Abstract
Cell fusion occurs throughout development, from fertilization to organogenesis. The molecular mechanisms driving plasma membrane fusion in these processes remain unknown. While yeast mating offers an excellent model system in which to study cell fusion, all genes previously shown to regulate the process act at or before cell wall breakdown; i.e., well before the two plasma membranes have come in contact. Using a new strategy in which genomic data is used to predict which genes may possess a given function, we identified PRM1, a gene that is selectively expressed during mating and that encodes a multispanning transmembrane protein. Prm1p localizes to sites of cell–cell contact where fusion occurs. In matings between Δprm1 mutants, a large fraction of cells initiate zygote formation and degrade the cell wall separating mating partners but then fail to fuse. Electron microscopic analysis reveals that the two plasma membranes in these mating pairs are tightly apposed, remaining separated only by a uniform gap of ∼8 nm. Thus, the phenotype of Δprm1 mutants defines a new step in the mating reaction in which membranes are juxtaposed, possibly through a defined adherence junction, yet remain unfused. This phenotype suggests a role for Prm1p in plasma membrane fusion.
Keywords: cell fusion, data mining, genomic expression analysis, membrane adherence, prezygote
Introduction
The question at the heart of membrane fusion is how to unite the hydrophobic lipid cores of two bilayers across a gulf of water. An answer has emerged from work on the fusion of viruses with host cells and the fusion of transport vesicles with plasma membrane and organelles. In these systems, a fusion protein, or fusase, drives the reaction. For influenza virus the fusase is the hemagglutinin protein; for vesicles, the fusase includes the SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor) complex (for recent reviews, see Hernandez et al. 1996; Jahn and Sudhof 1999). Although hemagglutinin and SNAREs differ in composition—hemagglutinin is a single viral surface protein capable of inserting directly into the host cell plasma membrane, whereas the SNARE complex assembles from subunits associated with different bilayers—their final structures bear remarkable similarities (Weber et al. 1998). In each case, the assembled fusase has domains inserted into each of two apposing bilayers and, between these domains, a remarkably stable coiled coil (Hughson 1995; Harbury 1998). According to current models, the energetics of forming this coiled coil are so favorable that they outweigh the cost of pulling together the negatively charged sheets of phosphate head groups and squeezing out the water in between, thus initiating bilayer fusion (Ramalho-Santos and de Lima 1998; Weber et al. 1998).
Although this relatively detailed mechanistic model accounts well for fusion by viruses and within the secretory pathway, it leaves unexplained a large and important class of membrane fusion—that of cell fusion. Cell fusion occurs between sperm and egg during fertilization, during development in syncytial tissues such as muscle where myoblast precursor cells fuse into a long tube that will differentiate into a muscle fiber, and in processes such as phagocytic engulfment of cells or debris by macrophages where widely separated regions of the immune cell's plasma membrane must fuse to complete engulfment (Hernandez et al. 1996; Shemer and Podbilewicz 2000). In each of these cases, a pair of plasma membrane bilayers fuses from the extracellular side. Do these cells therefore express a special kind of SNARE with a topology more like a viral fusase?
If so, it has not yet been found. The closest proteins identified so far are the ADAMs, integral membrane proteins that contain a peptide similar to the portion of hemagglutinin that inserts into a host cell's plasma membrane (Blobel et al. 1992; Bigler et al. 1997). During fertilization, ADAMs on the sperm bind to α6β1 integrins on the egg with the help of another egg membrane protein, CD9 (Almeida et al. 1995; Chen et al. 1999). Blocking this interaction or removing CD9 inhibits sperm–egg fusion (Chen et al. 1999; Le Naour et al. 2000; Miyado et al. 2000). As none of the proteins in this complex are known to contain coiled coils, how this structure might generate the force required to bring membranes close enough for fusion remains a mystery.
To identify novel proteins that mediate cell fusion, we turned to the model system of yeast mating, in which two haploid cells fuse to produce a diploid. The mating reaction proceeds, briefly, as follows. Haploids exist as one of two mating types, a or α, which secrete a pheromone (a factor or α factor, respectively) that cells of the opposite mating type can detect. When the pheromone concentration reaches a certain level, the mating reaction initiates. The first steps of mating include a cell cycle arrest, remodeling of the cell wall, and polarization of mating partners towards each other. When mating partners make contact, the cell walls knit together to form a continuous outer layer. At this point the mating partners are firmly attached but each is still surrounded completely by cell wall, the plasma membranes having not yet come in contact and the cells of course not yet having fused. Cells at this stage are said to have formed a “mating pair” or “prezygote.” To complete formation of a zygote, the cell wall separating the partners must be degraded, plasma membranes must come in contact and fuse, and finally the haploid nuclei must merge into a single diploid nucleus.
A number of genetic screens have identified mutants defective in these steps by looking for cells that can form mating pairs but not diploids. All of the mutants, however, arrest in the mating reaction at either the step of cell wall breakdown or nuclear fusion (reviewed in Berlin et al. 1991; Marsh and Rose 1997). Although these classes of genes have provided insight into cell polarization, cell wall reorganization, control of osmotic stability, and organelle positioning and dynamics, the genes that mediate the actual lipid bilayer fusion step that follows the proper juxtapositioning of plasma membranes have remained elusive. We have exploited the recently accumulating wealth of gene expression data to search for proteins that may govern the fusion of plasma membranes.
Materials and Methods
Informatics
Programs were written in the scripting language Perl. The source code for Webminer and a description of the database formats used are available at http://webminer.ucsf.edu. The Candida albicans and Schizosaccharomyces pombe homologues of PRM1 were identified by BLAST searches of the unfinished genome sequencing projects at the Stanford University DNA Sequencing Center at http://www-sequence.stanford.edu/group/candida, and the Sanger Centre at http://www.sanger.ac.uk/Projects/S_pombe, respectively. Multiple sequence alignments were performed with MultAlin (Corpet 1988; http://protein.toulouse.inra.fr/multalin.html). Transmembrane domain predictions were made using the SOSUI program (Hirokawa et al. 1998; http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0E.html), and then refined by discarding putative transmembrane segments not present in all homologues. Coiled coil predictions were made using LearnCoil-VMF as described by Singh et al. 1999(http://nightingale.lcs.mit.edu/cgi-bin/vmf).
Yeast Strains and Plasmids
Strains used in this study appear in Table . Gene replacements, epitope tagged constructs, and green fluorescent protein (GFP) fusions were generated with the PCR-transformation technique (Longtine et al. 1998): PCR was performed with unique primers and a standard set of template plasmids to generate linear DNA consisting of a pair of integration sequences targeted against PRM1, flanking a generic cassette. The cassette contained three copies of the hemagglutinin (HA)-epitope tag, the coding sequence of GFP, or neither, and a selectable marker. Transformation of wild-type diploids resulted in insertion of this construct such that it replaced the entire PRM1 coding sequence or, for gene tagging, inserted in frame at its 3′ end, replacing the natural stop codon. After selection, diploids were assayed by PCR for correct insertion of the construct and then sporulated to recover haploids of both mating types that carried the integrated DNA. The plasmid pDN291, as previously described, was used to express soluble cytosolic GFP (Ng and Walter 1996). The plasmids pRS314 and pRS316 are standard vectors containing the TRP1 and URA3 genes, respectively, which were used here to create a set of mating type-specific selectable markers (Sikorski and Hieter 1989).
Table 1.
