Abstract
Envoplakin and periplakin are two plakins that are precursors of the epidermal cornified envelope. We studied their distribution and interactions by transfection of primary human keratinocytes and other cells. Full-length periplakin localized to desmosomes, the interdesmosomal plasma membrane and intermediate filaments. Full length envoplakin also localized to desmosomes, but mainly accumulated in nuclear and cytoplasmic aggregates with associated intermediate filaments. The envoplakin rod domain was required for aggregation and the periplakin rod domain was necessary and sufficient to redistribute envoplakin to desmosomes and the cytoskeleton, confirming earlier predictions that the proteins can heterodimerize. The linker domain of each protein was required for intermediate filament association. Like the NH2 terminus of desmoplakin, that of periplakin localized to desmosomes; however, in addition, the periplakin NH2 terminus accumulated at cell surface microvilli in association with cortical actin. Endogenous periplakin was redistributed from microvilli when keratinocytes were treated with the actin disrupting drug Latrunculin B. We propose that whereas envoplakin and periplakin can localize independently to desmosomes, the distribution of envoplakin at the interdesmosomal plasma membrane depends on heterodimerization with periplakin and that the NH2 terminus of periplakin therefore plays a key role in forming the scaffold on which the cornified envelope is assembled.
Keywords: envoplakin, periplakin, desmosomes, cornified envelope, keratinocytes
Introduction
One important function of mammalian epidermis is to provide a physical barrier between the body and the environment. This function depends on a structure known as the cornified envelope that is formed in the outermost epidermal layers (Reichert et al. 1993; Simon 1994; Ishida-Yamamoto and Iizuka 1998; Nemes and Steinert 1999). The cornified envelope is assembled from a range of precursor proteins that become cross-linked to one another through the activity of transglutaminases. The resulting structure is an insoluble layer of protein with covalently attached lipid, ∼15-nm thick, that is deposited on the inner surface of the plasma membrane.
The most current model of how the cornified envelope is assembled (Steinert and Marekov 1999) is based on two types of data: immunoelectron microscopy of isolated envelopes using antibodies to known precursor proteins, and sequencing of peptides released from envelopes during controlled sequential proteolysis (Marekov and Steinert 1998; Steinert and Marekov 1995, Steinert and Marekov 1997, Steinert and Marekov 1999). An early step in envelope assembly is transglutaminase-mediated oligomerization of a precursor known as involucrin. Since involucrin is a cytosolic protein there must be a mechanism by which it becomes associated with the plasma membrane. It is believed that this occurs by cross-linking of involucrin to two proteins, envoplakin and periplakin, which are found at desmosomes and the interdesmosomal plasma membrane (Simon and Green 1984; Ma and Sun 1986; Ruhrberg et al. 1996, Ruhrberg et al. 1997). Subsequent to incorporation of involucrin, envoplakin, and periplakin into the envelope, further proteins are added, including small proline-rich proteins, loricrin, and keratin filaments.
Envoplakin and periplakin belong to the plakin family of cytolinker proteins that also includes desmoplakin, plectin, and BPAG1 (reviewed by Ruhrberg and Watt 1997; Fuchs and Cleveland 1998; Houseweart and Cleveland 1998; Fuchs and Yang 1999; Herrmann and Aebi 2000). In addition to being expressed in the epidermis, envoplakin and periplakin are found in other stratified epithelia and in two-layered and transitional epithelia such as the mammary gland and bladder (Ruhrberg et al. 1996, Ruhrberg et al. 1997; Aho et al. 1998). Periplakin is also highly expressed in brain (Aho et al. 1998). In differentiating keratinocytes that have not yet assembled a cornified envelope envoplakin and periplakin are found at desmosomal plaques, in close proximity to desmoplakin, in an interdesmosomal network at the plasma membrane, and associated with keratin filaments (Ruhrberg et al. 1996, Ruhrberg et al. 1997).
All conventional plakins comprise an NH2-terminal globular domain, a central rod domain and a COOH-terminal globular domain. More distantly related proteins include Drosophila kakapo (Stumpf and Volk 1998; Gregory and Brown 1998) and its mammalian homolgue ACF7 (Leung et al. 1999; Karakesisoglou et al. 2000), which share homology with plakins at the NH2 terminus and with dystrophin at the COOH terminus. The NH2-terminal domain is known to direct desmoplakin and plectin to membrane localization sites (desmosomes and hemidesmosomes, respectively) (Bornslaeger et al. 1996; Smith and Fuchs 1998; Wiche 1998; Geerts et al. 1999). Plectin and some alternatively spliced variants of BPAG1 have, in addition, an NH2-terminal actin binding domain (Yang et al. 1996; Liu et al. 1996; Andrä et al. 1998; Fuchs et al. 1999). One of the BPAG1 isoforms has an NH2-terminal microtubule binding site (Yang et al. 1999) and plectin interacts with microtubules, probably via microtubule-associated proteins (Svitkina et al. 1996; Wiche 1998). The central rod domain forms a coiled coil structure that mediates assembly of plakins into parallel homodimers (Green et al. 1990, Green et al. 1992; Wiche et al. 1991; Sawamura et al. 1991) and the COOH-terminal globular domains of desmoplakin, plectin, and BPAG1 bind intermediate filaments (Foisner et al. 1988; Stappenbeck and Green 1992; Stappenbeck et al. 1993; Wiche et al. 1993; Kouklis et al. 1994; Nikolic et al. 1996; Yang et al. 1996; Steinböck et al. 2000).
Although periplakin and envoplakin share the overall structure of desmoplakin, plectin, and BPAG1, their COOH-terminal domains, are considerably smaller, with envoplakin having only one of the globular subdomains (the C box) and periplakin none (Ruhrberg et al. 1996, Ruhrberg et al. 1997; Aho et al. 1998). The COOH termini of envoplakin and periplakin do, however, include the recently defined linker motif, which is the region of highest sequence homology between all family members (Ruhrberg et al. 1997; Ruhrberg and Watt 1997; Aho et al. 1998; Mahoney et al. 1998; Määttä et al. 2000). This motif comprises the entire COOH terminus of periplakin and in other plakins lies NH2-terminal to the final COOH-terminal globular subdomain (the C box); it includes all but 7 of the 50 amino acids that in plectin comprise the intermediate filament binding site (Nikolic et al. 1996; Mahoney et al. 1998). In addition to their small COOH-terminal domains a notable difference between periplakin, envoplakin, and the other plakins is that their rod domain sequences predict that heterodimer formation would be energetically favorable; indirect support for heterodimer formation comes from the observation that the two proteins coimmunoprecipitate under stringent buffer conditions (Ruhrberg et al. 1997).
Given the proposed role of envoplakin and periplakin as the scaffold on which the cornified envelope is assembled and their divergence (Ruhrberg et al. 1997; Määttä et al. 2000) from other members of the plakin family, we decided to investigate what determines their subcellular distribution and interactions. We have analyzed the full-length proteins and their individual domains by stable and transient transfection of primary human keratinocytes and other cell types. We demonstrate a key role for periplakin in directing the subcellular distribution of both proteins and, specifically, for the NH2 terminus of periplakin in targeting the proteins to the interdesmosomal plasma membrane.
Materials and Methods
Generation of cDNA Constructs
Plasmids containing partial cDNAs (Ruhrberg et al. 1996, Ruhrberg et al. 1997) for both envoplakin and periplakin were used to create full-length cDNAs by using unique restriction enzyme digests to allow fusion of multiple cDNA fragments and assembly into pBluescript II KS+/− (Stratagene). The full-length cDNAs were then tagged with either influenza virus hemagglutinin (HA; periplakin) or FLAG (envoplakin) protein coding sequences in a PCR reaction using primers that linked the epitope tags in frame with the plakin cDNAs at the COOH-terminal ends. Subsequently, the new COOH-terminal fragments were ligated to the cDNA in pBluescript and the resulting tagged cDNA was subcloned into pCI-neo, a mammalian expression vector from Promega. The primers used were: AGGCCCTCCTCTCCGGGATGATCAGTGAAGA (envoplakin COOH terminus, 5′ end); GCTCTAGAGCTCACTTGTCATCGTCGTCCTTGTAGTCGCGAAGGGAGCGCGGGACGGTGGGGGAGGCGGAGCGGTA (envoplakin COOH-terminal FLAG tag, 3′ end); AAGGTAACCCATACGCAGAAGGTGGTGCTGCA (periplakin COOH terminus, 5′ end); CCAAGCTTGGGTCTAGAGCGGCCGCCTAAGCGTAGTCTGGCACGTCGTATGGGTACTTCTGCCCAGATACCAAGACCGCA (periplakin COOH-terminal HA tag, 3′ end).
