Abstract
During limb development, chondrocytes located at the epiphyseal tip of long bone models give rise to articular tissue, whereas the more numerous chondrocytes in the shaft undergo maturation, hypertrophy, and mineralization and are replaced by bone cells. It is not understood how chondrocytes follow these alternative pathways to distinct fates and functions. In this study we describe the cloning of C-1-1, a novel variant of the ets transcription factor ch-ERG. C-1-1 lacks a short 27–amino acid segment located ∼80 amino acids upstream of the ets DNA binding domain. We found that in chick embryo long bone anlagen, C-1-1 expression characterizes developing articular chondrocytes, whereas ch-ERG expression is particularly prominent in prehypertrophic chondrocytes in the growth plate. To analyze the function of C-1-1 and ch-ERG, viral vectors were used to constitutively express each factor in developing chick leg buds and cultured chondrocytes. We found that virally driven expression of C-1-1 maintained chondrocytes in a stable and immature phenotype, blocked their maturation into hypertrophic cells, and prevented the replacement of cartilage with bone. It also induced synthesis of tenascin-C, an extracellular matrix protein that is a unique product of developing articular chondrocytes. In contrast, virally driven expression of ch-ERG significantly stimulated chondrocyte maturation in culture, as indicated by increases in alkaline phosphatase activity and deposition of a mineralized matrix; however, it had modest effects in vivo. The data show that C-1-1 and ch-ERG have diverse biological properties and distinct expression patterns during skeletogenesis, and are part of molecular mechanisms by which limb chondrocytes follow alternative developmental pathways. C-1-1 is the first transcription factor identified to date that appears to be instrumental in the genesis and function of epiphyseal articular chondrocytes.
Keywords: transcription factor ERG, articular chondrocytes, growth plate chondrocytes, limb development, tenascin-C
Introduction
During limb skeletogenesis, chondrocytes follow two distinct developmental pathways to two distinct fates and functions. The relatively few chondrocytes present at the epiphyseal extremity of long bone anlagen develop into permanent articular chondrocytes, which give rise to articular cartilage, produce abundant extracellular matrix, and maintain normal joint function throughout life (Mitrovic 1977, Mitrovic 1978; Archer et al. 1994; Pacifici 1995). In contrast, the much more numerous chondrocytes constituting the shaft of the long bone anlagen are transient, become organized into growth plates, undergo maturation, hypertrophy, mineralization and apoptosis, and are eventually replaced by bone cells via endochondral ossification (Howlett 1979; Hunziker 1994). Both populations of chondrocytes are of obvious crucial importance for skeletogenesis and skeletal function. By organizing and maintaining the articular tissue, articular chondrocytes allow the joints and associated structures to exert their important biomechanical roles and permit normal body movements throughout life. Likewise, by undergoing proliferation, maturation, and hypertrophy, the growth plate chondrocytes allow each skeletal element to grow in length and width, contribute to the determination of shape and orientation of the elements, and permit the normal replacement of hypertrophic mineralized cartilage with definitive bone and bone marrow.
Given the fundamental roles played by articular and growth plate chondrocytes in skeletal organization and function, it is not surprising that there is ever increasing interest in them. This has led to the identification of a number of molecules with important roles in the development, behavior, and function of growth plate chondrocytes, including signaling molecules, growth factors, and transcription factors (Colvin et al. 1996; Vortkamp et al. 1996; Enomoto-Iwamoto et al. 1998; Koyama et al. 1999). In contrast, we remain largely ignorant about the mechanisms leading to the development of articular chondrocytes, particularly at the molecular level. Thus, we know little about how the numerically few presumptive articular chondrocytes emerging at the epiphyseal tip of long bone anlagen: (a) acquire their permanent phenotype, (b) escape the maturation and ossification process followed by the more numerous growth plate chondrocytes, (c) transmit these properties to their progeny, and (d) remain functional throughout life. Detailed information on articular chondrocyte development and function would not only be of intrinsic biological interest, but is also likely to have important medical ramifications for joint diseases such as osteoarthritis, in which the articular chondrocyte phenotype is altered and joint function is progressively lost (Hamerman 1989).
It was shown recently that the transcription factor ERG is expressed at sites of future joint development in the early chick limb (Ganan et al. 1996; Macias et al. 1997). ERG belongs to the ets family of transcription factors, which are highly conserved and are involved in a variety of biological processes (Rao et al. 1987; Wasylyk et al. 1993, Wasylyk et al. 1998). Every family member contains the ETS domain, a conserved 85–amino acid region that mediates binding to ets motifs containing the core recognition sequence 5′-GGAA-3′. Three major splice variants of mammalian ERG have been cloned (Duterque-Coquillaud et al. 1993; Prasad et al. 1994). The longest variant, termed ERG-3 or p55erg, is ∼480 amino acids long. In comparison, ERG-2 lacks a short 24–amino acid segment in the central portion of the protein upstream of the ETS domain, and ERG-1/p49 erg lacks both the 24–amino acid segment and an adjacent 5′ 27–amino acid segment. All ERG variants bind DNA and transactivate reporter constructs containing ets response elements, but it is not known whether they have different functions in vivo (Duterque-Coquillaud et al. 1993; Prasad et al. 1994). One avian ERG, ch-ERG, has been cloned to date (Dhordain et al. 1995) and corresponds to mammalian ERG-3/p55erg.
Based on the finding that ch-ERG is expressed at sites of joint formation in early chick embryo limbs, we tested the hypothesis in this study that ch-ERG may be involved in articular chondrocyte development. We report here that in addition to ch-ERG, chick limb cartilages express a novel ERG variant we termed C-1-1, which lacks only the 27–amino acid segment. We found by in situ hybridization, immunohistochemistry and quantitative RT-PCR that C-1-1 is preferentially expressed in developing articular chondrocytes, whereas ch-ERG expression is prominent in prehypertrophic chondrocytes in the growth plate of developing long bones. Experimental analyses in vivo and in vitro reveal that C-1-1 and ch-ERG have very distinct biological properties and may thus play important roles in the behavior and functional diversification of limb chondrocytes.
Materials and Methods
Cartilage Tissue Isolation and RT-PCR
Samples of articular cartilage layer and underlying growth plate were microsurgically isolated from day 17 chick embryo tibiotarsus or femurs. Care was taken to remove adhering perichondrial and connective tissues and obtain as homogeneous cartilaginous tissue as possible. RNA was isolated by the guanidium isothiocyanate method (Iwamoto et al. 1993b) and processed for standard RT-PCR as described (Enomoto-Iwamoto et al. 1998). In brief, 1 μg of RNA was reverse transcribed with Superscript reverse transcriptase (GIBCO BRL) and random hexamers, and amplification was carried out with Elongase (GIBCO BRL) for 30 cycles (95°C for 10 s and 60°C for 1 min) with the following ch-ERG sense and antisense primers (indicated as primer A and primer B in Fig. 1 B, respectively): 5′-CGAAGAGTTATTGTGCCAGC and 3′-GGCACTGTTGAAGATGATGGT. PCR products were separated by 2% agarose gel electrophoresis. PCR products subcloned into pCRII vector (Invitrogen) were sequenced on both strands by an automated DNA sequencer.
Figure 1.
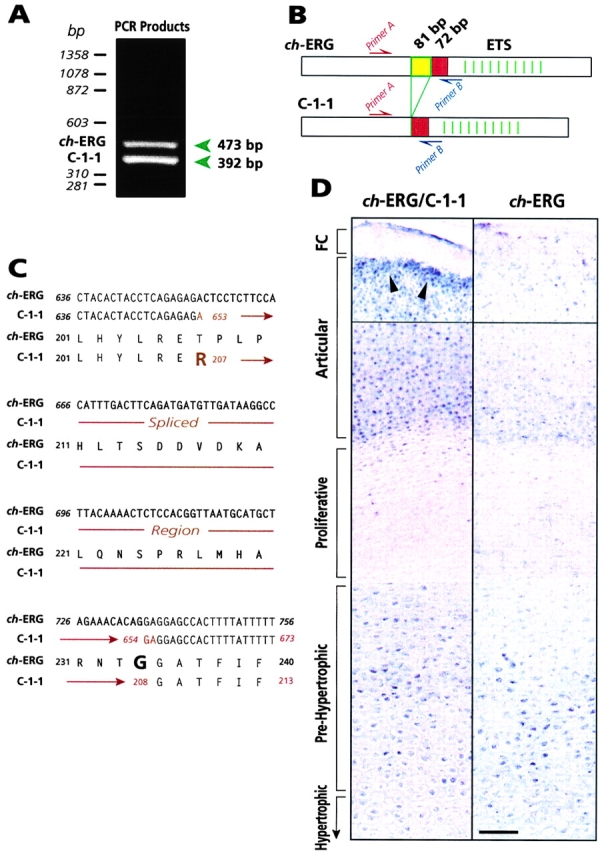
Cloning and analyses of C-1-1 and ch-ERG. (A) Electrophoretic pattern of RT-PCR products with RNA isolated from the entire day 17 chick embryo tibiotarsus. (B) Schematic representation of ch-ERG and C-1-1 showing the location of the alternatively spliced 81- and 72-bp segments located ∼200 bp upstream of the ETS DNA-binding domain. Location of primers used for RT-PCR is indicated. (C) Nucleotide and predicted amino acid sequences of the alternatively spliced region and flanking segments. Note that an amino acid substitution from G to R is predicted to occur in C-1-1 as a result of splicing. (D) In situ hybridization analysis of ch-ERG and C-1-1 gene expression in day 17 chick embryo tibiotarsus, using a common probe hybridizing to both ch-ERG and C-1-1 (left panel) or a probe specific for ch-ERG (right panel). Positive hybridization signal appears as dark blue-purple. Arrowheads point to strongly positive chondrocytes located along the cartilage-fibrocartilage (FC) border. Bar, 35 μm.