Strains Used in this Study
| Strain | Genotype |
|---|---|
| MHY200 | MATa, PRM1-HA:S.kluyveri HIS3 +, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS314 |
| MHY201 | MATα, PRM1-HA:S.kluyveri HIS3 +, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS316 |
| MHY153 | MATa, PRM1-GFP:S.kluyveri HIS3 +, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS314 |
| MHY154 | MATα, PRM1-GFP:S.kluyveri HIS3 +, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS316 |
| MHY209 | MATa, his3-Δ200, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS314 |
| MHY210 | MATα, his3-Δ200, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS316 |
| MHY198 | MATa, Δprm1::S.kluyveri HIS3+, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS314 |
| MHY199 | MATα, Δprm1::S.kluyveri HIS3+, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pRS316 |
| MHY189 | MATα, his3-Δ200, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pDN291 |
| MHY191 | MATα, Δprm1::S.kluyveri HIS3+, ura3-Δ99, leu2-Δ1, trp1-Δ99, ade2-101ochre, pDN291 |
All strains were constructed in the W303 background.
Preparation of Cell Lysates and Western Blotting
To detect expression of Prm1p-HA, 5 ml of an exponentially growing culture at optical density of 0.5 U A600 was either treated with 10 μl of a solution containing 5 mg/ml α factor (Sigma-Aldrich) in DMSO or mixed with an equal volume of cells of the opposite mating type, also from an exponentially growing culture. When mixing cells of both mating types, the maximal gene induction was seen when both cultures had grown continuously in log phase overnight from very low density, presumably to allow accumulation of pheromone in the medium; indeed, the conditioned media of these cultures alone had detectable inducing activity (not shown). At the relevant time point after mixing, cultures were briefly spun at 4°C, and the supernatant was aspirated. The cell pellet was resuspended in 50 μl SDS-PAGE sample buffer, added to a small volume of glass beads, and lysed by continuous vortexing at 4°C for 90 s. The entire procedure took <4 min. The lysates were boiled for 10 min and then spun to remove insoluble debris. Alternatively, for endoglycosidase H treatment, cells were lysed as above with the exception that sample buffer was replaced by 45 μl denaturation buffer as provided by the manufacturer (New England Biolabs, Inc.). Samples were then boiled 10 min, mixed with 5 μl G5 buffer as provided and 1 μl enzyme, incubated for 90 min at 37°C, and diluted 1:10 in SDS-PAGE sample buffer before loading. For Western blot analysis, lysates were run on a 12.5% SDS–polyacrylamide gel and transferred to nitrocellulose membrane using standard protocols. Membranes were blotted with a mouse monoclonal anti-HA primary antibody (HA.11; Covance) at 1:1,000 dilution, and a goat anti–mouse secondary antibody coupled to horseradish peroxidase (Bio-Rad Laboratories) at 1:2,000 dilution and developed with an enhanced chemiluminescence kit (Renaissance kit; NEN Life Science Products).
Fluorescence Microscopy of Prm1p-GFP
To visualize the localization of Prm1p-GFP in pheromone-treated haploids, cells were grown to log phase in defined media with twice the standard concentration of adenine to prevent accumulation of autofluorescent byproducts of adenine biosynthesis. The culture was then exposed to 10 μg/ml α factor. Samples were taken at 40, 70, and 100 min after pheromone addition, placed on a slide, and imaged on a confocal microscope (Leica). Alternatively, to inspect Prm1p-GFP's localization in zygotes, cells of opposite mating types that each carried the PRM1-GFP fusion were grown to log phase, mixed in equal numbers, spotted on a yeast extract/peptone/dextrose (YPD) plate, and incubated for 2 h at 30°C. Cells were then resuspended from the plate, spotted on a slide, and imaged. Because the Prm1p-GFP signal was faint, a single medial optical section was first taken by averaging four high-intensity laser scans, which bleached most of the fluorescence. Then, a stack of eight optical sections was collected to document the remaining fluorescence in the cells. This information was then used to deconvolve the high-intensity section, using OpenLab software (Improvision). Images were also smoothed and contrast enhanced with this software.
Quantitative Assay of Cell Fusion
Cells of opposite mating types, with the α strain expressing soluble cytosolic GFP, were grown to log phase, mixed, and vacuumed to a nitrocellulose filter. The filter was placed cell-side up on a YPD plate, and the plate was incubated for 3 h at 30°C. Cells were then scraped off the filter, fixed in 4% paraformaldehyde, and incubated at 4°C overnight. This mixture was then spotted on a slide and observed with a confocal microscope (Leica). First, a field was selected randomly using transmission optics. Then, groups of zygotes and mating pairs within that field were identified by bright-field microscopy and subsequently scored as fused zygotes or unfused mating pairs by switching between bright-field and fluorescence. This procedure was continued until all the zygotes and mating pairs in the field were scored, at which point a new field was chosen and the procedure begun again. To capture images, a single optical section was taken by both bright-field and fluorescence microscopy. These images were then superimposed and contrast enhanced.
Electron Microscopy
For mating reactions, equal numbers of Δprm1 cells of opposite mating types were mixed, spun down, spotted on a YPD plate, and allowed to mate for 3 h at 30°C. Cells were scraped off and fixed in EM fix (1% glutaraldehyde, 0.2% paraformaldehyde, 0.04 M KPO4, pH 7) for 5 min, spun, and incubated on ice in EM fix for 50 min. Cells were then washed twice with 0.9% NaCl, once with water, and once with 2% KMnO4 (Mallinckrodt). Cells were next incubated in 2% KMnO4 for 45 min at room temperature. They were then dehydrated through graded ethanol (10-min washes with 50, 70, 80, 90, 95, and 100% ethanol) and stored in a final wash of 100% ethanol overnight. To prepare for embedding, cells were washed five times for 10 min each with propylene oxide. For embedding, cells were stepped through graded concentrations of resin (32% Epon, 18% Araldite, 34% DDSA, 16% NMA; Ted Pella Inc.) mixed with propylene oxide, as follows: 2 h each with a 1:2 resin:propylene oxide mix, a 1:1, a 2:1, and a 3:1 mix, followed by a 1-h wash with pure resin and overnight infiltration with pure resin. The next day, cells were transferred to resin containing ∼2% BDMA (Ted Pella Inc.), incubated 4 h, and finally put in fresh resin with 2% BDMA, pelleted, and incubated at 60°C for several days for the resin to harden. Sections of ∼60-nm thickness were cut, stained with lead citrate (Ted Pella Inc.), and imaged with an electron microscope (EM400; Philips).
Results
A New Strategy for Identifying Genes that Regulate Cell Fusion
We devised a strategy to identify mating-specific genes that may have escaped earlier genetic screens due to functional redundancy within or between mating partners. Such redundancy often produces a weak phenotype that can be difficult to detect. For example, in the case of the redundant genes FUS1 and FUS2, a fus2 mutant displays little mating defect unless both mating partners also have deficiencies in FUS1 (Trueheart et al. 1987). To avoid overlooking functionally redundant genes in our search, we employed a reverse genetic strategy that did not depend initially on the strength of the mutant phenotype. Specifically, we asked, “What pheromone-induced membrane proteins have not yet been studied?”
To address this question, we compiled already-published databases of gene expression data and gene properties, restructured them in a common format, and wrote a program to search this composite database. We used the program, called Webminer (see Materials and Methods), to examine gene expression profiles of cells arrested in G1 by treatment with the pheromone α factor. These genomic expression datasets were originally collected in the course of another group's study of cell-cycle transcription and made available online (Spellman et al. 1998; http:// genome-www.stanford.edu/cellcycle). We reinterpreted the data to identify a number of strongly pheromone-induced proteins. As a second criterion, we demanded that potential target proteins have at least one hydrophobic domain, indicative of secretory or membrane proteins.