All truncated periplakin cDNAs were constructed by PCR amplification from the full-length cDNA linking the epitope tag to the mutant cDNA in frame at either the NH2-terminal or COOH-terminal end (see Fig. 1). Primers for generating the periplakin cDNA fused with the HA epitope tag were as follows: 5′-GACCGAGTCGACCTACGCATAGTCAGGAACATCGTATGGGTAATTCTCGGTCTTCAGCACCTC-ATACCGCTGCT (P-1/2N); 5′-AAGACGAATTCGCCGCCACCATGTACCCATACGATGTTCCTGACTATGCGCAGAAGCGCCTG-GGCTCC (P-C); 5′-GACCGAGTCGACCTACGCATAGTCAGGAACATCGTATGGGTAGACGGCCACGGAGCCCAGGCGCTT (P-R and P-NR); 5′- GTGCTCAAGAATTCAGCCACCATGTACCCATACGATGTTCCTGACTATGCGCAGGAATCGGTGGTGAGGAA- GGAGGTGCT (P-RC).
Figure 1.
Constructs used for transfection. Domain nomenclature is as described by Ruhrberg et al. 1996, Ruhrberg et al. 1997. L, linker domain; a.a, amino acid; HA, influenza virus hemagglutinin.
All the envoplakin partial cDNAs except for E-NR and E-RC (see Fig. 1) were assembled using PCR reactions to amplify targeted sequences which were then subcloned in frame into a modified pCI-neo vector (F9). F9 was created by inserting the FLAG epitope into the SmaI site of the pCI multicloning region, flanked by an ATG start site, a TAG termination codon sequence, and interior SmaI and EcoRV restriction sites. The FLAG oligomers were: 5′-GAGGATGACGCCACCATGGATATCGACTACAAGGACGACGATGACAAGCCCGGGTAGATTTCTGCT-3′ and 5′-AGCAGAAATCTACCCGGGCTTGTCATCGTCGTCCTTGTAGTCGATATCCATGGTGGCGGCGTCATCCTC-3′. The envoplakin primers were designed according to the human envoplakin cDNA sequence (GenBank/EMBL/DDBJ accession number U53786), and began and ended at the amino acid residues indicated in Fig. 1.
The envoplakin mutant containing the NH2 terminus and rod domain (E-NR) was constructed by digestion of the full-length envoplakin cDNA with EcoRI followed by partial digestion with BglII. The resultant frag ment was then subcloned into F9 using EcoI and EcoRV, placing the FLAG sequence at the COOH-terminal end.
The E-RC construct was assembled with PCR amplification of the envoplakin cDNA using the following primers: 5′-ATGGCCCAAGCAGGGAGAGAGTCAGAG (5′ end) and 5′-GCGATTTGGCTTTCTGGAGGACGTCAAT (3′ end). The resulting fragment was digested at the unique AatII restriction site and ligated to the full-length FLAG-tagged envoplakin cDNA, which had previously been digested with HindIII and AatII to remove sequences encoding the NH2 terminus. The resulting E-RC construct was then subcloned into pCI-neo.
Cells and Cell Lines
Normal human keratinocytes (strains km, kp, and kq) isolated from newborn human foreskin were maintained in the presence of mitomycin C–treated 3T3 feeder cells in FAD (one part Ham's F12 to three parts DME, supplemented with 1.8 × 10−4 M adenine) medium supplemented with 10% FCS (Imperial Laboratories) and HICE cocktail, consisting of 0.5 μg/ml hydrocortisone, 5 μg/ml insulin, 10−10 M cholera toxin, and 10 ng/ml epidermal growth factor (Ruhrberg et al. 1996, Ruhrberg et al. 1997). An HPV16-transformed line of human keratinocytes, vp (Pei et al. 1991), and a spontaneously immortalized line of adult mouse keratinocytes, MKneg (Romero et al. 1999), were grown in FAD + FCS + HICE without a feeder layer.
A431 (human epidermoid carcinoma), COS7 (African green monkey kidney), NIH and Swiss 3T3 (mouse embryonic fibroblasts), SCC-13 (human squamous cell carcinoma), and HeLa (human cervical carcinoma) cell lines were maintained in DME supplemented with 10% FCS. SW13-1.1 and 1.2 clones (Sarria et al. 1994) were cultured in FAD medium supplemented with 5% FCS; clone 1.2 was maintained under selection in 500 μg/ml G418 (Sigma-Aldrich).
Transient Transfection
Two different methods were used to transiently transfect cells, each of which gave the same results. In the first method primary human keratinocytes were seeded on coverslips at a density of 7.5 × 105 cells/well in a six-well plate in FAD + FCS + HICE for 3 h. Nonadherent cells were removed by washing twice in PBS and adherent cells were cultivated in keratinocyte serum-free medium (KSFM; GIBCO BRL) overnight. The next day, the medium was replaced with transfection reaction mix and incubated for 2 h. The cells were then cultivated in KSFM for 16 h, after which time the medium was replaced with FAD + FCS + HICE for a further 5 or 24 h. The transfection reaction mix consisted of Fugene 5 (Boehringer) or Superfect (QIAGEN) lipofection reagent, combined with serum-free DME and 2 μg of the relevant plasmid DNA, according the manufacturer's protocol. A431, SCC-13, 3T3, and HeLa cells were transfected in their normal culture media with Fugene 5 the day after plating at densities of 3–4 × 105 cells per well and fixed 24 h later.
In the second method primary human keratinocytes were adapted to KSFM without feeders for one passage before transfection. The day before transfection, keratinocytes were seeded on coverslips in 24-well plates at a density of 5 × 104 cells/well. The keratinocytes were treated with transfection reaction mixture for 1–2 h, washed with PBS, and transferred to FAD + FCS + HICE for 24 h. The transfection mixture consisted of Superfect reagent (QIAGEN) combined with KSFM and plasmid DNA, according the manufacturer's protocol. For transfection of HeLa, COS7 and NIH3T3 cells serum-free DME was used instead of KSFM.
Establishment of Stably Transfected Cell Lines
A well-differentiated, spontaneously immortalized line of keratinocytes derived from adult mouse epidermis (MKneg cells) (Romero et al. 1999) was adapted to growth in KSFM. The cells were transfected with Superfect containing full-length FLAG-tagged envoplakin or full-length HA-tagged periplakin in pCI-neo. 72 h after transfection cells were transferred to selection medium containing 0.5 mg/ml G418 (GIBCO BRL). G418-resistant colonies were isolated using cloning rings and transferred to 24-well plates. A total of 48 clones transfected with the envoplakin construct and 10 clones transfected with the periplakin construct were screened by immunofluorescence with anti-FLAG or anti-HA antibodies. Positive clones (five expressing envoplakin; two expressing periplakin) were expanded and maintained in KSFM or FAD + FCS + HICE containing 0.5 mg/ml G418.
Immunofluorescence Staining
The following primary antibodies were used: 11-5F (mouse monoclonal antibody to desmoplakin; gift of David Garrod, University of Manchester, Manchester, UK), DPI/II (mouse monoclonal antidesmoplakin; ICN Biomedicals), CR3 and CR5 (rabbit antibodies to periplakin and envoplakin, respectively; Ruhrberg et al. 1997), AE11 (mouse monoclonal antibody to periplakin; gift of Henry Sun, New York University Medical School, New York, NY; Ma and Sun 1986), LP34 (mouse monoclonal antikeratins 5, 6, 18; Lane et al. 1985), LL001 (mouse monoclonal antibody to keratin 14; Lane 1982), 13.2 (mouse monoclonal antivimentin; Sigma-Aldrich), V9 (mouse monoclonal antivimentin; Novocastra), rabbit antigiantin (a gift of David Shima, Imperial Cancer Research Fund), goat antitransferrin receptor (Santa Cruz Biotechnology, Inc.), AF8 (mouse monoclonal antibody to calnexin; gift of M.B. Brenner, Brigham and Women's Hospital, Harvard Medical School, Boston, MA; Hochstenbach et al. 1992), 1D3 (rabbit antiprotein disulphide isomerase; gift of S. Fuller, EMBO, Heidelberg, Germany; Huovila et al. 1992), TUB 1A2 (mouse monoclonal antibody to β-tubulin; Sigma-Aldrich), rabbit anti-HA (Y11; Santa Cruz Biotechnology, Inc.), rabbit anti-FLAG (Santa Cruz Biotechnology, Inc.), M2 (mouse monoclonal anti-FLAG; Kodak), and mouse monoclonal antilamin B2 (Novocastra). Alexa 488– or Alexa 594–conjugated goat anti–rabbit or –mouse IgG (Molecular Probes) were used as secondary antibody. In some experiments M2 and LP34 were directly conjugated to Alexa 488 and Alexa 594 fluorophores, respectively. When two different antibodies were used to detect the same protein by immunofluorescence they gave similar staining patterns.