Quantitative RT-PCR was carried out using the one-step TaqMan EZ RT-PCR kit and Prism 7700 sequence detection system according to manufacturer's protocols (PE Biosystems). 0.5-μg aliquots of RNA isolated from the above tissue samples were reverse transcribed and amplified using rTth DNA polymerase and the following primers and TaqMan probes: for ch-ERG, 5′-ACTCAGGCCGAGCTATAACG, 3′-TGATCTCCTCGCTTGCTCA, and TacMan probe 5′-FAM-CTCCACGGTTAATGCATGCTAGAAA-TAMRA-3′; for C-1-1, 5′-CGCAGATATCCTCCTGTCACA, 3′-TGATCTCCTCGCTTGCTCATA, and TacMan probe 5′-FAM-ACTACCTCAGAGAGAGAGGAGCCACTTT-TAMRA-3′. Data were normalized to 18S ribosomal RNA simultaneously measured in each sample with the following primers: 5′-CGGCTACCACATCCAAGGAA, 3′-GCTGGAATTACCGCGGCT, and TaqMan probe 5′-JOE-TGCTGGCACCAGACTTGCCCTC-TAMRA-3′.
In Situ Hybridization
This procedure was carried out as described (Koyama et al. 1995) with some modifications. Tissue sections were pretreated with 1 μg/ml proteinase K for 1 min at room temperature and acetylated. Sections were hybridized with 35S-UTP–labeled antisense or sense probes (∼1 × 106 DMP/section) at 50°C for 16 h, washed, treated with 20 μg/ml RNase A for 30 min, washed three times with 0.1× SSC at 50°C for 10 min/wash, coated with Kodak NTB3 photographic emulsion, and exposed for 7–14 d. Alternatively, sections were hybridized with digoxigenin-labeled riboprobes synthesized according to manufacturer's directions (Boehringer), washed as above, and developed with Nitroblue Tetrazolium, 5-bromo-4-chloro-3-indoyl-phosphate substrate solution. The chick type II and type X collagen cDNA clones used were described previously (Koyama et al. 1995). To prepare a ch-ERG–specific probe, oligomers containing an EcoRI site at 5′ end and HindIII site at 3′ end were used for PCR of the 81-bp region present in ch-ERG. The PCR product was then subcloned into pGEM3Z vector. A ch-ERG/C-1-1 probe was prepared by amplification of nucleotide 429–901 region and subcloning into pGEM-T easy vector (Promega).
Immunohistochemistry
This procedure was carried out as described with some modifications (Shimazu et al. 1996). For detection of ERG proteins, paraffin sections of limb long bone cartilaginous anlagen were deparaffinized, rehydrated and treated with 1 μg/ml proteinase K for 10 min at 37°C to unmask nuclear antigens. Sections were then incubated for 2 h at room temperature with different dilutions of primary antibodies which were: (a) rabbit affinity-purified antibodies to human ERG-1/ERG-2 (Santa Cruz Biotechnology; these antibodies cross-react with all variants of ERG and were originally raised against a peptide corresponding to COOH terminus amino acids 344–363 of human ERG-1 that is present in every ERG variant and is highly conserved among species); or (b) a rabbit antiserum specific for ch-ERG that we prepared. Peptide PLPHLTSDDVDKALQNSPRLMH corresponding to the 81-bp region present in ch-ERG only was injected three times in different rabbits. Specificity of the antiserum used in this study (antiserum no. 46200) was verified by ELISA assays. After extensive rinsing, bound antibodies were revealed by the biotin-avidin-peroxidase detection method, Histofine (Nichirei), or by the biotin-avidin-β-galactosidase method (Lim and Chae 1989; Koyama et al. 1999). Slides were counterstained with fast green or left unstained and were mounted and viewed by microscopy.
In experiments monitoring changes in tenascin-C distribution, concentrated RCAS-C-1-1 or RCAS-ch-ERG virus was first injected in the leg bud of stage 22–23 chick embryos; contralateral limb bud was left untreated and served as control. Embryos were allowed to develop until day 10–11, and both legs were dissected and fixed with 4% paraformaldehyde in PBS for 24 h. Serial 8-μm paraffin sections from infected and control legs were mounted on the same glass slides so that the immunostaining results were directly comparable. Sections were treated with 5 mg/ml hyaluronidase in blocking solution (10% normal goat serum) for 3 h at 37°C to remove matrix components and facilitate antibody penetration. Sections were then incubated in fresh blocking solution containing a 1:200 dilution of monoclonal antibody M1 against chick tenascin-C (Chiquet and Fambrough 1984) for 16 h at room temperature. After rinsing, sections were incubated with a 1:250 dilution of rhodamine-conjugated goat anti–mouse IgGs for 1 h, rinsed again, and mounted with 60% glycerol. Sections were viewed and photographed with a Zeiss microscope equipped with epifluorescence.
RCAS Constructs and Limb Implantation
Full-length ch-ERG and C-1-1 cDNAs were isolated from a day 17 chick embryo sternal cartilage λZAP (Stratagene) cDNA library that we prepared. Coding sequences were amplified by PCR (primers: 5′-ATCTTGATCACATTATGGCAAGC and 3′-CACTTAGTAGTAGGTGCCAAGATGG), resequenced to confirm coding fidelity, and subcloned into the ClaI site of RCAS(A) retroviral vector (Hughes et al. 1987). The resulting RCAS-C-1-1 and RCAS-ch-ERG plasmids were transfected into germ-free SPAFAS chick embryo fibroblasts by FuGENE6 transfection reagent (Boehringer) to produce viral particles; companion cultures were transfected with insert-less RCAS plasmid to produce insert-less control virus. Viral particles were isolated and concentrated from fibroblast conditioned media by ultracentrifugation (Enomoto-Iwamoto et al. 1998). For in vivo experiments, concentrated virus (RCAS, RCAS-C-1-1, or RCAS-ch-ERG) was injected in the vicinity of skeletal mesenchymal condensations in stage 22–23 germ-free chick embryo leg bud. Embryos were returned to the incubator and examined at later times. Alternatively, a small fragment of virally infected fibroblast monolayer was implanted into the mesenchyme of stage 22–23 chick embryo leg bud in the vicinity of mesenchymal condensations (Koyama et al. 1996), and embryos were examined at later time points. Viral injection and fibroblast implantation gave comparable results.
Chondrocyte Cultures, Immunoblots, and Northern Blots
Chondrocytes were isolated from the caudal one-third portions of day 17–18 chick embryo sterna (Gibson and Flint 1985; Iwamoto et al. 1993b). The freshly isolated cells were infected with ch-ERG, C-1-1 or insert-less RCAS viruses, and maintained in serum-containing monolayer cultures for up to 5 wk with weekly subculturing. To insure that the ch-ERG- and C-1-1 viruses had maintained coding fidelity throughout the culture period, RNA was isolated from cultures of different ages and subjected to RT-PCR, using primers encompassing the alternative-spliced 81-bp region. The sense primer used, 5′-CTCGCGTACCACTGTGGCAT, corresponds to a sequence between the splice site and the cloning site of the RCAS vector, and the antisense primer used, 3′-TCCGACAGAAGCTCCAGTAGGAA, corresponds to nucleotides 1,005–1,021 common to both ch-ERG and C-1-1. RNA isolation, Northern and Western blot procedures, cDNA probes, immunocytochemistry, and histochemical detection of mineral used for analyses of chondrocyte culture were carried out as described (Iwamoto et al. 1993b; Koyama et al. 1995). Nuclei were isolated from monolayer cultures by slight modifications of previous methods (Kawabe et al. 1999). In brief, cells were rinsed and scraped into PBS at 4°C, pelleted by centrifugation, resuspended in cold hypotonic buffer (10 mM Hepes, pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 5 mM EDTA, and 5 mM EGTA), passed through a 22 1/2–G needle 20 times, and finally spun at 3,500 rpm. Pellets were suspended in Hepes buffer containing 25% glycerol, shaken for 30 min at 4°C, and centrifuged; the resulting supernatants represented nuclear extracts and were processed for Western blot analysis of ch-ERG and C-1-1 content using the antisera described above.
RNA isolated from chondrocyte cultures was processed for Northern blot analysis using chick tenascin-C and aggrecan cDNA clones as described previously (Pacifici et al. 1993).
Results
Cloning of C-1-1 and Expression Patterns of ch-ERG and C-1-1
In the first set of experiments, we asked whether ch-ERG is expressed at late stages of chick limb skeletogenesis and whether ch-ERG variants homologous to mammalian variants occur in chick and are expressed in cartilage. To this end, RNA was isolated from the cartilaginous femur or tibiotarsus of day 17 chick embryos and was processed for reverse transcription polymerase chain reaction (RT-PCR; Fig. 1 A). At this stage of development, the femur and tibiotarsus contain (a) a well-formed epiphysis displaying a thick articular chondrocyte layer and an overlaying thin fibrocartilage layer facing the synovial cavity, and (b) a long metaphyseal-diaphyseal region with growth plate chondrocytes undergoing maturation, hypertrophy, and endochondral ossification (Howlett 1979; Pacifici 1995). The PCR primers used were from sequences upstream and downstream of the 81- and 72-bp segments alternatively spliced in mammals (Fig. 1 B). We obtained two products of 473 and 392 bp (Fig. 1 A). Sequence analysis revealed that the 473-bp product encoded the previously described ch-ERG (Fig. 1 C), while the 392-bp product encoded an alternative and novel variant lacking the 81-bp segment only (Fig. 1B and Fig. C). We named this avian variant C-1-1. No additional ch-ERG variants were obtained by 5′ and 3′ RACE analysis of cartilage samples (not shown).
To identify the chondrocyte population(s) expressing ch-ERG and/or C-1-1, longitudinal sections of day 17 chick embryo tibiotarsus were processed for in situ hybridization (Koyama et al. 1995). We used a common probe that hybridizes to both C-1-1 and ch-ERG, and a probe which is ch-ERG specific and encodes the 81-bp segment present only in ch-ERG. The common probe produced a strong hybridization signal in the articular chondrocyte layer and in the prehypertrophic zone of growth plate (Fig. 1 D, left panel). Signal was particularly strong in chondrocytes located along the border between the articular layer and overlaying fibrocartilage, but there was no signal in the fibrocartilage layer itself (Fig. 1 D, left panel, arrowheads). In comparison, the ch-ERG–specific probe produced a strong signal only in the prehypertrophic zone (Fig. 1 D, right panel), suggesting that the signal in articular cartilage layer was largely due to C-1-1. Low signal was seen in the proliferative and hypertrophic zones with either probe (Fig. 1 D). Sense probes produced no specific hybridization signal (not shown).