Specifically, we set an arbitrary cut-off to select genes that are induced more than threefold by mating pheromone. This criterion identified a set of 54 candidate open reading frames (ORFs) out of the 6,116 ORFs assayed in the genomic expression dataset (Fig. 1). We next assigned a score to every ORF to reflect its likelihood of encoding a membrane protein. To calculate these values, we wrote a program that scans predicted protein sequences in windows of 19 amino acid residues and assigns a hydrophobicity, or H, value to each window based on its amino acid composition. The hydrophobicity values we used are based on the empirically observed frequency of each amino acid's presence in known transmembrane domains (Boyd et al. 1998). The highest H value among all of a protein's windows has been defined as that protein's MaxH (Boyd et al. 1998). In most organisms, the MaxH values of all proteins fall into a bimodal distribution with a trough at 28.5 (Boyd et al. 1998). Lower values represent the set of cytosolic proteins (e.g., Tub1p, α tubulin, has a MaxH of 22.5), and higher values represent membrane proteins (e.g., Hxt1p, a hexose transporter, has a MaxH of 30.9). In Saccharomyces cerevisiae the bimodal distribution of MaxH values is present, but the overlap between the two sets is considerable. As a result, many known membrane proteins have MaxH values <28.5. We therefore set a less stringent threshold, by considering all ORFs with MaxH values >25 to be possible membrane proteins, yielding a set of 2,524 ORFs. This parameter narrowed our pool of 54 candidates to 20 genes, which we henceforth refer to as PRM genes (pheromone-regulated membrane proteins).
Figure 1.
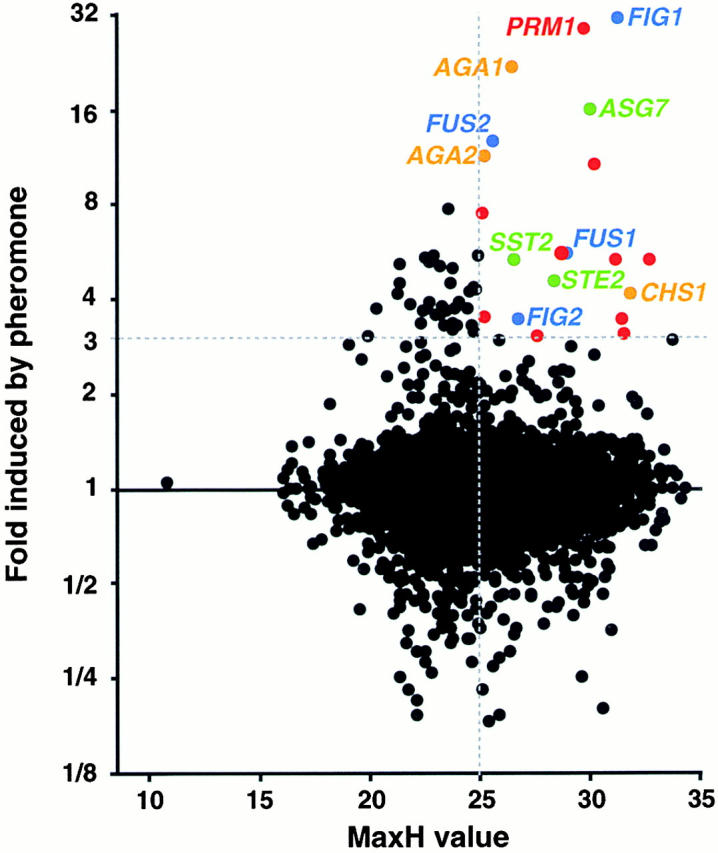
Identification of pheromone-induced putative membrane proteins by data mining. Dots represent the transcriptional induction in response to mating pheromone (y axis) and likelihood of coding for a membrane protein (x axis) of all 6,116 ORFs. ORFs induced more than threefold with a MaxH score >25 were investigated further. Green, mating-type specific; orange, cell wall remodeling; blue, involved in cell fusion; red, uncharacterized PRM genes.
Of these 20 genes, 10 have previously assigned functions (Fig. 1). Intriguingly, the identification of all 10 genes can be rationalized in light of roles they have in mating: four genes are involved in cell fusion (including the prototypical fusion genes FUS1 and FUS2; Trueheart et al. 1987) (Fig. 1, blue), three genes are involved in cell wall synthesis and remodeling (including AGA1 and AGA2, which encode the mating agglutinins; Cappellaro et al. 1991) (orange), and three genes are involved in other functions relevant to mating (including STE2, which encodes the a-specific pheromone receptor; Jenness et al. 1983) (green).
The remaining 10 ORFs had not been studied (Table ; Fig. 1, red). Based on the successful identification of other membrane proteins involved in mating, they have a high likelihood of also being players in the process. We describe here the characterization of the most highly induced ORF, YNL279w, which we call PRM1.
Table 2.
Characteristics of the PRM Genes
| Gene | ORF | Pheromone induction (fold) | Predicted protein size (amino acids) | Predicted transmembrane segments |
|---|---|---|---|---|
| PRM1 | YNL279W | 31 | 661 | 5 |
| PRM2 | YIL037C | 11 | 656 | 4 |
| PRM3 | YPL192C | 8 | 133 | 1 |
| PRM4 | YPL156C | 6 | 284 | 1 |
| PRM5 | YIL117C | 5 | 318 | 1 |
| PRM6 | YML047C | 5 | 352 | 2 |
| PRM7 | YDL039C | 4 | 115 | 1 |
| PRM8 | YGL053W | 4 | 237 | 2 |
| PRM9 | YAR031W | 3 | 298 | 3 |
| PRM10 | YJL108C | 3 | 383 | 5 |
Prm1p Is a Conserved Fungal Protein with Five Putative Transmembrane Domains
The predicted S. cerevisiae Prm1p has clearly identifiable homologues in other fungi, such as C. albicans (Contig 5-2425, position 7551–5680), S. pombe (GenBank/EMBL/DDBJ No. 7630122), and Kluyveromyces lactis (GenBank/EMBL/DDBJ No. AJ229977; Ozier-Kalogeropoulos et al. 1998) (Fig. 2 A), but contains no recognizable motifs to hint at its function.
Figure 2.
Comparison of Prm1p sequences from S. cerevisiae, C. albicans, and S. pombe. (A) Chemically similar, aligned amino acids are shaded. In the S. cerevisiae sequence, predicted transmembrane domains are overlined and potential glycosylation sites are boxed. (B) Schematic of proposed topology for Prm1p. All consensus glycosylation sites (S. cerevisiae) are marked with Y. The intensity of shading indicates the degree of sequence similarity between the three yeast homologues: the sequence is divided into 40 blocks, each 15 amino acids in length, and each block is shaded according to the number of conserved residues contained in a 45 amino acid window centered on it. Overall percent identity between sequences: S. cerevisiae and C. albicans, 20% identical; S. cerevisiae and S. pombe, 22% identical.