Cells on coverslips were fixed in 3–4% paraformaldehyde for 20 min at room temperature or in methanol/acetone 1:1 on ice or at −20°C for 5 min. Paraformaldehyde fixation gave optimal preservation of membrane-associated antigens, whereas cytoskeletal antigens were optimally visualized after methanol/acetone fixation. Paraformaldehyde-fixed cells were permeabilized with 0.1–0.2% Triton X-100 for 5 min at room temperature. In some experiments cells were incubated with 200 ng/ml Latrunculin B (Calbiochem-Novabiochem) for 3 h at 37°C before fixation. After fixation cells were washed thoroughly in PBS or PBSABC and blocked in 0.2% fish skin gelatin (Sigma-Aldrich) or a 1:500 dilution of goat serum in PBS. Cells were incubated with primary antibodies for 45 min at room temperature or with FITC-conjugated phalloidin (to visualize F-actin) for 10 min at room temperature, washed in PBS or PBSABC, and then incubated for a further 45 min at room temperature with the appropriate Alexa conjugated secondary antibodies. After further washing in PBS or PBSABC coverslips were mounted in Gelvatol (Monsanto) and examined using an Axiophot Microscope (Carl Zeiss, Inc.) or a laser scanning confocal microscope (LSM 510; Carl Zeiss, Inc.). Digital images were prepared using Adobe Photoshop 5.0.
Electron Microscopy and Immunogold Labeling
For conventional transmission EM, keratinocytes were grown to confluence on 6-cm tissue culture dishes, and then detached as an intact sheet by incubation with 50 mg/ml Dispase II (Boehinger) in serum-free FAD at 37°C for 5 min. The sheets were washed in PBS and collected as a pellet by centrifugation. The pellet was fixed in glutaraldehyde and processed as described previously (Gandarillas et al. 1999).
For immunogold labeling, cell pellets prepared as described above were transferred to a BalTec high pressure freezing apparatus for cryofixation at a pressure of 2,500 bar and temperature of −170°C (Studer et al. 1989). The frozen specimens were freeze-substituted in anhydrous acetone containing 0.5% uranil acetate and kept at −90°C in freeze-substitution medium for 72 h. The temperature was incrementally raised to −45°C using the Bal-Tec freeze-substitution apparatus. Specimens were washed in anhydrous acetone and embedded in Lowicryl HM20 resin at −45°C with UV polymerization for 48 h.
Ultrathin sections of cell sheets were mounted on carbon-coated grids and labeled as follows. After 10 min incubation on a drop of PBS, the grids were blocked in 0.05% fish skin gelatin in PBS (FGS/PBS) for 30 min, and then labeled with rabbit anti-FLAG or anti-HA antibody overnight at 4°C. The sections were washed with 0.05% FGS/PBS and then labeled with a 1:65 dilution of protein A conjugated to 10 nm gold (J.W. Slot, Cell Biology Department, University of Utrecht, Netherlands) in 0.05% FGS/PBS for 30 min. After further washing in PBS and distilled water the sections were air-dried and contrasted with saturated aqueous uranyl acetate and lead citrate. As a negative control the primary antibody was omitted. For double labeling, sections were first labeled with the anti-FLAG antibody followed by 5 nm protein A gold and then with LP34 antibody followed by 10 nm gold-conjugated goat anti–mouse IgG (Bio Cell International, Cardiff, UK) (Slot et al. 1991). Thin sections were examined with a Jeol EM1200 or EM1010 electron microscope.
Results
Full-length Envoplakin Forms Nuclear and Cytoplasmic Aggregates
Transfection of full-length, FLAG-tagged envoplakin cDNA (Fig. 1) into primary human epidermal keratinocytes resulted in the formation of prominent nuclear and cytoplasmic aggregates (Fig. 2, a–d, i, and j), with only a small proportion of the tagged protein accumulating at cell–cell borders. Aggregates were observed in both basal and involucrin-positive keratinocytes and in standard and low calcium medium (data not shown). Envoplakin aggregates were also observed when untagged full-length envoplakin was expressed, ruling out a potential contribution of the FLAG tag (data not shown). Aggregates formed in all cell types transfected: the human keratinocyte-derived tumor lines SCC13, HeLa, and A431, HPV16-immortalized human keratinocytes (vp), a well differentiated mouse keratinocyte line (MKneg), COS7 cells, and NIH and Swiss 3T3 mouse embryo fibroblasts (Fig. 2 and data not shown).
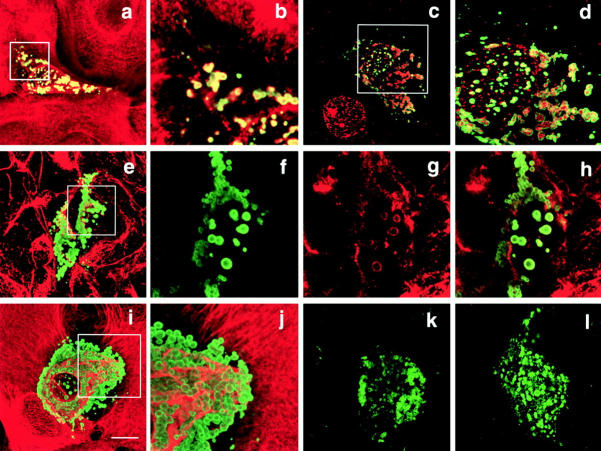
Figure 2.
Transient transfection of full-length FLAG-tagged envoplakin into primary human keratinocytes (a–d, i, and j), COS7 cells (e–h), SW13-1.1 (k), and SW13-1.2 (l) cells. Green fluorescence: anti-FLAG. Red fluorescence: antikeratin (LP34) (a, b, i, and j); antilamin B (c and d); antivimentin (e, g, and h). Bar, 10 μm. Areas boxed in a, c, e, and i are shown at higher magnification in b, d, f–h, and j, respectively.
Envoplakin aggregates were not labeled with antibodies to markers of the Golgi (giantin), endosomes (transferrin receptor), endoplasmic reticulum (calnexin), or lysosomes (protein disulphide isomerase) (data not shown). However, cytoplasmic aggregates stained positive with antibodies to keratin filaments in epithelial cells (Fig. 2, a and b) and to vimentin in fibroblasts and COS7 cells (Fig. 2, e–h). Nuclear aggregates were labeled with antibodies to lamin B2 (Fig. 2c and Fig. d). In all cell types except primary keratinocytes all aggregates labeled with antibodies to intermediate filaments; however, in keratinocytes heterogeneity was observed, with aggregates in occasional cells being unlabeled (compare Fig. 2, a and b with i and j). The absence of keratin labeling did not show a simple correlation with the differentiation status of the cells since aggregates were labeled with the pan-keratin antibody LP34 (which recognizes keratins 5, 6, and 18) and with monospecific antibodies to keratin 14 (expressed by basal keratinocytes) and keratin 6 (expressed by differentiating keratinocytes in hyperproliferative epidermis) (Fig. 2, a and b and data not shown).
To determine whether aggregate formation required the presence of cytoplasmic intermediate filaments full-length FLAG-tagged envoplakin was transfected into two clones of the human adrenal tumor cell line SW13 (Sarria et al. 1994). Aggregates were observed in both the vimentin-positive SW13-1.1 clone (Fig. 2 k) and the vimentin-negative SW13-1.2 clone (Fig. 2 l), indicating that aggregate formation occurred independently of an intermediate filament cytoskeleton.
To further analyze the envoplakin aggregates, transiently transfected primary human keratinocytes (Fig. 3, a and b) and stably transfected mouse keratinocytes (Fig. 3 c) were examined by conventional and immunoelectron microscopy. Transiently transfected cells were characterized by bundles of keratin filaments within which were trapped large numbers of cytoplasmic vesicles; in some cells the nuclear membrane appeared to be disrupted (Fig. 3, a and b). When ultrathin cryosections were incubated with anti-FLAG antibodies and protein A gold the keratin filaments were specifically labeled (data not shown).
Figure 3.