To obtain more quantitative data, RNA was isolated from the articular layer and from the underlying growth plate of day 17 tibiotarsus and processed for quantitative RT-PCR, using the single-tube single-enzyme system and internal normalization to 18S ribosomal RNA. We found that C-1-1 and ch-ERG transcripts were present in both samples (Table ). However, C-1-1 transcripts were several fold more abundant in the articular cartilage sample than growth plate sample, while ch-ERG transcript levels were similar in both samples. The C-1-1/ch-ERG ratio was ∼2.8 in articular cartilage and 0.8 in growth plate (Table ).
Table 1.
Determination of C-1-1 and ch-ERG Expression Levels by Quantitative RT-PCR
| Tissue | C-1-1 ± SD | ch-ERG ± SD | C-1-1/ch-ERG |
|---|---|---|---|
| Articular cartilage | 146 ± 33.0 | 52 ± 16.3 | 2.81 |
| Growth plate | 35 ± 7.5 | 44 ± 4.2 | 0.80 |
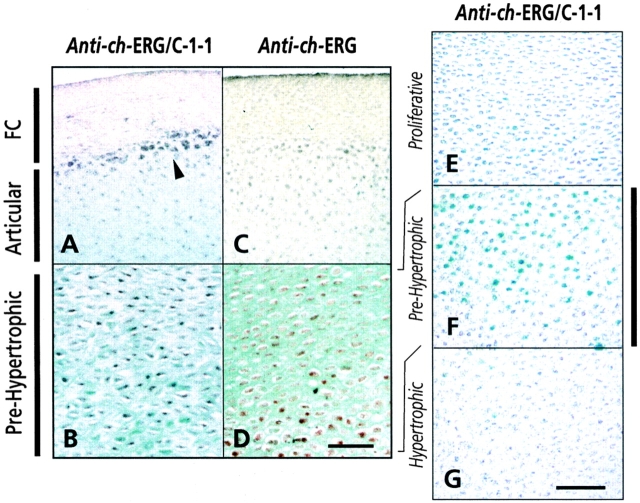
To verify and extend these findings, we processed longitudinal sections of day 17–18 tibiotarsus for immunohistochemistry (Fig. 2), using a commercially available antiserum that reacts with every ERG variant (anti–ch-ERG/C-1-1) and an antiserum specific for ch-ERG that we prepared (anti–ch-ERG). The common ch-ERG/C-1-1 antiserum produced specific nuclear staining in both articular (Fig. 2 A) and prehypertrophic chondrocytes (Fig. 2B and Fig. F); as also seen by in situ hybridization, expression was particularly strong in chondrocytes at the articular-fibrocartilage border (Fig. 2 A, arrowhead). This antiserum did not stain chondrocytes in the proliferative and hypertrophic zones (Fig. 2E and Fig. G) and fibrocartilage cells (Fig. 2 A). In comparison, the ch-ERG–specific antibodies produced strong nuclear staining in prehypertrophic chondrocytes (Fig. 2 D) but much less staining in articular chondrocytes (Fig. 2 C). Other growth plate zones exhibited barely visible staining (not shown).
Figure 2.
Immunohistochemical localization of ERG proteins. Longitudinal sections of day 17–18 tibiotarsus were processed for immunodetection of ERG proteins, using ch-ERG/C-1-1 antiserum (A, B, and E–G) or ch-ERG–specific antiserum (C and D). Histochemical detection of antibodies was by the peroxidase method followed by fast green counterstaining (A–D) or β-galactosidase method without counterstaining (E–G). Positively staining nuclei appear as dark blue or brown. See text for further details. Arrowhead in A points to strongly immunopositive chondrocytes located along the cartilage-fibrocartilage (FC) border. Bars: (A–D) 35 μm; (E–G) 30 μm.
Taken together, the above data show that C-1-1 expression characterizes chondrocytes in the developing articular cartilage layer, while ch-ERG is preferentially produced by prehypertrophic chondrocytes in the growth plate.
Biological Activities of ch-ERG and C-1-1 In Vivo
To determine the biological properties and possible functions of ch-ERG and C-1-1 in skeletogenesis, we ectopically expressed these factors in early chick limb buds using the replication-competent retroviral vector RCAS (Hughes et al. 1987) and determined the effects on long bone development at later stages. Accordingly, we injected concentrated ch-ERG or C-1-1 virus in the anterior mesenchymal region of the leg bud in stage 22–23 (day 3.5–4.0) chick embryos; alternatively, we implanted a small pellet of fibroblasts producing ch-ERG or C-1-1 viral particles in the same region. As control, companion embryos were implanted in the same position with insert-less RCAS virus or fibroblasts producing insert-less virus. Eggs were reincubated until day 10 (stage 36) of embryogenesis, and the effects of misexpression of ch-ERG or C-1-1 were determined by histology and in situ hybridization.
These procedures resulted in a high frequency of infection of the developing tibiotarsus. In control day 10 embryos, the tibiotarsus displayed characteristic histological features, including a proximal flat-shaped articulating epiphysis (Fig. 3 A, arrowhead), a distal round-shaped articulating epiphysis (Fig. 3 B), and a long metaphyseal-diaphyseal shaft with growth plate chondrocytes undergoing maturation and ossification (Fig. 3 A, arrows). The diaphysis contained hypertrophic mineralizing cartilage, endochondral bone, and invading marrow (Fig. 3 E), and was surrounded by a well-formed intramembranous bone collar (Fig. 3 E, arrowheads). When we examined ch-ERG virus-infected embryos, relatively few effects were seen. A slight longitudinal shortening of tibiotarsus was obtained in some cases (2/7). However, the overall histological organization of tibiotarsus was normal, and the diaphysis contained seemingly normal hypertrophic cartilage, endochondral and intramembranous bone (Fig. 3 G, arrowheads), and invading marrow. In sharp contrast, when we examined C-1-1 virus-infected embryos, we found that the tibiotarsus was significantly shorter and abnormally shaped compared with control (5/6) (Fig. 3C and Fig. D). The epiphyses were severely abnormal; paradoxically, the proximal epiphysis was elongated (Fig. 3 C) and the distal one was broad and flatter (Fig. 3 D); that is, the reverse of control epiphyses. In addition, the metaphyseal-diaphyseal shaft displayed no hypertrophic cartilage (Fig. 3 C, arrow), no endochondral bone, no marrow, and no intramembranous bone collar (Fig. 3 F, arrowheads).
Figure 3.
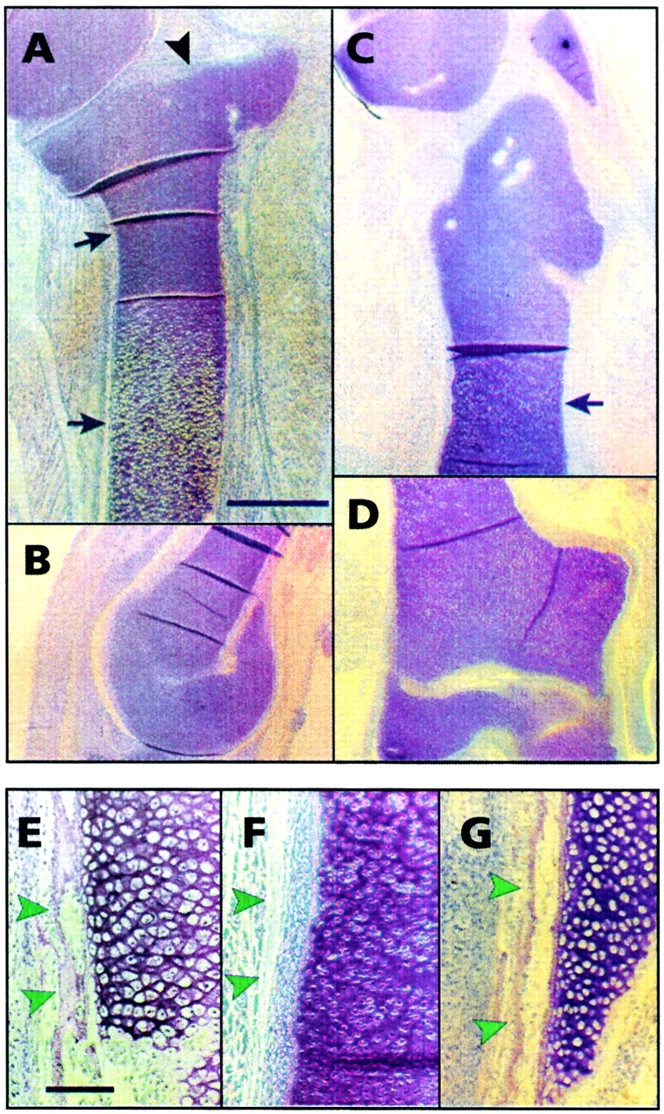
Functional analyses of C-1-1 and ch-ERG roles in chondrocyte and long bone development. Retroviral particles encoding C-1-1 or ch-ERG were implanted in stage 22–23 chick embryo leg buds in ovo; insert-less retroviral particles were implanted in companion embryos as control. Embryos were reincubated and examined on day 10 by histology. (A and B) Proximal one-third portion and distal epiphysis of control tibiotarsus, respectively. Note the characteristic flat proximal articular epiphysis (A, arrowhead) and the round distal epiphysis (B), and the long metaphyseal shaft with chondrocytes undergoing maturation and hypertrophy (arrows in A). (C and D) Entire C-1-1 virus-infected tibiotarsus. These pictures were taken at the same magnification as (A and B) and thus show that the C-1-1 tibiotarsus is nearly half the length of the control. Both proximal and distal epiphyses are severely malformed, and the metaphyseal-diaphyseal shaft (arrow in C) lacks a normal growth plate and hypertrophic chondrocytes. (E) Diaphyseal portion of control tibiotarsus viewed at higher magnification; it displays mineralizing post-hypertrophic chondrocytes, invading bone and marrow precursor cells, and a well-developed intramembranous bone collar (arrowheads). (F) Diaphyseal portion of C-1-1 tibiotarsus in which hypertrophic chondrocytes, marrow invasion and a bone collar (arrowheads) are all absent. (G) Diaphyseal portion of ch-ERG virus-infected tibiotarsus. Note that it closely resembles control tibiotarsus and contains hypertrophic chondrocytes and a bone collar (arrowheads). Bars: (A–D) 1.5 mm; (E–G) 60 μm.