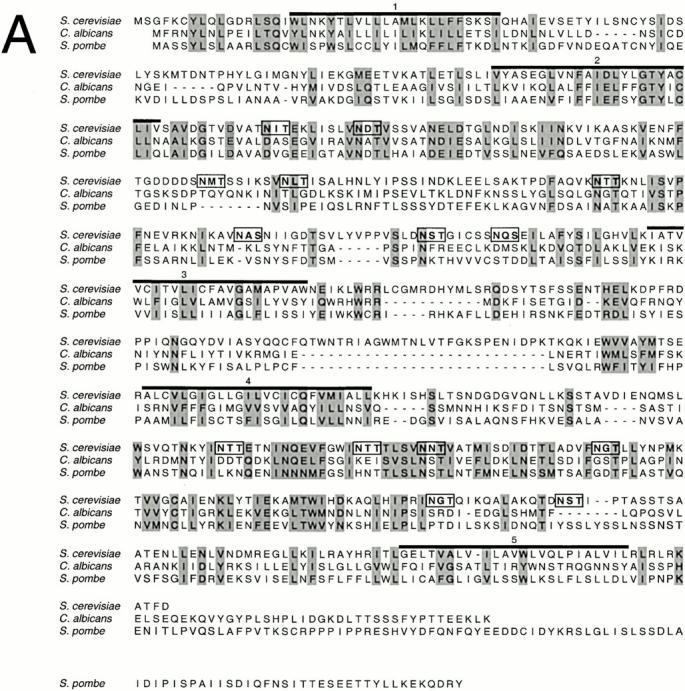
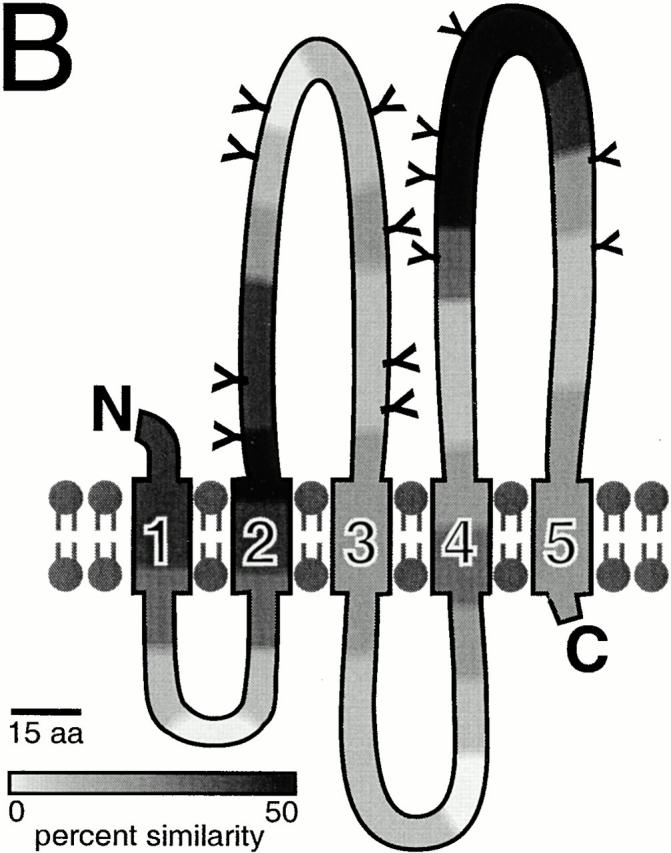
Prm1p has five conserved regions that, based on their hydrophobic character, are likely to span the membrane (Fig. 2 A, overlined). These putative transmembrane domains would divide the protein into two segments of ∼175 residues each on one side of the membrane and two 50–100 amino acid segments on the other side of the membrane (Fig. 2 B). Together, both of the larger segments harbor 14 potential N-glycosylation sites (Fig. 2a and Fig. b, boxed and Y symbol), whereas the smaller ones have none. The large segments display the greatest sequence similarity between the three homologues, with about two thirds of the residues conserved. Intriguingly, these segments are identified as potential coiled-coil–forming regions by LearnCoil-VMF, a program designed to recognize viral fusases (Singh et al. 1999). However, it is unlikely that a coiled-coil structure could assemble within a region of the protein that is anchored on both sides by transmembrane segments. The predicted overall picture of Prm1p, then, is that of a multispanning integral membrane protein presenting a large, evolutionarily conserved face on one side of the membrane and a smaller, less conserved face on the other (Fig. 2 B).
Pheromone Rapidly Activates Prm1p Expression in both Mating Types
To characterize Prm1p, we constructed strains carrying a fusion gene that appends an HA-epitope tag to the protein's COOH terminus (Prm1p-HA). We then assayed cells under mating or control regimes for the expression of Prm1p-HA by resolving total cell lysates with SDS-PAGE and visualizing Prm1p-HA by immunoblot.
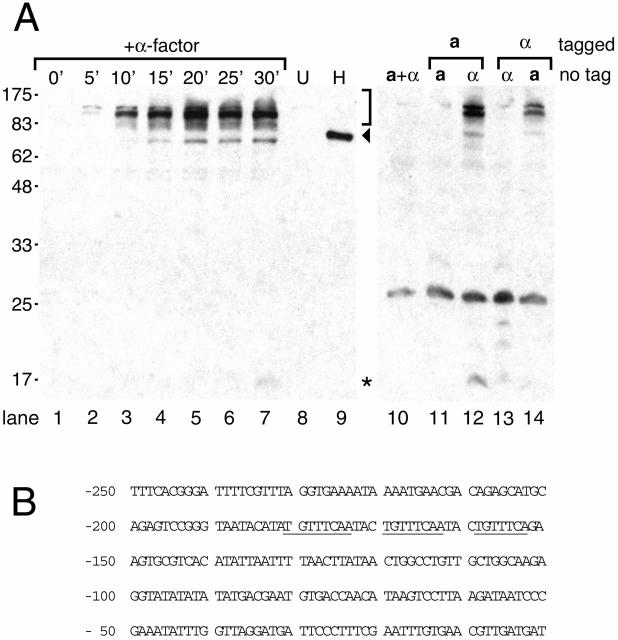
Vegetatively growing cells did not express Prm1p-HA at detectable levels (Fig. 3 A, lane 1), but initiated expression within 5 min after addition of α factor (lane 2). After 20 min of pheromone treatment, the Prm1p level reached a maximum and persisted at steady state (lanes 5–7).
Figure 3.
Expression profiles of Prm1p. (A, lanes 1–9) A strain of mating type a bearing a chromosomal copy of PRM1-HA (lanes 1–7 and 9), or a wild-type control strain (lane 8) was treated with 10 μg/ml alpha factor for 0–30 min, pelleted, and lysed by bead beating. Extracts were resolved by SDS-PAGE on a 12.5% gel and immunoblotted using an anti–HA antibody. For lane 9, the extract was treated with endoglycosidase H before analysis by SDS-PAGE. (A, lanes 10–14) The following strains were mixed: control wild-type strains of mating types a and α (A, lane 10), an a strain bearing PRM1-HA and an untagged strain of the same (A, lane 11) or the opposite (A, lane 12) mating type, an α strain bearing PRM1-HA and an untagged strain of the same (A, lane 13) or the opposite (A, lane 14) mating type. These mixtures were rotated for 30 min at 30°C, pelleted, lysed, and the extracts were analyzed as above. The 73-kD form of Prm1p, presumably corresponding to the primary, unglycosylated translation product, is indicated with the arrowhead. The glycosylated forms of Prm1p migrating as a broad band centered at 115 kD are indicated with the bracket. A 15-kD putative proteolytic fragment is indicated with the asterisk. (B) The PRM1 promoter sequence, beginning 250 nucleotides upstream of the translational start codon, is shown. Pheromone response element consensus sequences are underlined.
Western blot analysis identified Prm1p-HA (and by extension Prm1p) as several major forms: a sharp band migrating at 73 kD, the size predicted from the PRM1-HA open reading frame (Fig. 3 A, arrowhead), and a series of broad bands centered at roughly 115 kD (bracket). These species collapsed to a single band of ∼73 kD after treatment with endoglycosidase H, indicating that the larger bands are heterogeneously glycosylated (Fig. 3 A, lane 9). The presence of extensive oligosaccharide addition confirms our prediction that Prm1p is initially integrated into the membrane of the endoplasmic reticulum and, based on the proposed topology in Fig. 2 B, suggests that Prm1p may display its two large conserved segments on the lumenal or extracellular side of the membrane.
In addition to the newly synthesized and glycosylated forms, we also reproducibly observed a weaker band migrating at ∼15 kD, which appeared after 30 min of pheromone treatment (Fig. 3 A, lane 7, *). Based on the position of the HA epitope, this band is likely to represent a COOH-terminal fragment, indicating that Prm1p-HA may undergo proteolytic processing during its maturation.
Cells of both mating types induce Prm1p when challenged with partners of the opposite mating type. Cells of mating type a expressed Prm1p-HA when mixed with untagged α cells for 30 min, but not when mixed with cells of the same mating type (Fig. 3 A, lanes 11 and 12). The converse is also true (Fig. 3 A, lanes 13 and 14): α cells expressed Prm1p-HA when mixed with untagged a cells, but not when mixed with untagged α cells. Prm1p-HA induction in α cells was weaker than in a cells, perhaps due to reduced diffusion of the lipophilic a factor compared with the more hydrophilic α factor.