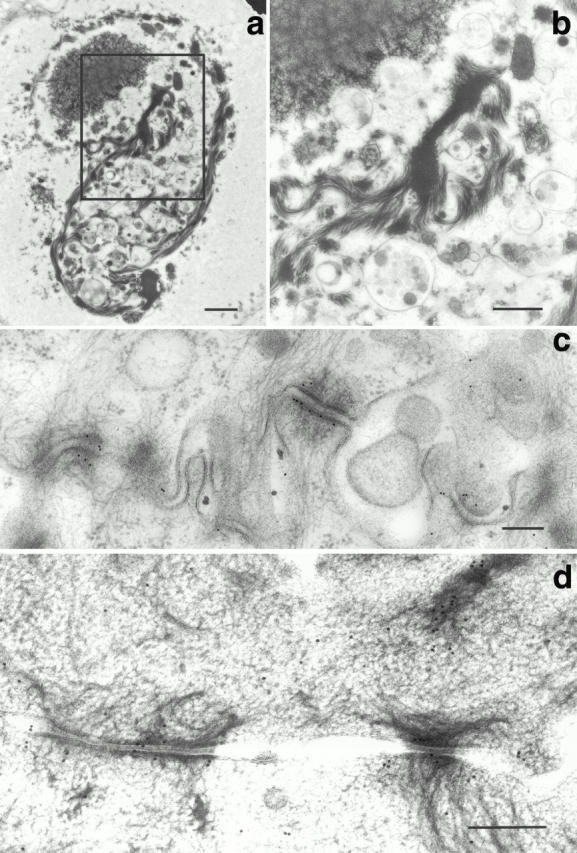
Electron micrographs of transiently transfected primary human keratinocytes (a and b) and stably transfected mouse keratinocytes (c and d). Cells were transfected with FLAG-tagged full-length envoplakin (a–c) or HA-tagged full-length periplakin (d). (a and b) Conventional transmission EM: (b) corresponds to boxed area in a. (c and d) Immunoelectron microscopy with antibodies to FLAG (c) or HA (d). Bars: (a) 1 μm; (b) 500 nm; (c and d) 200 nm.
In contrast to the aggregates in transiently transfected cells (Fig. 2; Fig. 3, a and b) most of the aggregates in stably transfected keratinocytes were small. It is therefore likely that cells containing large aggregates did not survive or failed to divide. In stably transfected keratinocytes full-length envoplakin was localized both to small aggregates and to the plaques of desmosomes (Fig. 3 c and data not shown). We conclude that although much of the envoplakin expressed in transfected cells formed aggregates full-length envoplakin did have the ability to incorporate into desmosomes, in the same location as the endogenous protein (Fig. 3 c; compare Ruhrberg et al. 1996).
The Envoplakin Rod Domain Is Required for Aggregate Formation
To investigate the domains of envoplakin responsible for aggregation we transiently transfected cells with the partial envoplakin constructs illustrated in Fig. 1. Nuclear and cytoplasmic aggregates formed when the COOH-terminal C box and all but 11 amino acids of the linker domain were deleted (Fig. 4, a and b) or when the NH2 terminal domain was deleted (Fig. 4c and Fig. d). In each case the aggregates stained positive with antibodies to intermediate filaments (keratins in epithelial cells; vimentin in 3T3 cells) (Fig. 4, a–d and data not shown). Thus the association of the aggregates with intermediate filaments occurred in the absence of either the NH2 terminus or the C box of envoplakin.
Figure 4.
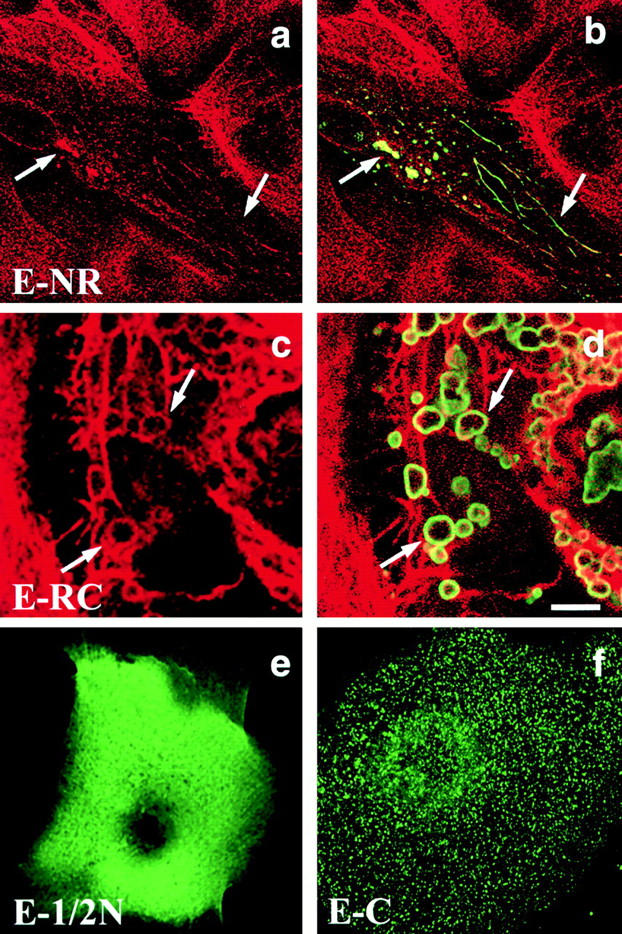
Transient transfection of primary human keratinocytes with different domains of envoplakin. E-NR (a and b); E-RC (c and d); E-1/2N (e); E-C (f). Green fluorescence: anti-FLAG. Red fluorescence: antikeratin (LP34). The same fields of cells are shown in a and b and in c and d. Arrows in a–d show the positions of individual envoplakin aggregates and associated intermediate filaments. Bar: (a and b) 9.3 μm; (c and d) 5 μm; (e) 4 μm; (f) 4.4 μm.
The first 500 amino acids of the NH2 terminus (E-1/2N) and the COOH-terminal C box of envoplakin did not form aggregates. The E-1/2N construct, corresponding to the region of desmoplakin required to target desmosomes (Bornslaeger et al. 1996), was diffusely distributed in the cytoplasm (Fig. 4 e). The C box was also distributed throughout the cytoplasm, but in a punctate pattern (Fig. 4 f). We conclude that aggregation required the rod domain of envoplakin. Neither the NH2 terminus nor C box in isolation showed any strong association with the cytoskeleton and there was no evidence that either construct accumulated at desmosomes (Fig. 4e and Fig. f).
Full-length Periplakin Associates with Desmosomes and Keratin Filaments
When HA-tagged full-length periplakin was transiently transfected into primary human keratinocytes the protein accumulated on the apical plasma membrane and at cell–cell contacts (Fig. 5, a and b). In cells expressing high levels of the protein there was additional filamentous cytoplasmic staining (Fig. 5c and Fig. d) with, in some cells, bundling and partial collapse of the keratin filament network (data not shown). Double labeling with antidesmoplakin revealed partial colocalization of periplakin with desmosomal junctions (Fig. 5e and Fig. f) and this was confirmed by immunoelectron microscopy of mouse keratinocytes stably transfected with full-length HA-tagged periplakin (Fig. 3 d). Cytoplasmic periplakin showed partial colocalization with keratin filaments in keratinocytes, particularly at points of insertion of keratin filaments into desmosomes (Fig. 5c and Fig. d; Fig. 3 d). In COS7 cells periplakin showed partial colocalization with vimentin filaments (Fig. 5g and Fig. h). The distribution of untagged full-length periplakin in transiently transfected cells was the same as that of the HA tagged protein (data not shown).
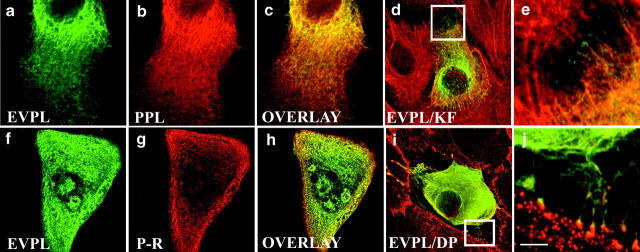
Figure 5.
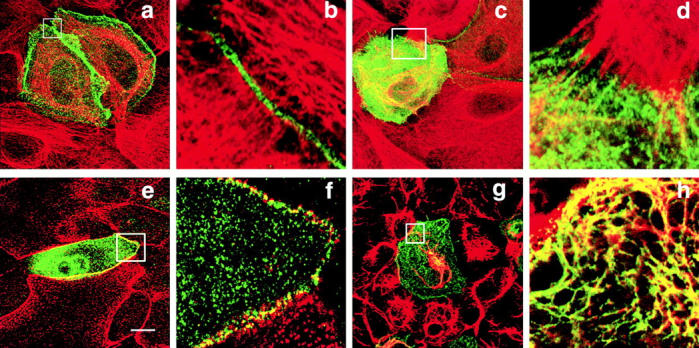
Transient transfection of full-length HA-tagged periplakin into primary human keratinocytes (a–f) and COS7 cells (g and h). Green fluorescence: anti-HA. Red fluorescence: antikeratin (LP34) (a–d); antidesmoplakin (e and f); antivimentin (g and h). Bar: 10 μm. Areas boxed in a, c, e, and g (composite images) are shown at higher magnification as 0.2–0.5μm slices in b, d, f, and h, respectively.
