubMed] [Google Scholar]