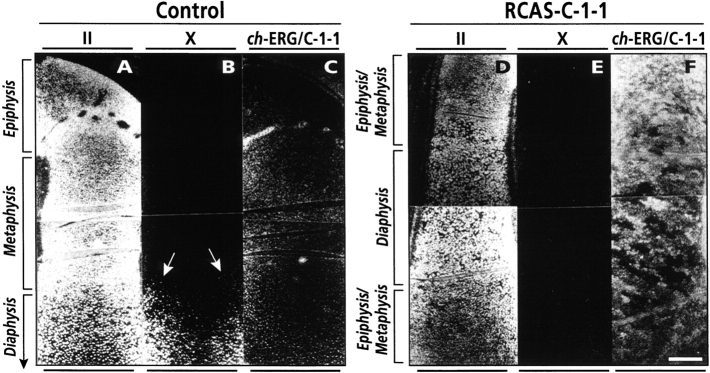
To further assess the effects of C-1-1 misexpression, longitudinal sections of day 10 control and C-1-1–infected legs were processed for in situ hybridization. In control tibiotarsus, expression of the cartilage characteristic type II collagen gene was strong throughout the cartilaginous tissues (Fig. 4 A) and type X collagen gene expression was restricted to hypertrophic chondrocytes (Fig. 4 B, arrows). In C-1-1–infected tibiotarsus, type II collagen expression was strong as in control (Fig. 4 D), but type X collagen gene expression was undetectable (Fig. 4 E), confirming that chondrocyte maturation had been halted. To correlate these gene expression patterns to viral distribution and C-1-1 misexpression, companion longitudinal sections were hybridized with radiolabeled ch-ERG/C-1-1 probe and exposed for a short and identical period of time so that hybridization signal levels were directly comparable. In control specimens, hybridization signal was relatively weak (given the short exposure; Fig. 4 C), whereas in C-1-1–infected specimens signal was extremely strong and present throughout the tibiotarsus and adjacent connective tissues (Fig. 4 F).
Figure 4.
In situ hybridization analysis of gene expression in day 10 control (A–C) and C-1-1 virus-infected (D–F) tibiotarsus. (A and D) Strong type II collagen expression is present in both control (A) and C-1-1–infected (D) tibiotarsus, indicating that infection had no obvious deleterious effects on the differentiated phenotype of chondrocytes. (B and E) Type X collagen expression is present only in control tissue (B, arrows) but not in C-1-1 tissue (E), indicating that chondrocyte hypertrophy had been blocked. (C and F) Relatively weak hybridization signal for endogenous ch-ERG/C-1-1 RNA is seen in control tissue (C), owing to the short exposure time used; in comparison, extremely strong signal is seen in C-1-1–infected tissue (F), indicating presence of C-1-1–encoding virus throughout the cartilaginous tissue and adjacent connective tissues. Note that about one-third of control tibiotarsus is shown in (A–C), whereas nearly the entire RCAS-C-1-1 tibiotarsus is shown in (D–F). Bar, 200 μm.
The severe phenotypic changes in the tibiotarsus of C-1-1–infected embryos were limited to the tibiotarsus itself, did not reflect systemic changes, and did not occur in neighboring uninfected skeletal elements. For example, the fibula next to the C-1-1–infected tibiotarsus shown in Fig. 3C and Fig. D, above had remained uninfected and was morphologically and histologically normal (Fig. 5 A). It displayed abundant type II collagen transcripts in each region (Fig. 5 B) and abundant type X collagen transcripts in its hypertrophic diaphysis (Fig. 5 C), and contained low normal levels of ch-ERG/C-1-1 transcripts, whereas these transcripts were extremely abundant in the surrounding virus-infected perichondrial-mesenchymal tissues (Fig. 5 D).
Figure 5.
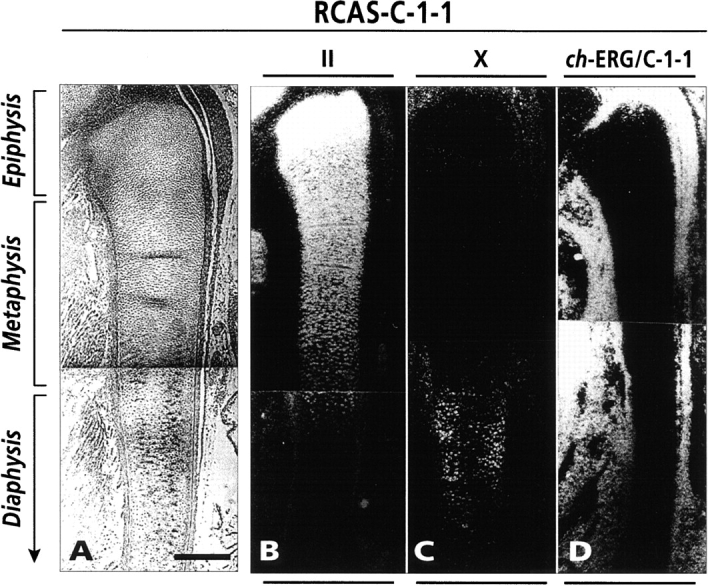
In situ hybridization analysis of gene expression in day 10 chick embryo fibula. Longitudinal sections of fibula and surrounding tissues from C-1-1 virus-infected embryos were processed for in situ hybridization with indicated probes. (A) Phase micrograph; (B) type II collagen hybridization, showing a characteristic strong signal in epiphysis and metaphysis and a much reduced signal in hypertrophic diaphysis; (C) type X collagen hybridization, showing strong signal in hypertrophic diaphysis; and (D) hybridization with common ch-ERG/C-1-1 probe, showing lack of signal in fibula but extremely strong signal in surrounding perichondrial and connective tissues. Bar, 200 μm.
Together, the results show that the biological activities of C-1-1 and ch-ERG are very different. C-1-1 appears to be a potent inhibitor of chondrocyte maturation and hypertrophy; this indicates that one function of this factor is to maintain chondrocytes in an immature and developmentally stable state, such as that displayed by articular chondrocytes. Instead, ch-ERG appears to have modest biological effectiveness.
C-1-1, but Not ch-ERG, Induces Tenascin-C Gene Expression In Vivo and In Vitro
Given the above conclusion, one would predict that C-1-1 should regulate and induce expression of phenotypic traits which are characteristic of articular and immature chondrocytes, while ch-ERG should not. Thus, we determined whether C-1-1 and ch-ERG differentially affect gene expression of tenascin-C, an extracellular matrix molecule produced by articular and immature chondrocytes but not produced by maturing growth plate chondrocytes (Pacifici et al. 1993; Savarese et al. 1996). In this respect, tenascin-C is a unique and rare phenotypic marker, given that articular and growth plate chondrocytes otherwise share numerous phenotypic traits, including collagens II, IX, and XI, link protein, aggrecan, hyaluronan, and others.
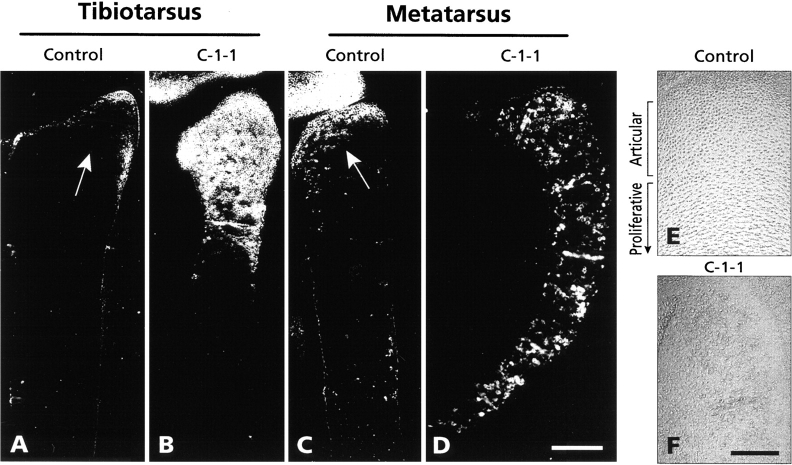
To test the above prediction in vivo, C-1-1 or ch-ERG virus was again injected in the leg bud of stage 22–23 chick embryos; contralateral leg bud was not injected and served as control. On day 10–11 of embryogenesis, legs were dissected, fixed, and embedded in paraffin; the resulting longitudinal serial sections were processed for immunohistochemistry, using the chick tenascin-C monoclonal antibody M1 (Chiquet and Fambrough 1984). To compare the results directly, sections from infected and control legs were mounted on the same microscopic slides and simultaneously processed for immunohistochemistry. In this series of virus injections, both the tibiotarsus and metatarsal elements were infected. In control uninfected elements, tenascin-C displayed a characteristic distribution; it was confined to most epiphyseal developing articular chondrocytes (Fig. 6A and Fig. C, arrows) and was largely absent from the long underlying growth plate (Pacifici et al. 1993). A similar distribution was seen in ch-ERG virus-infected elements (not shown). In sharp contrast, tenascin-C staining in C-1-1 virus-infected elements was both stronger and more widely distributed. In the abnormally shaped tibiotarsus, tenascin-C staining occupied a large portion of the epiphysis-metaphysis and was quite strong; it then decreased in the remainder of the shaft (Fig. 6 B). In the similarly abnormal metatarsal element, tenascin-C staining was actually detectable in each portion and was continuously present from proximal epiphysis to distal epiphysis (Fig. 6 D).
Figure 6.
Immunohistochemical analysis of tenascin-C distribution. Longitudinal sections from control (A and C) and C-1-1 virus-infected (B and D) day 10 chick embryo tibiotarsus and metatarsus were reacted with chick tenascin-C monoclonal antibody and processed for immunofluorescence. Arrows in (A) and (C) point to tenascin-C present in epiphyseal articular layer in control tissue. (E and F) Phase micrographs of epiphyseal portion of control (E) and C-1-1 virus-infected (F) metatarsus. Bars: (A–D) 250 μm; (E and F) 100 μm.