Periplakin Rescues Envoplakin from Aggregates
Envoplakin and periplakin are coimmunoprecipitated from keratinocyte lysates and analysis of their rod domain sequences suggests that heterodimerization would be energetically favorable (Ruhrberg et al. 1997). To examine potential interactions between the two proteins we investigated whether cotransfection of envoplakin with periplakin would prevent envoplakin from forming aggregates. The following molar ratios of periplakin to envoplakin cDNAs were transfected into keratinocytes: 10:1, 5:1, 2:1, 1:1, 1:2, 1:5, and 1:10. When coexpressed at a molar ratio of 1:1 or greater the two proteins showed extensive colocalization (Fig. 6, a–c and data not shown). No envoplakin aggregates were observed and instead both proteins had a filamentous distribution throughout the cytoplasm (Fig. 6, a–e, i, and j), with some concentration at cell–cell borders (Fig. 6d, Fig. e, Fig. i, and Fig. j). The cytoplasmic staining showed partial colocalization with keratin filaments (Fig. 6d and Fig. e) and the cell–cell border staining showed partial colocalization with desmoplakin (Fig. 6i and Fig. j). The distribution of periplakin in cells cotransfected with envoplakin was the same as that in cells transfected with periplakin alone (compare Fig. 5 and Fig. 6; and data not shown). When the molar ratio of periplakin to envoplakin fell below 1 envoplakin aggregates were observed (data not shown). We conclude that the two proteins interact and that the 1:1 ratio of cDNAs required for periplakin to rescue envoplakin would be consistent with envoplakin–periplakin heterodimer formation.
Figure 6.
Transient cotransfection of primary human keratinocytes with full-length FLAG-tagged envoplakin and full-length HA-tagged periplakin (a–e, i, and j) or the periplakin rod domain (P-R; f–h). Green fluorescence: anti-FLAG. Red fluorescence: anti-HA (b, c, g, and h); antikeratin (LP34; d and e); antidesmoplakin (i and j). Panels a–c and f and g show the same cells. Bar: (a–c) 7.5 μm, (d and i) 10 μm, (f–h) 12 μm. Areas boxed in panels d and i are shown at higher magnification in e and j, respectively.
To map the domain of periplakin that was required to prevent full-length envoplakin from aggregating a series of partial periplakin constructs were cotransfected into primary keratinocytes in combination with envoplakin. Transfection of a 1:1 molar ratio of envoplakin with the periplakin NH2 terminus + rod (P-NR), rod + COOH terminus (P-RC), or rod alone (P-R) prevented aggregate formation (Fig. 1; Fig. 6, f–h and data not shown). However, aggregates did form when envoplakin was cotransfected with the periplakin 1/2 NH2 terminus (data not shown). We conclude that the rod domain of periplakin is necessary and sufficient to prevent envoplakin from forming aggregates.
Periplakin NH2 Terminus Localizes at the Plasma Membrane and COOH Terminus Associates with Keratin Filaments
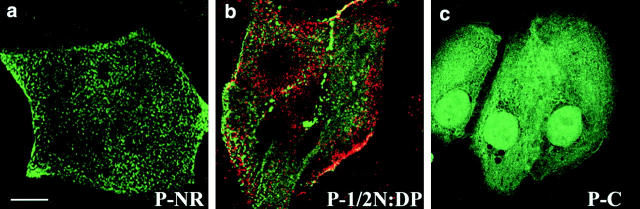
To examine which domains of periplakin mediated the association with the plasma membrane and cytoskeleton we tested a series of periplakin deletion constructs (Fig. 1) by transient transfection of primary human keratinocytes. A construct in which the COOH terminus was deleted localized to the plasma membrane in a punctate distribution (Fig. 7 a). The same distribution was observed when the 1/2 NH2 terminus of periplakin (again corresponding to the region of desmoplakin shown to target to desmosomes; Bornslaeger et al. 1996) was transfected (Fig. 7 b). In contrast, the COOH terminus of periplakin showed a filamentous distribution throughout the cytoplasm (Fig. 7 c). The 1/2 NH2 terminus of periplakin showed limited colocalization with desmoplakin at cell–cell borders, but periplakin was also extensively localized to the free apical membranes of keratinocytes, where desmosomes are absent (Fig. 7 b). When the 1/2 NH2 terminus of periplakin and the 1/2 NH2 terminus of desmoplakin (Bornslaeger et al. 1996) were cotransfected there was colocalization of both constructs at individual desmosomes and no evidence that either construct affected the distribution of the other (data not shown). We conclude that the periplakin NH2 terminus targets the protein to the plasma membrane, including desmosomes, whereas the COOH terminus mediates the interaction with keratin filaments.
Figure 7.
Transient transfection of primary human keratinocytes with different HA-tagged domains of periplakin. (a and c) Green fluorescence: anti-HA. (b) Red fluorescence: anti-HA; green fluorescence: antidesmoplakin. Bar: (a and b) 9 μm; (c) 11 μm.
Periplakin NH2 Terminus Associates with the Cortical Actin Cytoskeleton
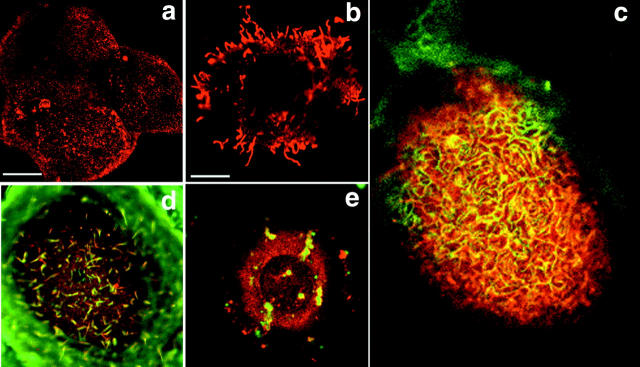
The punctate apical distribution of the periplakin 1/2 NH2-terminal construct was reminiscent of the short microvilli of keratinocytes in medium with a calcium ion concentration of >1 mM (Morrison et al. 1988) (Fig. 8 a). The microvillar association was more pronounced in A431 cells, which have longer microvilli than keratinocytes (Fig. 8 b). There was extensive colocalization of the 1/2 NH2 terminus of periplakin and actin at the apical plasma membrane (Fig. 8 c). When cells were treated with Latrunculin B to depolymerize F-actin the periplakin construct redistributed with the actin (Fig. 8d and Fig. e).
Figure 8.
Transient transfection of the NH2-terminal domain of periplakin (P-1/2N, HA tagged) into primary human keratinocytes (a and c–e) or A431 cells (b). Red fluorescence: anti-HA. Green fluorescence: phalloidin-FITC. Cell in panel e was treated with Latrunculin B (200 ng/ml, 3 h) before fixation. Bars: (a) 13 μm; (b) 6.3 μm; (c) 4.3 μm; (d and e) 4.5 μm.
We next examined whether endogenous periplakin was associated with the actin cytoskeleton. Periplakin in keratinocytes was concentrated both at cell–cell borders, as reported previously (Ruhrberg et al. 1997), and in a punctate distribution on free apical membranes (Fig. 9 a). Endogenous periplakin showed extensive colocalization with polymerized actin (Fig. 9, a–c). In keratinocytes, Latrunculin B caused collapse of the actin cytoskeleton into actin-rich spots and resulted in a corresponding redistribution of endogenous periplakin (Fig. 9, d–f), even though the keratin filament network remained intact (data not shown; Spector et al. 1999). A different actin depolymerizing drug, cytochalasin D, also caused redistribution of periplakin (data not shown). In contrast, the distribution of desmoplakin was not affected by Latrunculin B treatment (Fig. 9, g–i).
Figure 9.
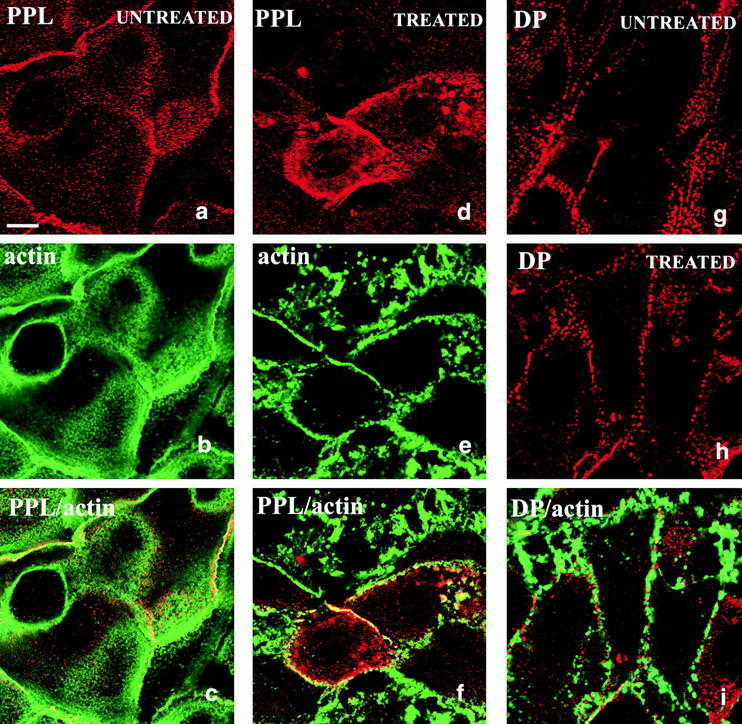
Distribution of endogenous periplakin and desmoplakin in primary human keratinocytes. Cell were untreated (a–c and g) or treated with Latrunculin B (d–f, h, and i). Red fluorescence: periplakin (PPL, detected with AE11; a, c, d, and f), desmoplakin (DP; g–i). Green fluorescence: actin. a–c, d–f, h, and i show the same fields of cells. Bar, 6.3 μm.