The speed and extent of Prm1p expression during mating probably resulted from the presence of pheromone-responsive elements (PREs) upstream of the gene's coding sequence, as is true for many other mating-specific genes. The promoter of PRM1 contains three head-to-tail repeats closely matching the consensus PRE, TGTTTCAa t (Fig. 3 B) (Yuan and Fields 1991). The repeats are separated by a trinucleotide spacer TAC. These sequences appear 150–180 nucleotides upstream of the PRM1 coding sequence and probably serve as binding sites for the transcription factor Ste12p, a target of the MAP kinase cascade that links gene expression to the presence of extracellular pheromone (Herskowitz 1995).
Prm1p Localizes to the Site of Cell Fusion
As a first step towards elucidating the function of Prm1p, we asked in what cellular compartment(s) the protein resides. To this end, we constructed strains bearing a chromosomal copy of a PRM1-GFP fusion gene driven by its own promoter, which allowed us to detect the Prm1p-GFP gene product by fluorescence microscopy.
Prm1p-GFP first became visible after 40 min of pheromone treatment as two rings, one encompassing the nucleus and one at the cell periphery (Fig. 4 A). This staining pattern is typical of the endoplasmic reticulum in yeast, consistent with Prm1p entering the secretory pathway.
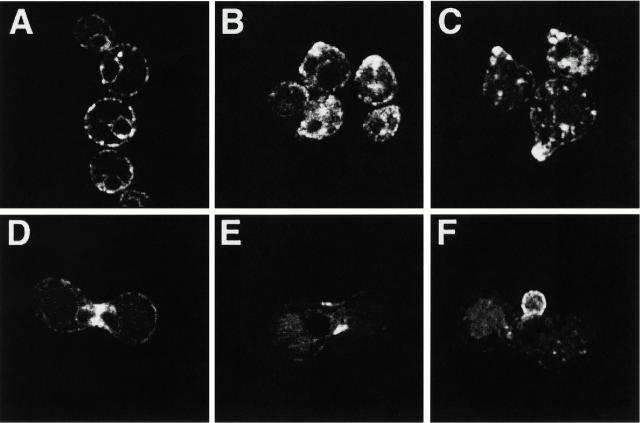
Figure 4.
Localization of Prm1p. (A–C) A strain of mating type a bearing a PRM1-GFP fusion gene was treated with 10 μg/ml α factor. Samples were taken and imaged on a confocal microscope after 40, 70, and 100 min of incubation, respectively. Apparent loss of ER staining in B and C is due primarily to differences in signal gain used to collect each image. (D–F) Strains of opposite mating types, each bearing the PRM1-GFP fusion gene, were mixed, concentrated, and spotted on a YPD plate. After ∼2 h at 30°C, cells were resuspended and imaged as above. Images of representative cells are shown.
70 min after addition of α factor most cells have arrested in the G1 phase of the cell cycle, evidenced by their large unbudded state, and have begun to polarize. Prm1p accumulated in the “potbelly” formed by this polarization in addition to its persistent staining of the endoplasmic reticulum (Fig. 4 B).
By 100 min of pheromone treatment, most cells have formed mating projections, or shmoos. These shmoos would, in a more physiological setting, orient towards the greatest pheromone concentration and serve as the site where mating partners first make contact. Prm1p localized to the tip of the shmoo, where cell fusion would occur (Fig. 4 C).
We next mixed a and α cells, both bearing the PRM1-GFP fusion gene. In such physiological mating mixes, Prm1p-GFP localized at the midpoint of recently formed mating pairs, or zygotes, where two cells have met and initiated the steps required to degrade the intervening cell wall and fuse their plasma membranes (Fig. 4 D). In mating pairs that have already completed this fusion step, Prm1p-GFP formed a collar around the neck of the zygote (Fig. 4 E).
When the resulting diploid began to bud, Prm1p-GFP localized to the growing daughter (Fig. 4 F). Since diploids no longer express Prm1p (data not shown), the protein staining the first daughter was probably inherited from the parental cells.
More than Half of All Mating Pairs Deficient in PRM1 Fail to Fuse
To test whether Prm1p participates in cell fusion during mating as its expression profile and localization suggested, we constructed strains in which PRM1 was deleted by gene replacement (see Materials and Methods). When both mating partners lacked PRM1, we observed morphologically aberrant mating pairs by phase contrast microscopy. The most common aberration was the presence of a pronounced dark band at the mating pair neck, reflecting the undegraded cell wall between mating partners suggestive of a defect in cell fusion.
To monitor this phenotype more decisively, we constructed a Δprm1 α strain expressing a soluble, cytosolic form of GFP that marks its cytoplasm. This strain allowed us to readily distinguish fused zygotes from unfused mating pairs by scoring whether GFP had spread to both cells (indicating successful cell fusion) or remained restricted to one mating partner (indicating a failure to fuse). Using this assay, we observed unambiguously that matings between Δprm1 partners produced a mixture of fused zygotes and unfused mating pairs (Fig. 5A and Fig. B).
Figure 5.
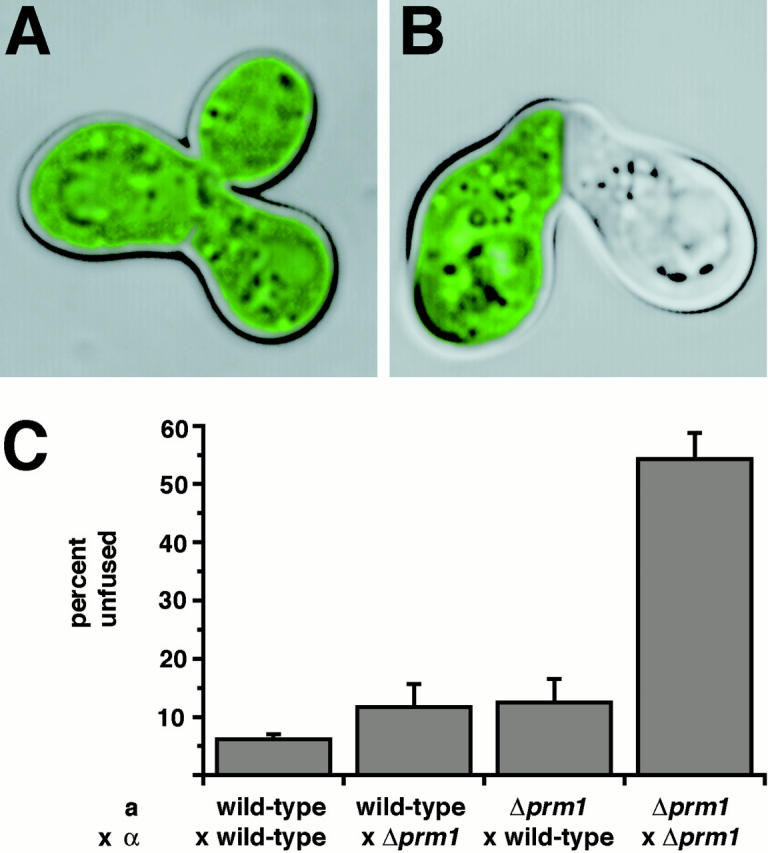
Δprm1 cells exhibit a fusion defect during mating. (A and B) Δprm1 a cells were mixed with Δprm1 α cells expressing soluble cytosolic GFP as a reporter of cytoplasmic mixing between mating partners. This mixture was applied to a nitrocellulose filter and incubated for 3 h on a YPD plate. Fluorescent micrographs that show the GFP-stained cytoplasm of the α partner were super-positioned over bright-field images that depict the entire zygote/mating pair. (C) Mating mixes in which either the a partner, the α partner, both, or neither carry a deletion of PRM1 were prepared as described above. In all cases the α partner carried soluble cytosolic GFP. Zygotes/mating pairs were visually identified, and then scored with regard to cell fusion by microscopy. Bars represent the average percent of zygotes/mating pairs that scored as unfused in four independent experiments. During each experiment, 300 zygote/mating pairs per mating mix were counted: WT a × WT α, 6.2 ± 0.8%; WT a × Δprm1 α, 11.7 ± 4.0%, Δprm1 a × WT α, 12.5 ± 4.0%; Δprm1 a × Δprm1 α, 54.3 ± 4.5%.