Tenascin-C can affect cell adhesion and morphology (Erickson 1993). It is noteworthy that the wider tenascin-C distribution in C-1-1 virus-infected elements was accompanied by disturbances in the normal patterns of characteristic chondrocyte morphologies. For example, in control metatarsus the epiphyseal tenascin-C–positive incipient articular chondrocytes were round in shape (Fig. 6 E) and were followed by tenascin-C–negative proliferative growth plate chondrocytes exhibiting their typical flat and variously shaped morphology (Fig. 6 E; Howlett 1979; Pacifici et al. 1993). In the C-1-1 virus-infected element, a clear transition from round to flat chondrocytes was absent; instead, most chondrocytes exhibited a roundish-to-oval morphology (Fig. 6 F) and most of them were tenascin-C rich (Fig. 6 D).
For an in vitro test of the hypothesis that C-1-1, but not ch-ERG, regulates expression of tenascin-C, we isolated resting chondrocytes from the caudal portion of day 17 chick embryo sternum, infected them with ch-ERG, C-1-1 or insert-less virus, and maintained them in culture for up to 5 wk with weekly subculturing (passages 1–5). Sternal chondrocytes were used because they can be isolated as homogenous populations, whereas chondrocyte populations isolated from limb skeletal elements usually contain contaminating perichondrial-fibroblastic cells (Gibson and Flint 1985; Iwamoto et al. 1993a). We found that virally driven C-1-1 overexpression did lead to increased tenascin-C mRNA levels at each passage, but ch-ERG overexpression did not (Fig. 7 A). Gene expression of aggrecan, a chondrocyte characteristic matrix component, was not affected by any of the viruses (Fig. 7 A, AG); the slight variations seen with passage number were not significant and may reflect partial decrease in phenotypic expression over time in culture. In good agreement with the RNA analysis, immunocytochemistry on passage 2 cultures showed that tenascin-C was hardly detectable in control cultures but readily apparent in C-1-1–overexpressing cultures (Fig. 7 B). Previous studies showed that factors such as PTH/PTHrP, Indian hedgehog (Ihh) and fibroblast growth factor-2, act as negative modulators of maturation in growth plate chondrocytes (Iwamoto et al. 1995; Colvin et al. 1996; Vortkamp et al. 1996). Interestingly, neither treatment of sternal chondrocytes with any of these factors nor virally driven Ihh overexpression led to increased expression of tenascin-C (not shown).
Figure 7.
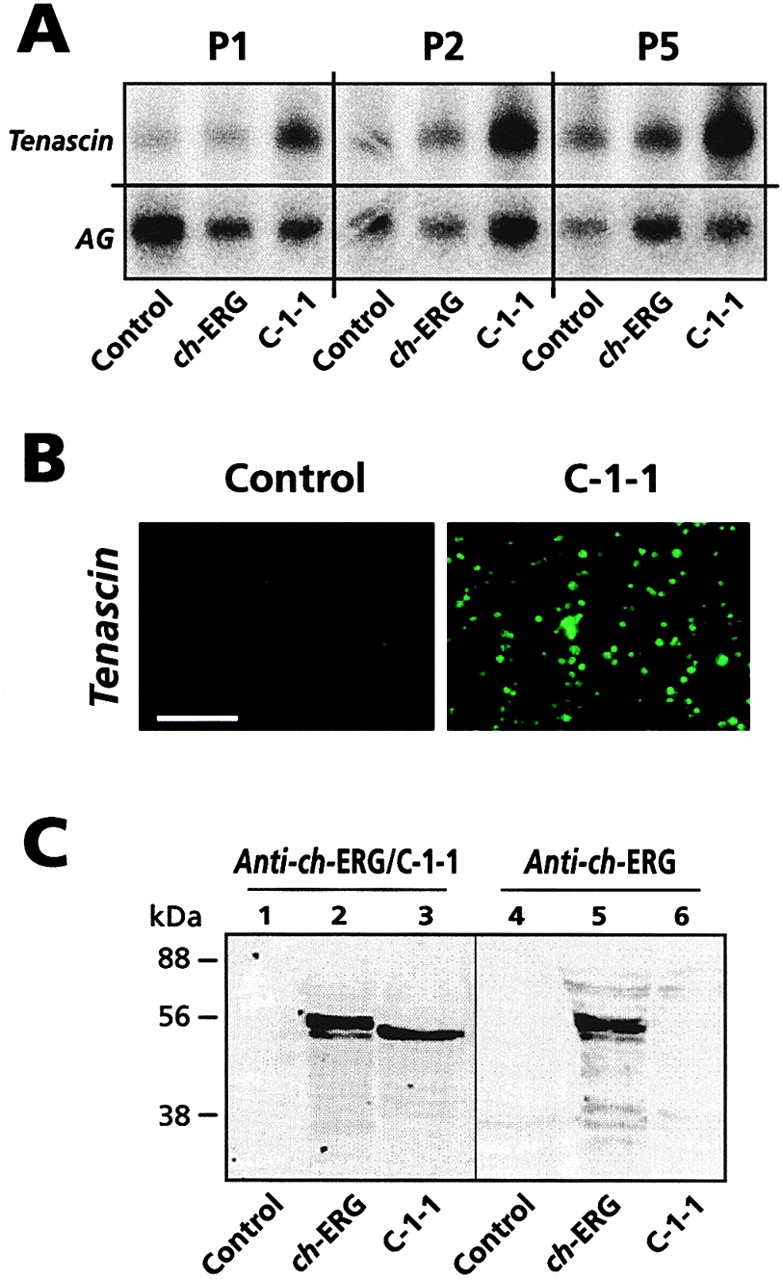
C-1-1 and ch-ERG effects on chondrocyte behavior and development in culture. (A) Northern blot analysis of tenascin and aggrecan (AG) gene expression in passage 1 (P1) to passage 5 (P5) caudal sternal chondrocyte cultures infected with insert-less virus (Control), ch-ERG virus (ch-ERG) or C-1-1 virus (C-1-1). (B) Immunofluorescence micrographs of control (left) and C-1-1 virus-infected (right) P2 cultures stained with tenascin-C monoclonal antibody. (C) Western blots to determine ch-ERG and C-1-1 content in nuclei from control, ch-ERG virus-infected and C-1-1 virus-infected cultures. Affinity-purified antibodies reacting to all ERG proteins (lanes 1–3) or antibodies specific for ch-ERG (lanes 4–6) were used. The major stained protein band in lanes 2 and 5 displays the expected ch-ERG size of ∼55 kD (see Dhordain et al. 1995), and the major band in lane 3 corresponds to C-1-1 and thus migrates slightly faster than ch-ERG. Note that the minor faster migrating band in lanes 2 and 5 likely represents a degradation product of ch-ERG. Bar, 60 μm.
To verify that the ch-ERG and C-1-1 viruses had caused the expected increases in ch-ERG and C-1-1 protein content, nuclear extracts were prepared from each of the above cultures and processed for Western blot analysis; we used the commercially available antiserum that reacts with every ERG variant (anti–ch-ERG/C-1-1), and the antiserum specific for ch-ERG that we prepared (anti–ch-ERG). We found that passage 1 chondrocytes infected with ch-ERG or C-1-1 virus already contained several fold more ch-ERG or C-1-1 protein than control cultures infected with insert-less virus (Fig. 7 C). Note that endogenous levels of ERG proteins in control cultures, though difficult to document in Fig. 7 C, were appreciable by visual inspection and became readily apparent after longer development of the blots (not shown).
Together, the above data show that C-1-1 can selectively induce expression of tenascin-C, a unique product of articular and immature chondrocytes, but ch-ERG can not. In addition, C-1-1 misexpression and the concurrent increases in tenascin-C distribution are accompanied by disturbances in chondrocyte cytoarchitecture.
Opposite Effects of ch-ERG and C-1-1 on Alkaline Phosphatase and Mineralization
In a final set of experiments, we further compared the biological properties of ch-ERG and C-1-1. We reasoned that if C-1-1 maintains chondrocytes in an immature state, it should inhibit alkaline phosphatase activity that is strongly expressed by mature hypertrophic chondrocytes (Leboy et al. 1989); however, ch-ERG should have no effect. To test this prediction, chondrocytes were isolated from the cephalic maturing portion of day 17 chick embryo sterna and grown in culture up to passage 4 by weekly subculturing. APase activity was monitored at each passage. We found that, as expected, APase activity had increased about threefold in control cultures between passage 2 and 3, a reflection of the ongoing and advancing maturation process (Fig. 8 A). In line with our hypothesis, we found that such increase had also occurred in ch-ERG but not in C-1-1–infected cultures (Fig. 8 A). In older passage 4 cultures, we again observed that APase activity had increased further in control and that no detectable APase was seen in C-1-1 cultures. Unexpectedly, however, APase activity in ch-ERG cultures now exceeded that in control by nearly twofold (Fig. 8 A).
Figure 8.
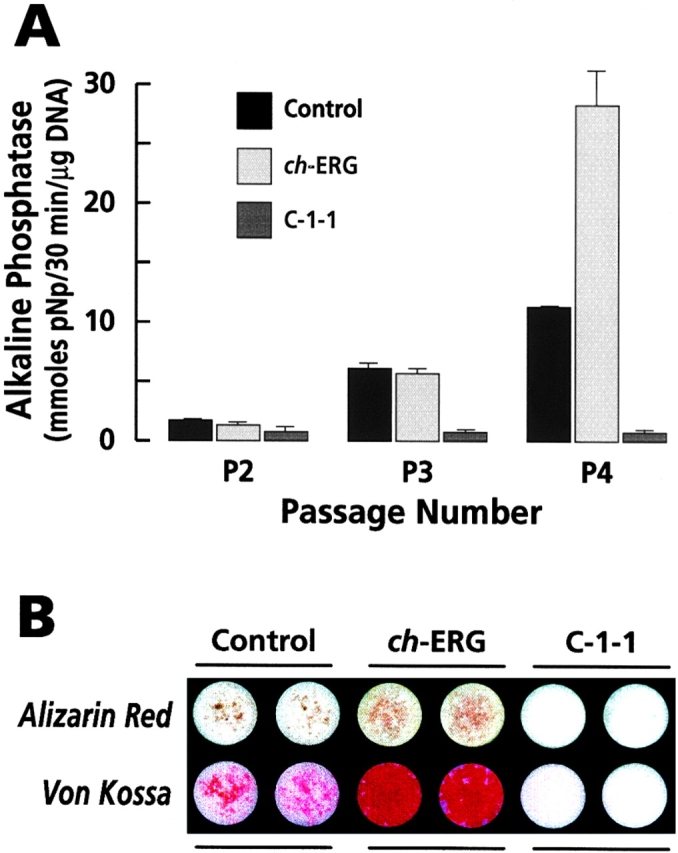
C-1-1 and ch-ERG effects on alkaline phosphatase activity and mineralization. (A) Histograms showing changes in alkaline phosphatase activity in control, ch-ERG virus-infected and C-1-1 virus-infected cultures at passage 2, 3, and 4. (B) Histochemical detection of mineral. Duplicate P4 microwell control and infected cultures were stained with mineral-specific alizarin red or von Kossa stains.