Discussion
Our earlier immunofluorescence and immunoelectron microscopy studies placed envoplakin and periplakin at the desmosomal plaque, at the interdesmosomal plasma membrane and in association with keratin filaments in terminally differentiating keratinocytes (Ruhrberg et al. 1996, Ruhrberg et al. 1997). We now show that on transfection full-length periplakin goes to all of these locations independently of envoplakin. In the absence of transfected periplakin some envoplakin localizes to desmosomes, but most ends up in large cytoplasmic and nuclear aggregates. We confirm our previous prediction that the two proteins can interact via their rod domains and show that periplakin associates with cortical actin via its NH2 terminus, thereby suggesting how envoplakin and periplakin can localize to the interdesmosomal plasma membrane, a key requirement for their scaffolding role during cornified envelope assembly (Steinert and Marekov 1999).
All plakins are believed to form two-stranded, in register parallel homodimers (O'Keefe et al. 1989; Tang et al. 1996; Ruhrberg and Watt 1997), but envoplakin and periplakin are unique in that both homo- and heterodimers are predicted to be energetically stable (Ruhrberg et al. 1997). We found that cotransfection of equimolar concentrations of full-length periplakin and envoplakin prevented envoplakin from accumulating in aggregates. Aggregation was dependent on the presence of the envoplakin rod domain, and the rod domain of periplakin was sufficient to prevent envoplakin from aggregating. Aggregates still formed when the NH2 or COOH terminus of envoplakin was deleted and this would imply a role for the rod domain in higher order complex formation, as reported for other plakins. Plectin, for example, may assemble as a homotetramer composed of two parallel dimers arranged antiparallel and overlapping along their entire length (Wiche et al. 1991; Svitkina et al. 1996; Ruhrberg and Watt 1997; but see also Wiche 1998).
The envoplakin aggregates are most likely an artifact of the high levels of expression achieved in transiently transfected cells, but do, nevertheless, provide useful information about the interaction of envoplakin with intermediate filaments. The aggregates were labeled with antibodies to keratins characteristic of simple and stratified epithelia, to vimentin and lamin B2, thus demonstrating an ability to interact with diverse intermediate filament types, as reported for plectin (Wiche et al. 1993; reviewed by Wiche 1998). Part of the intermediate filament binding domain of plectin is a functional nuclear localization signal (Nikolic et al. 1996) and there is a potential nuclear localization signal in the linker domain of envoplakin (data not shown). Desmoplakin can also interact with a range of intermediate filaments, but shows a preferential association with type II epidermal keratins (Kouklis et al. 1994; Meng et al. 1997). Envoplakin aggregates did not associate with microtubules (data not shown), although such interactions have now been established for plectin and certain BPAG1 isoforms (Svitkina et al. 1996; Wiche 1998; Yang et al. 1999). Envoplakin aggregates were not dependent on intermediate filaments and aggregate formation by truncated constructs (E-NR, E-RC) was independent of full-length envoplakin or periplakin since it occurred in 3T3 cells. Full-length periplakin associated with keratin and vimentin filaments.
The intermediate filament binding region of plectin has been mapped to 50 amino acids linking the COOH-terminal repeat domains 5 and 6 (box B and C in nomenclature of Green et al. 1992) (Nikolic et al. 1996). The first seven amino acids lie in the B box and the rest are part of the linker domain that is highly conserved between all plakins (Mahoney et al. 1998; Määttä et al. 2000). The E-RC and E-NR aggregates both associated with intermediate filaments; E-RC includes the entire linker domain and E-NR the first 11 amino acids. The distribution of the COOH terminus of periplakin, corresponding to the intact linker, was filamentous (Fig. 7 c), but that of envoplakin, comprising the C box without the linker, was diffuse and punctate (Fig. 4 f). It is therefore attractive to propose a role for the linker in intermediate filament binding. Yeast two-hybrid analysis of the COOH terminus of desmoplakin is consistent with a role for the linker domain in the interaction of desmoplakin with vimentin; however, for interaction with keratins a sequence within the C box is implicated and indeed there is evidence from peptide competition experiments that the equivalent region in envoplakin and plectin could also be involved (Meng et al. 1997). Although more work is required to clarify the sequences within envoplakin and periplakin that interact with intermediate filaments, the fact that periplakin lacks a C box and yet can associate with intermediate filaments does make the linker domain a likely candidate. In the autoimmune disorder paraneoplastic pemphigus autoantibodies to all the conventional plakins recognize the linker domain and it is interesting to speculate that this might contribute to the pathology of the disease by disrupting interactions between keratin filaments and adhesive junctions (Kiyokawa et al. 1998; Mahoney et al. 1998).
It was previously shown that although envoplakin and periplakin colocalize with desmoplakin at the desmosomal plaque they are also present between desmosomes (Ma and Sun 1986; Ruhrberg et al. 1997). This location is noteworthy because the interdesmosomal location is predicted to be of central importance for nucleating cornified envelope assembly (Steinert and Marekov 1999). Endogenous periplakin at the interdesmosomal plasma membrane was redistributed by treatment with Latrunculin B. The transfected NH2 terminus of periplakin localized to this site and was also sensitive to the drug. Although actin binding sites have been identified in the NH2 terminus of plectin and some isoforms of BPAG1 (Liu et al. 1996; Yang et al. 1996; Fuchs et al. 1999) the predicted amino acid sequence of periplakin, like that of desmoplakin and envoplakin, lacks an actin binding domain. An alternative possibility is that the interaction of periplakin with actin is indirect, possibly via ERM proteins such as ezrin. ERM proteins play a key role in microvillar formation (Yonemura et al. 1999 and references cited therein) and there is a striking concentration of the NH2-terminal periplakin construct at microvilli.
The first half of the NH2 terminus of periplakin was able to localize to desmosomes and the equivalent region of desmoplakin has the same properties (Bornslaeger et al. 1996). In contrast, the 1/2 NH2 terminus of envoplakin had a diffuse distribution and further experiments will therefore be required to identify the region of envoplakin required for targeting to desmosomes. The NH2 terminus of desmoplakin associates with itself, with desmocollin 1a, plakoglobin, and plakophilin 1, interactions that are important for desmosome assembly (Smith and Fuchs 1998; Kowalczyk et al. 1997, Kowalczyk et al. 1999). Plakophilin 1 contains two functionally distinct domains, a head domain that is involved in organizing the desmosomal plaque and an Arm repeat domain that could regulate the dynamics of the actin cytoskeleton (Hatzfeld et al. 2000). There is no evidence that the NH2 terminus of desmoplakin competes with that of periplakin for desmosomal association. It will be interesting to carry out more refined mapping of the periplakin NH2 terminus to discover whether there are distinct or overlapping sequences within the NH2 terminus that target to desmosomes and microvilli.
In summary, the overall organization of periplakin and envoplakin, as far as we have determined, is like other plakins, with the NH2 terminus mediating membrane association, the rod domain dimerization and the COOH terminus intermediate filament association. What makes periplakin and envoplakin unique is their ability to heterodimerize, and this may be of major importance for their role in cornified envelope assembly. Envoplakin and periplakin can independently localize to desmosomes, but the presence of envoplakin in the interdesmsomal membrane appears to be dependent on heterodimerization with periplakin.