We next quantitated the degree of the Δprm1 fusion defect using GFP-expressing wild-type and Δprm1 α strains. To do so, we mixed exponentially growing cultures of each of these strains with an appropriate partner strain, concentrated them on a filter, and placed the filter on a YPD plate where the cells were allowed to mate for 3 h. We then fixed the cultures for microscopy. At this point, zygotes produced by wild-type control cells were abundant but most were still freshly formed, having just begun to grow their first diploid bud.
In such mating mixes between wild-type control strains, 6% of zygotes/mating pairs scored as unfused (Fig. 5 C). Presumably, this baseline level reflects a kinetic intermediate in the mating reaction, and these cells would have eventually fused if the reaction were allowed to continue. Characteristically, these unfused mating pairs had a narrow neck. In contrast, when both mating partners lacked PRM1, 55% of zygotes/mating pairs were unfused, a ninefold increase over the number observed for wild-type strains (Fig. 5 C). These mating pairs may reflect either a kinetic delay in the fusion reaction, or they may represent a dead end in which some step in mating has gone awry and fusion cannot occur. At later time points the ratio of fused zygotes to unfused mating pairs did not appreciably change (data not shown), contrary to what a kinetic delay would predict. Moreover, in many of the mating pairs from a Δprm1 × Δprm1 mating, the neck diameter was significantly increased, indicating that these unfused mating pairs differed qualitatively from the ones observed at low frequency in the wild-type control reactions.
Is Prm1p required in both partners to promote efficient cell fusion? When one mating partner lacked PRM1 and the other was wild-type, we consistently observed a slight but significant fusion defect, with 12% of all mating pairs failing to fuse (Fig. 5 C). This defect was similar regardless of which partner carried the wild-type PRM1 allele. These results suggest that Prm1p functions symmetrically and can perform its duty even if present in only one mating partner, albeit at a consistently reduced efficiency.
Δprm1 Mating Pairs Form “Bubbles,” and Other Strange Shapes
In addition to the simple unfused phenotype shown in Fig. 4 B typical of all fusion mutants, we observed more unusual morphologies in Δprm1 × Δprm1 matings. Notably, some mating pairs displayed intercellular bubbles, pockets of GFP-labeled or unlabeled cytoplasm from one mating partner that appeared to have invaded the other (Fig. 6, A–G). These bubbles appeared with approximately equal frequency in either direction: “innies” invading the α partner (Fig. 6A and Fig. B), and “outies” extending from the α cell into the a cell (Fig. 6, C–G). Bubbles varied in size and shape, ranging from tiny bulges in an otherwise straight cell–cell interface to large rounded pockets or, rarely, serpentine extensions that stretched across the entire length of the other mating partner.
Figure 6.
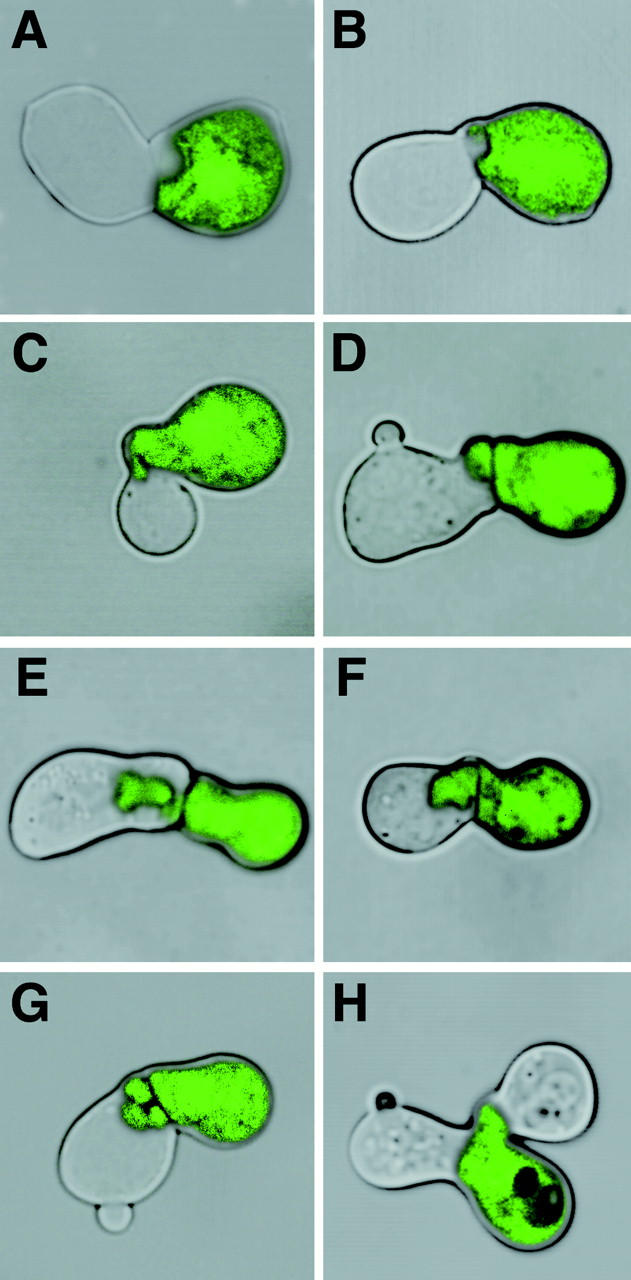
The Δprm1 cells' failure to fuse sometimes results in intercellular bubbles. Mating mixes were prepared and imaged as described in Fig. 5. Representative images are shown. (A and B) “Innies” intruding from the a cell (nonfluorescent) to the α cell (fluorescent). (C–F) “Outies” protruding from the α cell to the a cell. Note that the a cell in D escaped G1 arrest and started budding. (G) A multilobed outie. (H) An α cell, bottom right, simultaneously adhered to two a partners. The partner on the left has begun to bud.
Additionally, we observed one or both mating partners having budded a new daughter cell (Fig. 6D, Fig. G, and Fig. H). Budding indicates that a cell has escaped from the G1 arrest induced by exposure to mating pheromone and has reentered the cell cycle, committing itself to a new round of division. Apparently this release from pheromone arrest can occur even when surrounded by cells of the opposite mating type that are secreting pheromone and, in fact, even while adhered to one of them.
Lastly, some cells appeared to give up on the failed mating and, instead of budding, began to mate with another nearby partner. For instance, in Fig. 6 H, the GFP-expressing cell in the bottom right seemed to have attempted to mate with the partner on the left and failed. It then went on to try anew with the cell on the right, while its original mating partner exited the mating arrest and began to bud.
The ability of these cells to exit G1 or to polarize towards a new partner and reinitiate mating suggests that Δprm1 mutants do not simply fuse more slowly than wild-type. Rather, the fact that they abandon their attempt at fusion indicates they have reached a dead end and would not form normal diploid zygotes even if given more time.
The Δprm1 Defect Results in Closely Apposed, Unfused Plasma Membranes
The bubbles suggested a breach of the cell wall between the mating partners, a phenotype unlike other fusion mutants. We used thin-section electron microscopy to examine this aspect of the Δprm1 defect more closely.
Many mating pairs exhibited an apparent dissolution of their cell wall at the center of the interface between the mating partners (Fig. 7, A–D). In most cases we found it necessary to examine serial sections through a single mating pair to find the point where a breakthrough occurred. The region of cell wall degradation almost invariably included the center of the cell–cell interface. In some cases it appeared restricted to the center (Fig. 7B and Fig. D), whereas in others it seemed to have spread asymmetrically to one edge of the mating pair (A and C).