Because APase activity is associated with the mineralization phase of the maturation process (Ali et al. 1970), we asked whether the very large increase in APase activity in late passage ch-ERG cultures may be associated with mineral deposition in the extracellular matrix. Confluent passage 4–5 cultures of control, ch-ERG, and C-1-1 chondrocytes in microwell plates were grown for four additional days in medium containing 25 μg/ml ascorbate, an additive needed for mineralization in culture (Leboy et al. 1989). Cultures (in duplicate) were then processed for histochemical detection of mineral, using alizarin red or von Kossa stains. Indeed, we found that while some mineral was present in control cultures, much larger amounts of it were present in ch-ERG cultures (Fig. 8 B). In agreement with the antimaturation ability of C-1-1 demonstrated above, we found that mineral was completely absent in C-1-1 cultures (Fig. 8 B).
Discussion
In this study, we have demonstrated that developing chick limb skeletal elements express two variants of the transcription factor ERG, the full-length ch-ERG previously described by Dhordain et al. 1995 and a novel shorter variant (C-1-1) lacking a 27–amino acid segment located ∼80 amino acid upstream of the DNA-binding ETS domain. We demonstrate that the gene expression patterns and biological activities of ch-ERG and C-1-1 are distinct. C-1-1 characterizes developing epiphyseal articular chondrocytes, whereas ch-ERG is preferentially expressed by prehypertrophic chondrocytes in the growth plate and to a lesser extent by articular chondrocytes. When constitutively expressed, C-1-1 maintains chondrocytes in an immature and phenotypically stable phenotype, blocks their progression into type X collagen- and APase-rich hypertrophic chondrocytes, and induces expression of tenascin-C, a unique trait of articular and immature chondrocytes. In addition, C-1-1 blocks endochondral ossification and marrow invasion and the formation of an intramembranous bone collar. In comparison, when constitutively expressed in vitro, ch-ERG promotes chondrocyte development and expression of functions associated with the terminal phases of maturation, namely APase activity and mineral deposition in the extracellular matrix. The data indicate that C-1-1 and ch-ERG participate, and have distinct roles, in limb skeletogenesis. Because of its expression patterns and its powerful and distinct biological activity, C-1-1 may be instrumental in the genesis of articular chondrocytes and in the acquisition and maintenance by these cells of their most important feature, a stable and immature phenotype. On the other hand, given its ability to promote chondrocyte maturation in vitro and its weak activity in vivo, ch-ERG may have auxiliary roles in skeletogenesis and may be one among several factors that facilitate completion of chondrocyte maturation and transition from mineralized hypertrophic cartilage to endochondral bone.
Development of Articular and Growth Plate Chondrocytes
As pointed out in the introduction, while the understanding of articular chondrocyte development is quite limited at present, there is a large body of information on the development of growth plate chondrocytes. Thus, it has become apparent that the progression of growth plate chondrocytes through the resting, proliferative, prehypertrophic, hypertrophic, and mineralizing phases of maturation is regulated by a number of powerful factors and molecules, including insulin-like growth factors, fibroblast growth factors, bone morphogenetic proteins, hedgehog proteins, and retinoids (Colvin et al. 1996; Vortkamp et al. 1996; Enomoto-Iwamoto et al. 1998; Koyama et al. 1999). It is of interest that these various factors and molecules have contrasting effects on chondrocyte maturation. For instance, fibroblast growth factors and their tyrosine kinase receptors are found to exert a negative role on both chondrocyte proliferation and progression toward the hypertrophic stage (Colvin et al. 1996; Naski et al. 1998; Sahni et al. 1999). On the other hand, we have shown that endogenous retinoids and their nuclear receptors play a positive role on maturation and are required for chondrocyte hypertrophy, mineralization, and endochondral ossification (Koyama et al. 1999). Together, these studies have clearly indicated that the maturation process of growth plate chondrocytes is regulated by a fine balance between negative and positive mechanisms; that is, mechanisms that oppose maturation and mechanisms that favor it.
It follows that the development of articular chondrocytes may require a shift in such balance in favor of negative mechanisms, so that the maturation process would be effectively and irreversibly blocked and the cells would maintain a stable and immature phenotype through life (Pacifici et al. 1990). C-1-1 and ch-ERG may represent one example of such a shift. We show that these factors are expressed in both the articular layer and growth plate, but at very different ratios. In the articular layer the C-1-1/ch-ERG expression ratio is ∼2.8, whereas in the growth plate it is ∼0.8. Thus, by being more strongly expressed in the articular layer, C-1-1 may be able to exert an effective antimaturation action, maintain the cells in an immature stage, and participate in the regulation of traits characteristic of that stage; these traits include a small cell size, low APase activity, lack of type X collagen synthesis, lack of matrix mineral deposition, and strong tenascin-C synthesis. On the other hand, a shift in the C-1-1/ch-ERG expression ratio to ∼0.8 in the growth plate, combined with preferential expression of these factors at such a ratio in the prehypertrophic zone (see Fig. 1 D and 2 F), may produce a change in biological activity, with ch-ERG becoming preponderant and exerting a promaturation effect.
An important cytological characteristic of developing articular chondrocytes is that the cells are round in shape, whereas the underlying proliferative growth plate chondrocytes are flat and variously shaped (Howlett 1979; Lutfi 1974; Shimazu et al. 1996). In addition, articular chondrocytes are surrounded by abundant amounts of tenascin-C, whereas the growth plate chondrocytes are tenascin-C negative (Pacifici et al. 1993; Savarese et al. 1996). We have shown here that C-1-1 misexpression induces broader and higher expression of tenascin-C; this is accompanied by alterations in chondrocyte morphology, with most cells exhibiting a roundish-to-oval morphology. Thus, C-1-1 appears to be able to directly or indirectly induce two features, i.e., a round cell shape and tenascin-C synthesis, which normally distinguish articular chondrocytes from underlying growth plate chondrocytes and which may be critical for the genesis of articular chondrocytes themselves. In other cell types, tenascin-C plays an antiadhesive role and cells plated onto tenascin-C–coated substrates remain round (Erickson 1993). The protein could have a similar role in articular chondrocytes; if so, induction of tenascin-C and a round cell configuration would be causally linked and dependent on C-1-1 action.
An additional point deserves to be made with regard to tenascin-C induction by C-1-1. Other known negative modulators of the maturation process we tested, including Ihh, PTHrP and FGF-2, failed to induce tenascin-C gene expression in cultured chondrocytes. This finding is actually not surprising in view of the fact that Ihh and PTHrP, but not tenascin-C, are normally expressed in the growth plate (Vortkamp et al. 1996). Thus, our finding suggests an important implication: the antimaturation mechanisms such as those due to C-1-1, which allow developing epiphyseal articular chondrocytes to acquire and maintain a stable immature phenotype, may be different from those serving as negative modulators of maturation rates in growth plate chondrocytes.
Ets Factors and Limb Development
Our results complement well and extend previous work on the roles of ets factors in limb skeletogenesis. In situ hybridization studies in chick showed that ERG transcripts are first present in the prechondrogenic mesenchymal cell condensations in the early limb (Dhordain et al. 1995) and subsequently at sites of future joint development (Ganan et al. 1996; Macias et al. 1997). Related studies showed that in mouse embryo limbs another member of the ets family, ETS-2, is expressed throughout the mesenchyme and then becomes restricted to differentiated chondrocytes (Maroulakou et al. 1994), and that ETS-2 misexpression in transgenic mice leads to severe skeletal defects (Sumarsono et al. 1996). It was not established in these studies whether the ERG expressed in the mesenchymal condensations is ch-ERG, C-1-1 or another variant, whether ERG or its variants are expressed by interzone cells, which constitute the primordial joint and give rise to several joint structures (Mitrovic 1978), and whether ETS-2 is expressed by every chondrocyte present in long bone anlagen or is expressed only by specific populations. Nonetheless, it is clear from these studies and our data that at least two members of the ets family, ETS-2 and ERG, participate in limb skeletogenesis and that their functions are likely to be important and exerted at multiple stages of the process. Thus, the early, partially overlapping expression of ERG and ETS-2 in limb mesenchyme suggests that these factors may first cooperate in the cytodifferentiation process of condensed mesenchymal cells into chondrocytes. The subsequent, more distinct patterns of expression indicate that ERG and ETS-2 may start acting more independently, with ERG having a role in the onset of joint formation and ETS-2 operating in differentiated chondrocytes. Finally, our results indicate that ERG variants may ultimately play regionally specialized roles within each skeletal anlage.