Although our results lead us to propose that the NH2 terminus of periplakin could play a key role in forming the scaffold on which the cornified envelope is assembled, the transient transfection approach does not allow us to test the hypothesis further. It should, instead, be possible to use retroviral vectors to achieve sustained expression of the periplakin and envoplakin constructs with 100% transduction efficiency (Levy et al. 1998) and thereby directly examine their role in cornified envelope assembly. One priority will be to investigate the relationship between assembly of the protein and lipid components of the envelope. The lipid layer of the cornified envelope is formed when the contents of lamellar bodies, including ceramides, are extruded into the intercellular spaces in the upper layers of the epidermis, but it is not known whether this precedes transglutaminase cross-linking of the proteins or occurs simultaneously (Nemes et al. 1999). Envoplakin and periplakin undergo covalent attachment of ceramides (Marekov and Steinert 1998) in addition to isopeptide cross-linking and transglutaminase 1 can potentially catalyze both reactions (Nemes et al. 1999). By examining the relative timing of these events and whether the proteins associate with lamellar bodies in addition to the plasma membrane we should gain more insight into the earliest events in cornified envelop formation.
Acknowledgments
We are most grateful to everyone who provided us with reagents, in particular Kathy Green and Robert Evans. We thank Nasser Hajibagheri, Ken Blight, and Stephen Gschmeissner for carrying out the EM, and Daniel Zicha and Peter Jordan for advice on confocal microscopy.
Arto Määttä was funded by a European Molecular Biology Organization long-term fellowship and Tadashi Karashima was funded in part by a European Union Biotech Network grant.
Footnotes
T. DiColandrea and T. Karashima are joint first authors.
T. Karashima is on leave from Department of Dermatology, Kurume University School of Medicine, 67 Asahimachi, Kurume, Fukuoka 830, Japan.
Abbreviations used in this paper: FAD, Ham's F12 medium + adenine+ DME; FGS, fish skin gelatin; HA, hemagglutinin; HICE, hydrocortisone + insulin + cholera toxin + EGF; KSFM, keratinocyte serum-free medium.
References
- Aho S., McLean W.H., Li K., Uitto J. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin genes. Genomics. 1998;48:242–247. doi: 10.1006/geno.1997.5188. [DOI] [PubMed] [Google Scholar]
- Andrä K., Nikolic B., Stöcher M., Drenckhahn D., Wiche G. Not just scaffoldingplectin regulates actin dynamics in cultured cells. Genes Dev. 1998;12:3442–3451. doi: 10.1101/gad.12.21.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornslaeger E.A., Corcoran C.M., Stappenbeck T.S., Green K.J. Breaking the connectiondisplacement of the desmosomal plaque protein desmoplakin from cell–cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 1996;134:985–1001. doi: 10.1083/jcb.134.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R., Leichtfried F.E., Herrmann H., Small J.V., Lawson D.I., Wiche G. Cytoskeleton-associated plectinin situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J. Cell Biol. 1988;106:723–733. doi: 10.1083/jcb.106.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Cleveland D.W. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Yang Y. Crossroads on cytoskeletal highways. Cell. 1999;98:547–550. doi: 10.1016/s0092-8674(00)80041-0. [DOI] [PubMed] [Google Scholar]
- Fuchs P., Zörer M., Rezniczek G.A., Spazierer D., Oehler S., Castañón M.J., Hauptmann R., Wiche G. Unusual 5′ transcript complexity of plectin isoformsnovel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet. 1999;8:2461–2472. doi: 10.1093/hmg/8.13.2461. [DOI] [PubMed] [Google Scholar]
- Gandarillas A., Goldsmith L.A., Gschmeissner S., Leigh I.M., Watt F.M. Evidence that apoptosis and terminal differentiation of epidermal keratinocytes are distinct processes. Exp. Dermatol. 1999;8:71–79. doi: 10.1111/j.1600-0625.1999.tb00350.x. [DOI] [PubMed] [Google Scholar]
- Geerts D., Fontao L., Nievers M.G., Schaapveld R.Q., Purkis P.E., Wheeler G.N., Lane E.B., Leigh I.M., Sonnenberg A. Binding of integrin α6β4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J. Cell Biol. 1999;147:417–434. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.J., Parry D.A., Steinert P.M., Virata M.L., Wagner R.M., Angst B.D., Nilles L.A. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J. Biol. Chem. 1990;265:11406–11407. [PubMed] [Google Scholar]
- Green K.J., Virata M.L., Elgar G.W., Stanley J.R., Parry D.A. Comparative structural anaysis of desmoplakin, bullous pemphigoid antigen and plectinmembers of a new gene family involved in organization of intermediate filaments. J. Biol. Macromol. 1992;14:145–153. doi: 10.1016/s0141-8130(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Gregory S.L., Brown N.H. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J. Cell Biol. 1998;143:1271–1282. doi: 10.1083/jcb.143.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M., Haffner C., Schulz K., Vinzens U. The functions of plakophilin 1 in desmosome assembly and actin filament organization. J. Cell Biol. 2000;149:209–222. doi: 10.1083/jcb.149.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H., Aebi U. Intermediate filaments and their associatesmulti-talented structural elements specifying cytoarchitecture and cytodynamics. Curr. Opin. Cell Biol. 2000;12:79–90. doi: 10.1016/s0955-0674(99)00060-5. [DOI] [PubMed] [Google Scholar]
- Hochstenbach F., David V., Watkins S., Brenner M.B. Endoplasmic reticulum resident protein of 90 kD associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc. Natl. Acad. Sci. USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseweart M.K., Cleveland D.W. Intermediate filaments and their associated proteinsmultiple dynamic personalities. Curr. Opin. Cell Biol. 1998;10:93–101. doi: 10.1016/s0955-0674(98)80091-4. [DOI] [PubMed] [Google Scholar]
- Huovila A.P., Eder A.M., Fuller S.D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida-Yamamoto A., Iizuka H. Structural organization of cornified cell envelopes and alterations in inherited skin disorders. Exp. Dermatol. 1998;7:1–10. doi: 10.1111/j.1600-0625.1998.tb00295.x. [DOI] [PubMed] [Google Scholar]
- Karakesisoglou I., Yang Y., Fuchs E. An epidermal plakin that integrates actin and microtubule networks at cellular junctions. J. Cell Biol. 2000;149:195–208. doi: 10.1083/jcb.149.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa C., Ruhrberg C., Nie Z., Karashima T., Mori O., Nishikawa T., Green K.J., Anhalt G.J., DiColandrea T., Watt F.M., Hashimoto T. Envoplakin and periplakin are components of the paraneoplastic pemphigus antigen complex. J. Invest. Dermatol. 1998;111:1236–1238. doi: 10.1046/j.1523-1747.1998.00449.x. [DOI] [PubMed] [Google Scholar]
- Kouklis P.D., Hutton E., Fuchs E. Making a connectiondirect binding between keratin intermediate filaments and desmosomal proteins. J. Cell Biol. 1994;127:1049–1060. doi: 10.1083/jcb.127.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., Bornslaeger E.A., Borgwardt J.E., Palka H.L., Dhaliwal A.S., Corcoran C.M., Denning M.F., Green K.J. The amino-terminal domain of desmoplakin binds to plakoglobin and clusters desmosomal cadherin-plakoglobin complexes. J. Cell Biol. 1997;139:773–784. doi: 10.1083/jcb.139.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., Hatzfeld M., Bornslaeger E.A., Kopp D.S., Borgwardt J.E., Corcoran C.M., Settler A., Green K.J. The head domain of plakophilin-1 binds to desmoplakin and enhances its recruitment to desmosomes. Implications for cutaneous disease. J. Biol. Chem. 1999;274:18145–18148. doi: 10.1074/jbc.274.26.18145. [DOI] [PubMed] [Google Scholar]
- Lane E.B. Monoclonal antibodies provide specific molecular markers for the study of epithelial tonofilament organization. J. Cell Biol. 1982;92:665–673. doi: 10.1083/jcb.92.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane E.B., Bartek J., Purkis P.E., Leigh I.M. Keratin antigens in dfferentiating skin. Ann. NY Acad. Sci. 1985;455:241–258. doi: 10.1111/j.1749-6632.1985.tb50415.x. [DOI] [PubMed] [Google Scholar]
- Leung C.L., Sun D., Zheng M., Knowles D.R., Liem R.K.H. Microtubule actin cross-linking factor (MACF)a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 1999;147:41–48. doi: 10.1083/jcb.147.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L., Zhu A.J., Carroll J.M., Khazaal I., Péault B., Watt F.M. Optimised retroviral infection of human epidermal keratinocyteslong-term expression of transduced integrin gene following grafting on to SCID mice. Gene Ther. 1998;5:913–922. doi: 10.1038/sj.gt.3300689. [DOI] [PubMed] [Google Scholar]