Figure 7.
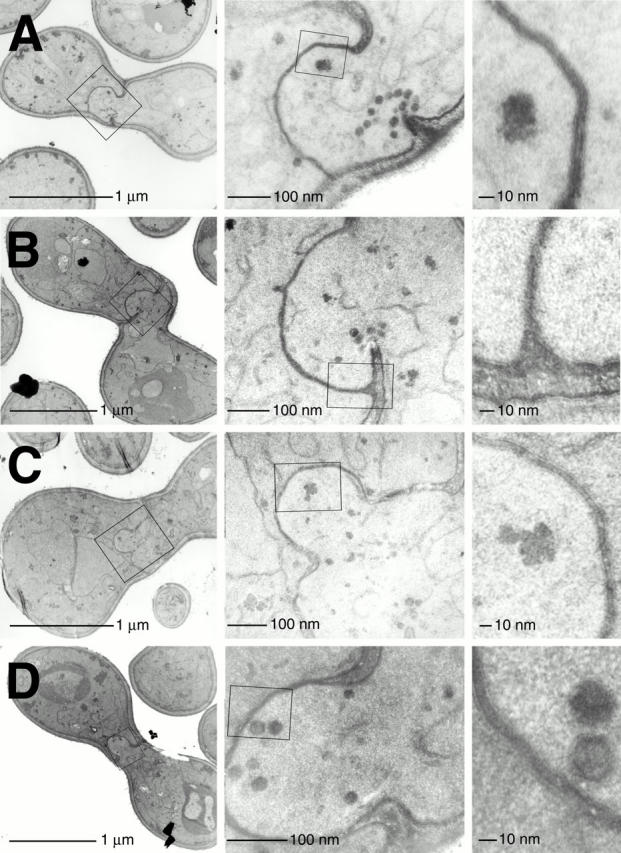
Δprm1 cells successfully degrade their cell wall and juxtapose plasma membranes, but then fail to fuse. Mating mixes of Δprm1 partners were prepared as described above. The cells were then fixed, stained, and imaged by electron microscopy. Three different magnifications are shown for each image (A–D). (Left) Unfused mating pairs. The fuzziest outermost layer of the depicted cells is the cell wall; the dark line underlying it is the plasma membrane. (Middle) Magnification of the box from the left-side panels, showing detail of the bubble. (Right) Magnification of the box from the middle panels, showing tightly juxtaposed membranes at a set distance.
Wild-type matings involve a similar local disruption of the cell wall at the center of this interface, followed by plasma membrane fusion and continued cell wall remodeling until the cytoplasmic bridge between the cells spans the entire width of the zygote and the cell wall becomes restricted to the periphery (Gammie et al. 1998). Details of the intermediates after cell wall breakdown but preceding membrane fusion are unknown because they have not been captured by electron microscopy, presumably because these steps occur rapidly.
Δprm1 cells appeared to complete successfully the initial cell wall breakdown but then failed to perform plasma membrane fusion and continued cell wall remodeling. At the site where cell wall was removed in Δprm1 matings, the two plasma membranes came into close apposition (Fig. 7). Additional membrane appeared to be added to this region equally by both partners, generating bulges that are likely to correspond to the bubbles seen by fluorescence microscopy. Thus, the volume of one mating partner must have grown while the volume of the other one shrank by the same amount. Meanwhile their surface areas must have increased coordinately.
Vesicles of ∼20-nm diameter were usually present in the bulge, often aligned in single-file rows oriented along a mating pair's long axis (Fig. 7 A), suggesting cytoskeletal attachment. These vesicles were packed with a densely staining material similar to that intervening between the two mating partners. These vesicles may deliver new membrane causing growth of the bulge.
Interestingly, the juxtaposed plasma membranes of the bulge were equidistant, consistently remaining separated by a gap of ∼8 nm along their entire length (Fig. 7). A thin layer of densely staining material was seen between them, reminiscent of membrane adherence junctions found between mammalian cells.
Discussion
PRM1 Encodes a Mating-specific Transmembrane Protein that Promotes Cell Fusion at a Very Late Step
We identified a novel protein with several traits expected of a factor involved in cell fusion during mating. First, Prm1p is expressed by cells of both mating types only in response to pheromone. Second, it localizes to the tips of mating projections in shmooing cells and to the necks of mating pairs and zygotes. Third, in its apparent topology it would present two large domains to the plasma membrane of a mating partner, domains that are conserved between widely divergent fungi. Lastly, deletion of PRM1 results in a significant defect in cell fusion, resulting in a ninefold increase in the number of unfused mating pairs compared with wild-type matings. Thus, in some respects, PRM1 resembles many genes described already. In one key regard, though, it differs dramatically from genes found to date. The unfused zygotes produced by a Δprm1 mating do not arrest with an intact cell wall as other fusion mutants do (Kurihara et al. 1994; Elia and Marsh 1996, Elia and Marsh 1998; Gammie et al. 1998; Santos et al. 1997; Erdman et al. 1998). Instead, Δprm1 mutants successfully degrade the cell wall and bring the mating partners' plasma membranes into close proximity. Nevertheless, the membranes remain unfused. This intermediate in the mating reaction has not been trapped before and defines a new step in the pathway. Upstream of membrane fusion, downstream of cell wall breakdown, Prm1p stands in a unique position to help us understand how the bilayers associate and what drives their fusion.
What Does Prm1p Do?
At present, we have insufficient information to distinguish among various models of how Prm1p may facilitate membrane fusion. In principle, Prm1p could either act directly at the fusion step, as a novel fusase, or indirectly, at a step upstream of fusion.
The simplest interpretation of the Δprm1 phenotype is that Prm1p participates directly in the fusion reaction. Yet this model must be reconciled with two observations. First, mutants lacking a fusase would be expected to display an absolute mating defect. On the contrary, almost half of all Δprm1 × Δprm1 mating pairs still fused successfully, and, using classical plate-based mating assays that measure diploid formation among thousands of cells at a time, the Δprm1 mating defect appeared negligible (data not shown). Thus, if Prm1p plays a direct role in membrane fusion, an alternative fusion machine (or other subunits in a Prm1p-containing complex) must exist and take over, albeit inefficiently, upon removal of Prm1p. Second, the suggested multi–membrane-spanning topology of Prm1p does not readily conform to the paradigms developed for viral or SNARE-containing fusases. In particular, full-length Prm1p offers no extracellular free ends that could easily be envisioned to function either as classical fusion peptides or to engage in coiled-coil interactions. Thus, it will be important to define the biochemistry of Prm1p in more depth: does Prm1p associate with other subunits? is it proteolytically processed when expressed on the cell surface (as hinted at by the preliminary observation of the COOH-terminal fragment in Fig. 3 A)? Proteolytic processing could generate protein fragments with a different—and, in light of existing models, more appealing—topology. Ultimate proof of a direct role of Prm1p in membrane fusion, of course, would only come from a biochemical demonstration that Prm1p, possibly with associated subunits, is sufficient for lipid bilayer fusion.
The alternative notion is that Prm1p acts upstream of the fusion event, in either a signaling or a structural capacity. For instance, Prm1p could act in a pathway that senses the proximity of mating partners and responds by activating the fusion machinery. Experiments with mutants weakly deficient for pheromone production have suggested the existence of such a pathway (Brizzio et al. 1996; Elia and Marsh 1996). Indeed, one of these mutants was noted to produce structures resembling Δprm1 bubbles, albeit at low frequency (see Figure 3 D in Elia and Marsh 1996). Another possibility is that the Δprm1 defect may be a structural problem rather than a signaling one. The densely staining matter separating plasma membranes may represent cell wall debris that a Δprm1 mutant cannot clear. However, since any remaining cell wall debris would have to bend and thin as the bubbles grew, the membranes of large bubbles should be closer together than those of small bubbles. In fact that is not true: the gap is consistently ∼8-nm wide along the entire membrane interface regardless of the bubble's size. This observation argues that the gap is occupied not by undegradable cell wall debris but by a specific structural element deposited uniformly as the bubble grows, possibly an adhesion complex fastening membranes in a prefusion state. Without Prm1p the adhesion complex might still assemble but function poorly. Consequently, membranes would stick together but not fuse.