Ets factors regulate transcription by forming heterodimeric complexes with family members or other transcription factors (Wasylyk et al. 1993, Wasylyk et al. 1998; Graves 1998). ETS-2 and ERG strongly interact with each other and can individually interact with c-Fos/c-Jun, producing heterotypic complexes with diverse biological activities and specificities (Butticé et al. 1996; Basuyaux et al. 1997). Interestingly, c-Fos, c-Jun, and other AP-1 complex members are expressed by both immature and hypertrophic chick chondrocytes, with c-Jun expression levels higher in immature than mature cells; in addition, virally driven misexpression of c-Fos or c-Jun in cultured chondrocytes and in the limb inhibits maturation and long bone development (Kameda et al. 1997; Watanabe et al. 1997), precisely as C-1-1 misexpression does. Thus, it is plausible that the biological functions of C-1-1 and ch-ERG in skeletogenesis involve interactions with ETS-2 and c-Fos/c-Jun. In particular, given the similar phenotypic consequences of their misexpression, C-1-1 and c-Fos/c-Jun may interact to maintain chondrocytes in an immature state and induce expression of traits characteristic of that state, such as tenascin-C. In preliminary studies (to be reported elsewhere), we have found that virally driven C-1-1 expression induces strong activity of a tenascin-C gene promoter-reporter construct, indicating that C-1-1 acts on the promoter directly. We have also found that the tenascin-C promoter contains a small element around position –300 that includes an ets consensus site adjacent to an AP-1 site; this element is reminiscent of biologically active ERG/AP-1–responsive element previously identified in other genes, such as that in the mammalian collagenase 1 gene (Butticé et al. 1996). Thus, such an element may similarly regulate tenascin-C gene expression in articular chondrocytes via interaction with C-1-1/c-Fos/c-Jun complexes.
Structural Basis of ERG Function
Work in mammalian systems has shown that ERG is most closely related to certain members of the ets gene family, including ETS-1, ETS-2, FLI 1, and GABPα (Wasylyk et al. 1997, Wasylyk et al. 1998). All these factors are found to contain domains upstream and downstream of the ETS DNA-binding domain (see schematic in Fig. 9 A) that modulate the factors' transcription activation abilities. The ETS domain, termed domain E, is located in the carboxyl half of the protein and contains a winged helix-loop-helix responsible for DNA binding. Domains A and C are transcriptional activator domains, domain B contains a helix-loop-helix thought to mediate protein–protein interactions (Seth and Papas 1990), and domains D and F are regulators of DNA binding. For example, deletion of domain F reduces the ability of human ERG-1 and ERG-2 to activate ets motif-containing reporter constructs by >65% (Siddique et al. 1993).
Figure 9.
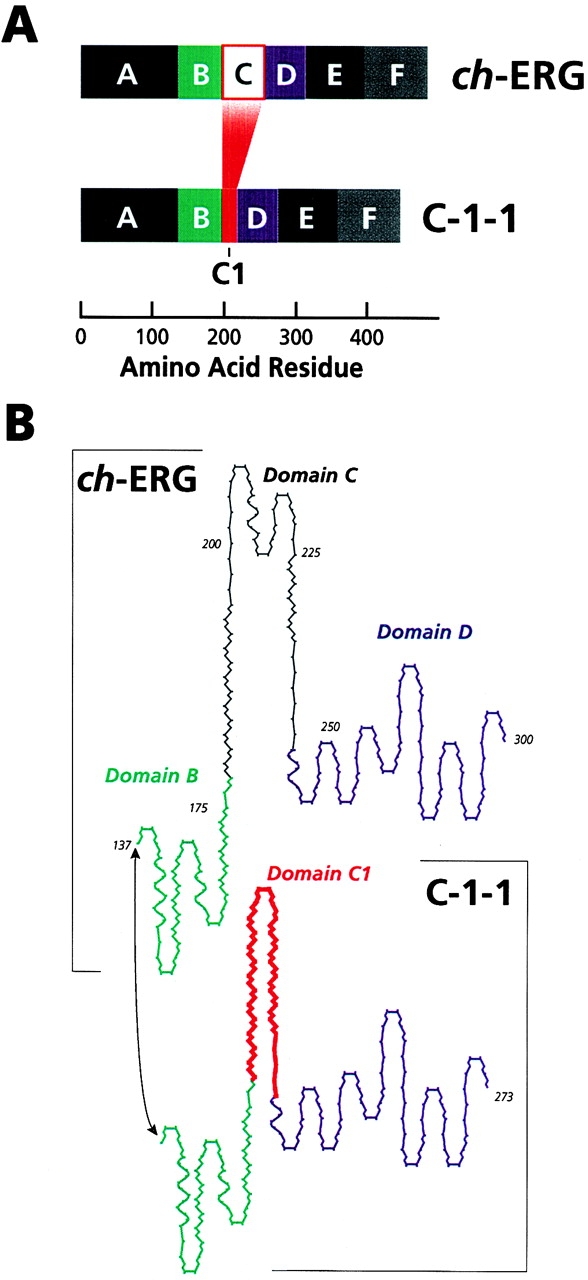
(A) Schematic of the domain structure of ch-ERG and C-1-1, modeled after Wasylyk and Nordheim 1997. (B) Predicted secondary structure of domains B, C and D in ch-ERG and C-1-1. The structurally different domain C in C-1-1 is indicated as domain C1.
ch-ERG shares >95% identity in amino acid sequence with its human counterpart ERG-3 (Dhordain et al. 1995) and thus, the structural organization of ch-ERG is undoubtedly quite similar to that of mammalian ERG as well as that of the other ets members above. What could then be the possible structural reasons for the significantly different biological properties of ch-ERG and C-1-1 we demonstrate here? The 27–amino acid segment present in ch-ERG and absent in C-1-1 represents the amino half of domain C (Fig. 9 A). Preliminary computer-based models of the secondary structure of domains B, C, and D in ch-ERG and C-1-1 are shown in Fig. 9 B. Both domains B and D are predicted to have a globular structure, whereas domain C has an extended configuration. In ch-ERG, domain C protrudes away from domains B and D and displays two close turns. Instead, domain C in C-1-1 (indicated as C1 in Fig. 9 B) is obviously much smaller and contains a readily apparent antiparallel β-sheet with a single turn. In addition, the overall spatial arrangement of domains B, C, and D seems to be different in C-1-1 and ch-ERG; the domains appear to be closer to, and more aligned with, each other in C-1-1 than ch-ERG. Obviously, these models await experimental validation. Nonetheless, they suggest that the predicted significant differences in domain C could have short- and long-range repercussions on the proteins, possibly altering protein-protein and protein-DNA properties and leading to the diverse biological properties of ch-ERG and C-1-1. Domains of ets factors, including domain C, have been shown to be targets of phosphorylation (Rabault and Ghysdael 1994; Wasylyk et al. 1997). If domain C in ERG turns out to be a similar target and if the presence or absence of the 27–amino acid segment affects phosphorylation, this pathway could also contribute to the diverse biological properties of ch-ERG and C-1-1.
We should mention that previous studies on human ERG-1, ERG-2 and ERG-3 found no major functional differences among these alternatively spliced variants (Duterque-Coquillaud et al. 1993; Prasad et al. 1994). The proteins exhibited similar DNA-binding affinity and similar ability to transactivate ets motif-containing reporter constructs. These observations would appear to disagree with our findings, but they probably do not. First, since the mammalian counterpart of C-1-1 has not yet been cloned, it was not included in the comparative analysis of ERG-1, ERG-2, and ERG-3. Second, it is possible that ERG-1, ERG-2, and ERG-3 may actually possess distinct biological properties, which, however, are not revealed by DNA binding and transactivation of reporter constructs. Biological assays more faithful to the in vivo situation may be required to reveal functional differences among variants, such as those used in our study or in studies of other ets variants (Melet et al. 1996).
In conclusion, the data in our study provide evidence that C-1-1 may have an important role in the genesis and function of articular chondrocytes. As far as we know, this is the first transcription factor that, by virtue of its expression patterns and distinct biological activity, has been linked to the development of these cells. Ongoing work should provide details on how C-1-1 exerts this important cellular role and in turn, contributes to the formation and life-long function of diarthrodial joints.
Acknowledgments
We thank Dr. Ellis Golub for help with ProtPlot protein modeling program, Ms. Eleanor Golden for help with immunostaining, and Drs. S. Adams, P. Billings, P. Leboy, and I. Shapiro for comments and suggestions.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan to M. Iwamoto, and National Institutes of Health grant RO1 AR46000-01 to M. Pacifici and J. Rosenbloom.
Footnotes
Abbreviations used in this paper: APase, alkaline phosphatase; Ihh, Indian hedgehog; PTHrP, parathyroid hormone related protein; and RT-PCR, reverse transcriptase polymerase chain reaction.