- Liu C.G., Maercker C., Castanon M.J., Hauptmann R., Wiche G. Human plectinorganization of the gene, sequence analysis, and chromosome localization (8q24) Proc. Natl. Acad. Sci. USA. 1996;93:4278–4283. doi: 10.1073/pnas.93.9.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A.S., Sun T.-T. Differentiation-dependent changes in the solubility of a 195-kD protein in human epidermal keratinocytes. J. Cell Biol. 1986;103:1275–1285. doi: 10.1083/jcb.103.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttä A., Ruhrberg C., Watt F.M. Structure and regulation of the envoplakin gene. J. Biol. Chem. 2000;275:19857–19865. doi: 10.1074/jbc.M001028200. [DOI] [PubMed] [Google Scholar]
- Mahoney M.G., Aho S., Uitto J., Stanley J.R. The members of the plakin family of proteins recognized by paraneoplastic pemphigus antibodies include periplakin. J. Invest. Dermatol. 1998;111:308–313. doi: 10.1046/j.1523-1747.1998.00279.x. [DOI] [PubMed] [Google Scholar]
- Marekov L.N., Steinert P.M. Ceramides are bound to structural proteins of the human foreskin epidermal cornified cell envelope. J. Biol. Chem. 1998;273:17763–17770. doi: 10.1074/jbc.273.28.17763. [DOI] [PubMed] [Google Scholar]
- Meng J.J., Bornslaeger E.A., Green K.J., Steinert P.M., Ip W. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J. Biol. Chem. 1997;272:21495–21503. doi: 10.1074/jbc.272.34.21495. [DOI] [PubMed] [Google Scholar]
- Morrison A.I., Keeble S., Watt F.M. The peanut lectin-binding glycoproteins of human epidermal keratinocytes. Exp. Cell Res. 1988;177:247–256. doi: 10.1016/0014-4827(88)90459-4. [DOI] [PubMed] [Google Scholar]
- Nemes Z., Steinert P.M. Bricks and mortar of the epidermal barrier. Exp. Mol. Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Nemes Z., Marekov L.N., Fésüs L., Steinert P.M. A novel function for transglutaminase 1attachment of long-chain ω-hydroxyceramides to involucrin by ester bond formation. Proc. Natl. Acad. Sci. USA. 1999;96:8402–8407. doi: 10.1073/pnas.96.15.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic B., MacNulty E., Mir B., Wiche G. Basic amino acid residue cluster within nuclear targeting sequence motif is essential for cytoplasmic plectin-vimentin network junctions. J. Cell Biol. 1996;134:1455–1467. doi: 10.1083/jcb.134.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe E.J., Erickson H.P., Bennett V. Desmoplakin I and desmoplakin II. Purification and characterization. J. Biol. Chem. 1989;264:8310–8318. [PubMed] [Google Scholar]
- Pei X.F., Gorman P.A., Watt F.M. Two strains of human keratinocytes transfected with HPV16 DNAcomparison with the normal parental cells. Carcinogenesis. 1991;12:277–284. doi: 10.1093/carcin/12.2.277. [DOI] [PubMed] [Google Scholar]
- Reichert U., Michel S., Schmidt R. The cornified envelopea key structure of terminally differentiating keratinocytes. In: Darmon M., Blumenberg M., editors. Molecular Biology of the Skin. Academic Press; New York: 1993. pp. 107–150. [Google Scholar]
- Romero M.R., Carroll J.M., Watt F.M. Analysis of cultured keratinocytes from a transgenic mouse model of psoriasiseffects of suprabasal integrin expression on keratinocyte adhesion, proliferation and terminal differentiation. Exp. Dermatol. 1999;8:53–67. doi: 10.1111/j.1600-0625.1999.tb00348.x. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C., Watt F.M. The plakin familyversatile organizers of cytoskeletal architecture. Curr. Opin. Genet. Dev. 1997;7:392–397. doi: 10.1016/s0959-437x(97)80154-2. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C., Hajibagheri M.A.N., Simon M., Dooley T.P., Watt F.M. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J. Cell Biol. 1996;134:715–729. doi: 10.1083/jcb.134.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C., Hajibagheri M.A.N., Parry D.A.D., Watt F.M. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J. Cell Biol. 1997;139:1835–1849. doi: 10.1083/jcb.139.7.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria A.J., Lieber J.G., Nordeen S.K., Evans R.M. The presence or absence of a vimentin-type intermediate filament network affects the shape of the nucleus in human SW-13 cells. J. Cell Sci. 1994;107:1593–1607. doi: 10.1242/jcs.107.6.1593. [DOI] [PubMed] [Google Scholar]
- Sawamura D., Li K., Chu M.L., Uitto J. Human bullous pemphigoid antigen (BPAG1). Amino acid sequences deduced from cloned cDNAs predict biologically important peptide segments and protein domains. J. Biol. Chem. 1991;266:17784–17790. [PubMed] [Google Scholar]
- Simon M. The epidermal cornified cell envelope and its precursors. In: Leigh I.M., Lane E.B., Watt F.M., editors. The Keratinocyte Handbook. Cambridge University Press; Cambridge: 1994. pp. 275–292. [Google Scholar]
- Simon M., Green H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell. 1984;36:827–834. doi: 10.1016/0092-8674(84)90032-1. [DOI] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J., Gigengback S., Lienhard G.E., James D.E. Immuno-localization of the insulin regulatable glucose transport in brown adipose tissue of rat. J. Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.A., Fuchs E. Defining the interactions between intermediate filaments and desmosomes. J. Cell Biol. 1998;141:1229–1241. doi: 10.1083/jcb.141.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Braet F., Shochet N.R., Bubb M.R. New anti-actin drugs in the study of the organization and function of the actin cytoskeleton. Microscopy Research and Technique. 1999;47:18–37. doi: 10.1002/(SICI)1097-0029(19991001)47:1<18::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Green K.J. The desmoplakin carboxyl terminus coaligns with and specficially disrupts intermediate filament networks when expressed in cultured cells. J. Cell Biol. 1992;116:1197–1209. doi: 10.1083/jcb.116.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck T.S., Bornslaeger E.A., Corcoran C.M., Luu H.H., Virata M.L.A., Green K.J. Functional analysis of desmoplakin domainsspecification of the interaction with keratin versus vimentin intermediate filement networks. J. Cell Biol. 1993;123:691–705. doi: 10.1083/jcb.123.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinböck F.A., Nickolic B., Coulombe P.A., Fuchs E., Traub P., Wiche G. Dose-dependent linkage, assembly inhibition and disassembly of vimentin and cytokeratin 5/14 filaments through plectin's intermediate filament-binding domain. J. Cell Sci. 2000;113:483–491. doi: 10.1242/jcs.113.3.483. [DOI] [PubMed] [Google Scholar]
- Steinert P.M., Marekov L.N. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin and SPRs are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J. Biol. Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- Steinert P.M., Marekov L.N. Involucrin is an early component in the assembly of the epidermal cornified cell envelope. J. Biol. Chem. 1997;270:2021–2030. doi: 10.1074/jbc.272.3.2021. [DOI] [PubMed] [Google Scholar]
- Steinert P.M., Marekov L.N. Initiation of assembly of the cell envelope barrier structure of stratified squamous epithelia. Mol. Biol. Cell. 1999;10:4247–4261. doi: 10.1091/mbc.10.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer D., Michel M., Muller M. High pressure freezing comes of age. Scanning Microsc. Suppl. 1989;3:253–269. [PubMed] [Google Scholar]
- Stumpf D., Volk T. Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, vein, at the muscle-tendon junction site. J. Cell Biol. 1998;143:1259–1270. doi: 10.1083/jcb.143.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., Borisy G.G. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 1996;134:715–729. doi: 10.1083/jcb.135.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.Y., Chaffotte A.F., Thacher S.M. Structural analysis of the predicted coiled-coiled rod domain of the cytoplasmic bullous pemphigoid antigen (BPAG1). Empirical localization of the N-terminal globular domain-rod boundary. J. Biol. Chem. 1996;271:9716–9722. doi: 10.1074/jbc.271.16.9716. [DOI] [PubMed] [Google Scholar]
- Wiche G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998;111:2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- Wiche G., Becker B., Luber K., Weitzer G., Castanon M.J., Hauptmann R., Stratowa C., Stewart M. Cloning and sequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J. Cell Biol. 1991;114:83–99. doi: 10.1083/jcb.114.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G., Gromov D., Donovan A., Castanon M.J., Fuchs E. Expression of plectin mutant cDNA in cultured cells indicates a role of COOH-terminal domain in intermediate filament association. J. Cell Biol. 1993;121:607–619. doi: 10.1083/jcb.121.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Dowling J., Yu Q.C., Kouklis P., Cleveland D.W., Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Yang Y., Bauer C., Strasse G., Wollman R., Julien J.-P., Fuchs E. Integrators of the cytoskeleton that stabilze microtubules. Cell. 1999;98:229–238. doi: 10.1016/s0092-8674(00)81017-x. [DOI] [PubMed] [Google Scholar]
- Yonemura S., Tsukita S., Tsukita S. Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. J. Cell Biol. 1999;145:1497–1509. doi: 10.1083/jcb.145.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]