References
- Ali S.Y., Saidera S.W., Anderson H.C. Isolation and characterization of calcifying matrix vesicles from epiphyseal cartilage. Proc. Natl. Acad. Sci. USA. 1970;67:1513–1520. doi: 10.1073/pnas.67.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer C.W., Morrison H., Pitsillides A.A. Cellular aspects of the development of diathrodial joints and articular cartilage. J. Anat. 1994;184:447–456. [PMC free article] [PubMed] [Google Scholar]
- Basuyaux J.P., Ferreira E., Stehelin D., Butticé G. The ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 1997;272:26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- Butticé G., Duterque-Coquillaud M., Basuyaux J.P., Carrere S., Kurkinen M., Stehelin D. Erg, an Ets-family member, differentially regulates human collagenase 1 (MMP1) and stromelysin (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- Chiquet M., Fambrough D.M. Chick myotendinous antigen. I. A monoclonal antibody as a marker for tendon and muscle morphogenesis. J. Cell Biol. 1984;98:1926–1936. doi: 10.1083/jcb.98.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin J.S., Bohne B.A., Harding G.W., McEwen D.G., Ornitz D.M. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nature Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- Dhordain P., Dewitte F., Desbiens X., Stehelin D., Duterque-Coquillaud M. Mesodermal expression of the chicken erg gene associated with precartilaginous condensation and cartilage differentiation. Mech. Dev. 1995;50:17–28. doi: 10.1016/0925-4773(94)00322-e. [DOI] [PubMed] [Google Scholar]
- Duterque-Coquillaud M., Niel C., Plaza S., Stehelin D. New human erg isoforms generated by alternative splicing are transcriptional activators. Oncogene. 1993;8:1865–1873. [PubMed] [Google Scholar]
- Enomoto-Iwamoto M., Iwamoto M., Mukudai Y., Kawakami Y., Nohno T., Higuchi Y., Takemoto S., Ohuchi H., Noji S., Kurisu K. Bone morphogenetic protein signaling is required for maintenance of differentiated phenotype, control of proliferation, and hypertrophy in chondrocytes. J. Cell Biol. 1998;140:409–418. doi: 10.1083/jcb.140.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P. Tenascin-C, tenascin-R and tenascin-Xa family of talented proteins in search of functions. Curr. Opin. Cell Biol. 1993;5:869–876. doi: 10.1016/0955-0674(93)90037-q. [DOI] [PubMed] [Google Scholar]
- Ganan Y., Macias D., Duterque-Coquillaud M., Ros M.A., Hurle J.M. Role of TGFβs and BMPs as signals controlling the position of the digits and the areas of interdigital cell death in the developing chick limb autopod. Development. 1996;122:2349–2357. doi: 10.1242/dev.122.8.2349. [DOI] [PubMed] [Google Scholar]
- Gibson G.J., Flint M.H. Type X collagen synthesis by chick sternal cartilage and its relationship to endochondral ossification. J. Cell Biol. 1985;101:277–284. doi: 10.1083/jcb.101.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B.J. Inner workings of a transcription factor partnership. Science. 1998;279:1000–1002. doi: 10.1126/science.279.5353.1000. [DOI] [PubMed] [Google Scholar]
- Hamerman D. The biology of osteoarthritis. New Engl. J. Med. 1989;320:1322–1330. doi: 10.1056/NEJM198905183202006. [DOI] [PubMed] [Google Scholar]
- Howlett C.R. The fine structure of the proximal growth plate of the avian tibia. J. Anat. 1979;128:377–399. [PMC free article] [PubMed] [Google Scholar]
- Hughes S.H., Greenhouse J.J., Petropoulos C.J., Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker E.B. Mechanism of longitudinal bone growth an its regulation by growth plate chondrocytes. Microsc. Res. Tech. 1994;28:505–519. doi: 10.1002/jemt.1070280606. [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Shapiro I.M., Yagami K., Boskey A.L., Leboy P.S., Adams S.L., Pacifici M. Retinoic acid induces rapid mineralization and expression of mineralization-related genes in chondrocytes Exp. Cell Res. 207 1993. 413 420a [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Yagami K., Lu Valle P., Olsen B.R., Petropoulos C.J., Ewert D.L., Pacifici M. Expression and role of c-myc in chondrocytes undergoing endochondral ossification J. Biol. Chem. 268 1993. 9645 9652b [PubMed] [Google Scholar]
- Iwamoto M., Shimazu A., Pacifici M. Regulation of chondrocyte maturation by fibroblast growth factor-2 and parathyroid hormone. J. Orthop. Res. 1995;13:838–845. doi: 10.1002/jor.1100130606. [DOI] [PubMed] [Google Scholar]
- Kameda T., Watanabe H., Iba H. c-Jun and JunD suppress maturation of chondrocytes. Cell Growth Diff. 1997;8:495–503. [PubMed] [Google Scholar]
- Kawabe Y., Suzuki T., Hayashi M., Hamakubo T., Sato R., Kodama T. The physiological role of sterol regulatory element-binding protein-2 in cultured human cells. Biochim. Biophys. Acta. 1999;1436:307–318. doi: 10.1016/s0005-2760(98)00119-2. [DOI] [PubMed] [Google Scholar]
- Koyama E., Leatherman J.L., Shimazu A., Nah H.-D., Pacifici M. Syndecan-3, tenascin-C and the development of cartilaginous skeletal elements and joints in chick limbs. Dev. Dyn. 1995;203:152–162. doi: 10.1002/aja.1002030204. [DOI] [PubMed] [Google Scholar]
- Koyama E., Leatherman J.L., Noji S., Pacifici M. Early chick limb cartilaginous elements possess polarizing activity and express Hedgehog-related morphogenetic factors. Dev. Dyn. 1996;207:344–354. doi: 10.1002/(SICI)1097-0177(199611)207:3<344::AID-AJA11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Koyama E., Golden E.B., Vaias L., Kirsch T., Adams S.L., Chandraratna R.A.S., Michaille J.-J., Pacifici M. Retinoid signaling is required for chondrocyte maturation and endochondral bone formation during limb skeletogenesis. Dev. Biol. 1999;208:375–391. doi: 10.1006/dbio.1999.9207. [DOI] [PubMed] [Google Scholar]
- Leboy P.S., Vaias L., Uschmann B., Golub E., Adams S.L., Pacifici M. Ascorbic acid induces alkaline phosphatase, type X collagen, and calcium deposition in cultured chick chondrocytes. J. Biol. Chem. 1989;264:17281–17286. [PubMed] [Google Scholar]
- Lim K., Chae C.B. A simple assay for DNA transfection by incubation of the cells in culture dishes with substrates for beta-galactosidase. Biotechniques. 1989;7:576–579. [PubMed] [Google Scholar]
- Lutfi A.M. The ultrastructure of cartilage cells in the epiphyses of long bones in the domestic fowl. Acta Anat. 1974;87:12–21. doi: 10.1159/000144152. [DOI] [PubMed] [Google Scholar]
- Macias D., Ganan Y., Sampath T.K., Piedra M.E., Ros M.A., Hurle J.M. Role of BMP-2 and OP-1 (BMP-7) in programmed cell death and skeletogenesis during chick limb development. Development. 1997;124:1109–1117. doi: 10.1242/dev.124.6.1109. [DOI] [PubMed] [Google Scholar]
- Maroulakou I.G., Papas T.S., Green J.E. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–1565. [PubMed] [Google Scholar]
- Melet F., Motro B., Rossi D.J., Zhang L., Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol. Cell. Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic D. Development of the diathrodial joints in the rat embryo. Am. J. Anat. 1978;151:475–485. doi: 10.1002/aja.1001510403. [DOI] [PubMed] [Google Scholar]
- Mitrovic D.R. Development of the metatarsalphalangeal joint in the chick embryoMorphological, ultrastructural and histochemical studies. Am. J. Anat. 1977;150:333–348. doi: 10.1002/aja.1001500207. [DOI] [PubMed] [Google Scholar]
- Naski M.C., Colvin J.S., Coffin J.D., Ornitz D.M. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Pacifici M. Tenascin-C and the development of articular cartilage. Matrix Biol. 1995;14:689–698. doi: 10.1016/s0945-053x(05)80011-3. [DOI] [PubMed] [Google Scholar]
- Pacifici M., Golden E.B., Oshima O., Shapiro I.M., Leboy P.S., Adams S.L. Hypertrophic chondrocytes. The terminal stage of differentiation in the chondrogenic cell lineage. In: Pepe F.A., Boettiger D.E., Pacifici M., Rubinstein N.A., Sanger J.W., editors. Cell Lineages in Development. Vol. 599. New York Academy of Sciences; New York: 1990. pp. 45–57. [DOI] [PubMed] [Google Scholar]
- Pacifici M., Iwamoto M., Golden E.B., Leatherman J.L., Lee Y.-S., Chuong C.-M. Tenascin is associated with articular cartilage development. Dev. Dyn. 1993;198:123–134. doi: 10.1002/aja.1001980206. [DOI] [PubMed] [Google Scholar]
- Prasad D.D.K., Rao V.N., Lee L., Reddy E.S.P. Differentially spliced erg-3 product functions as a transcriptional activator. Oncogene. 1994;9:669–673. [PubMed] [Google Scholar]
- Rabault B., Ghysdael J. Calcium-induced phosphorylation of ETS 1 inhibits its specific DNA binding activity. J. Biol. Chem. 1994;269:143–151. [PubMed] [Google Scholar]
- Rao V.N., Papas T.S., Reddy E.S. Erg, a human ets-related gene on chromosome 21alternative-splicing, polyadenylation, and translation. Science. 1987;237:635–639. doi: 10.1126/science.3299708. [DOI] [PubMed] [Google Scholar]
- Sahni M., Ambrosetti D.-C., Mansukhani A., Gertner R., Levy D., Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese J.J., Erickson H., Scully S.P. Articular chondrocyte tenascin-C production and assembly into de novo extracellular matrix. J. Orthop. Res. 1996;14:273–281. doi: 10.1002/jor.1100140216. [DOI] [PubMed] [Google Scholar]
- Seth A., Papas T.S. The c-ets-1 proto-oncogene has oncogenetic activity and is positively autoregulated. Oncogene. 1990;5:1761–1767. [PubMed] [Google Scholar]
- Shimazu A., Nah H.-D., Kirsch T., Koyama E., Leatherman J.L., Golden E.B., Kosher R.A., Pacifici M. Syndecan-3 and the control of chondrocyte proliferation during endochondral ossification. Exp. Cell Res. 1996;229:126–136. doi: 10.1006/excr.1996.0350. [DOI] [PubMed] [Google Scholar]
- Siddique H.R., Rao V.N., Lee L., Reddy E.S.P. Characterization of the DNA binding and transcriptional activation domains of the erg protein. Oncogene. 1993;8:1751–1755. [PubMed] [Google Scholar]
- Sumarsono S.H., Wilson T.J., Tymms M.J., Venter D.J., Corrick C.M., Kola R., Lahoud M.H., Papas T.S., Seth A., Kola I. Down's syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379:534–537. doi: 10.1038/379534a0. [DOI] [PubMed] [Google Scholar]
- Vortkamp A., Lee K., Lanske B., Segre G.V., Kronenberg H.M., Tabin C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Nordheim A. Ets transcription factorspartners in the integration of signal responses. In: Papavassiliou A.G., editor. Transcription Factors in Eukaryotes. Vol. 1. Chapman & Hall; New York: 1997. pp. 253–286. [Google Scholar]
- Wasylyk B., Hahn S.L., Giovane A. The Ets family of transcription factors. Eur. J. Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Bradford A.P., Gutierrez-Hartmann A., Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets 1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Hagman J., Gutierrez-Hartmann A. Ets transcription factorsnuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sciences. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Saitoh K., Kameda T., Murakami M., Niikura Y., Okazaki S., Morishita Y., Mori S., Yokouchi Y., Kuroiwa A., Iba H. Chondrocytes as a specific target of ectopic Fos expression in early development. Proc. Natl. Acad. Sci. USA. 1997;94:3994–3999. doi: 10.1073/pnas.94.8.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]