Stalking the Elusive Fusase
We present here the results of a new kind of gene hunt, one that is likely to become increasingly prevalent as genomic databases grow. Previously the problem of membrane fusion during yeast mating had proven refractory to genetic approaches. Despite attacks from several directions and the identification of many interesting genes that act during zygote formation, PRM1 is the first gene that clearly has some role at the level of membrane fusion. Why have mutants in this step been so difficult to find?
Most successful screens have recognized and in some way circumvented the central challenge of mating genetics. Specifically, in order to achieve an appreciable deficit in diploid formation it is usually necessary to impair a pathway not just in one cell but in both mating partners. Three kinds of strategies have solved this problem. First, some groups have taken on the formidable challenge of performing random mutagenesis in a way that generates each mutation in both mating types with complementary selectable markers (Berlin et al. 1991; Kurihara et al. 1994). This approach allowed a direct assay of the mating efficiency of any given mutant crossed to itself. This strategy has the advantage of not biasing toward a particular pathway—indeed, genes controlling not only cell fusion but nuclear fusion were found with it—but, perhaps due to its complexity, it was burdened by a high background of false-positive mutants that discouraged pursuing this approach to saturation.
A second approach uses a preexisting defect in a known fusion pathway, for example fus1 fus2 mutants (both of which genes were found serendipitously in existing lab strains), and asks for new mutants that mate poorly with the enfeebled strain but not with a wild-type strain (Berlin et al. 1991; Chenevert et al. 1994). This strategy has the advantage of being straightforward to set up and execute, although it is probably biased towards the pathway of the starting mutation and may not effectively find components of new pathways.
Lastly, pheromone-regulated genes have been identified and then mutants in these genes assayed for mating defects. This approach was originally carried out using a randomly integrated reporter construct (Erdman et al. 1998). We have here expanded and simplified this latter approach using preexisting genomic datasets combined with a computer-aided search for hydrophobic proteins. This technology let us begin examining candidate gene disruptions without ever doing a traditional screen. The Webminer software makes this approach readily adaptable to many studies that seek proteins expressed under certain conditions, not expressed under other conditions, and containing specific structural features.
The identification of Prm1p's role in cell fusion underscores the sensitivity of this computer-aided approach. Although the penetrance of the Δprm1 mating defect is probably at the limit of what traditional screens can detect, by identifying a small group of candidate genes and examining individual mating pairs in which both partners carried the relevant mutation we could witness a unique phenotype that now offers an opportunity to examine the mechanisms of bilayer association and fusion in molecular detail.
Acknowledgments
We thank Mei-Lie Wong for expert training and assistance with electron microscopy and Sandra Huling for advice on sample preparation. We also thank Ira Herskowitz, Thea Tlsty, Ursula Rüegsegger, Gustavo Pesce, Arash Komeili, and Ted Powers for helpful comments on the manuscript, members of the Walter lab for their support, and Ms. Gabriela Walter-Caldera for her help in scoring fusion defects.
This work was supported by grants from the National Institutes of Health. M.G. Heiman is a predoctoral fellow, and P. Walter is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; HA, hemagglutinin; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor; ORF, open reading frame; YPD, yeast extract/peptone/glucose.
References
- Almeida E.A.C., Huovila A.P.J., Sutherland A.E., Stephens L.E., Calarco P.G., Shaw L.M., Mercurio A.M., Sonnenberg A., Primakoff P., Myles D.G., White J.M. Mouse egg integrin alpha-6-beta-1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- Berlin V., Brill J.A., Trueheart J., Boeke J.D., Fink G.R. Genetic screens and selections for cell and nuclear fusion mutants. Methods Enzymol. 1991;194:774–792. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- Bigler D., Chen M., Waters S., White J.M. A model for sperm-egg binding and fusion based on ADAMs and integrins. Trends Cell Biol. 1997;7:220–225. doi: 10.1016/S0962-8924(97)01058-1. [DOI] [PubMed] [Google Scholar]
- Blobel C.P., Wolfsberg T.G., Turck C.W., Myles D.G., Primakoff P., White J.M. A potential fusion peptide and an integrin ligand domain in a protein active in sperm–egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- Boyd D., Schierle C., Beckwith J. How many membrane proteins are there? Prot. Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V., Gammie A.E., Nijbroek G., Michaelis S., Rose M.D. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J. Cell Biol. 1996;135:1727–1739. doi: 10.1083/jcb.135.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C., Hauser K., Mrsa V., Watzele M., Watzele G., Gruber C., Tanner W. Saccharomyces cerevisiae a- and alpha-agglutinincharacterization of their molecular interaction. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:4081–4088. doi: 10.1002/j.1460-2075.1991.tb04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.S., Tung K.S., Coonrod S.A., Takahashi Y., Bigler D., Chang A., Yamashita Y., Kincade P.W., Herr J.C., White J.M. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha-6-beta-1implications for murine fertilization. Proc. Natl. Acad. Sci. USA. 1999;96:11830–11835. doi: 10.1073/pnas.96.21.11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J., Valtz N., Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae . Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M.C., Hogan J.D., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD)comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L., Marsh L. Role of the ABC transporter Ste6 in cell fusion during yeast conjugation. J. Cell Biol. 1996;135:741–751. doi: 10.1083/jcb.135.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L., Marsh L. A role for a protease in morphogenic responses during yeast cell fusion. J. Cell Biol. 1998;142:1473–1485. doi: 10.1083/jcb.142.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A.E., Brizzio V., Rose M.D. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P.A. Springs and zipperscoiled coils in SNARE-mediated membrane fusion. Structure (Lond.) 1998;6:1487–1491. doi: 10.1016/s0969-2126(98)00147-6. [DOI] [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Virus–cell and cell–cell fusion. Annu. Rev. Cell Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeastfor mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. SOSUIclassification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hughson F.M. Structural characterization of viral fusion proteins. Curr. Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- Jahn R., Sudhof T.C. Membrane fusion and exocytosis. Annu. Rev. Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jenness D.D., Burkholder A.C., Hartwell L.H. Binding of alpha-factor pheromone to yeast a cellschemical and genetic evidence for an alpha-factor receptor. Cell. 1983;35:521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Kurihara L.J., Beh C.T., Latterich M., Schekman R., Rose M.D. Nuclear congression and membrane fusiontwo distinct events in the yeast karyogamy pathway. J. Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F., Rubinstein E., Jasmin C., Prenant M., Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marsh L., Rose M.D. The pathway of cell and nuclear fusion during mating in S. cerevisiae . In: Pringle J., Broach J.R., editors. Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 827–888. [Google Scholar]
- Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Ng D.T.W., Walter P. ER membrane protein complex required for nuclear fusion. J. Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozier-Kalogeropoulos O., Malpertuy A., Boyer J., Tekaia F., Dujon B. Random exploration of the Kluyveromyces lactis genome and comparison with that of Saccharomyces cerevisiae . Nucleic Acids Res. 1998;26:5511–5524. doi: 10.1093/nar/26.23.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos J., de Lima M.C. The influenza virus hemagglutinina model protein in the study of membrane fusion. Biochim. Biophys. Acta. 1998;1376:147–154. doi: 10.1016/s0304-4157(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Santos B., Duran A., Valdivieso M.H. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae . Mol. Cell. Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., Podbilewicz B. Fusomorphogenesiscell fusion in organ formation. Dev. Dyn. 2000;218:30–51. doi: 10.1002/(SICI)1097-0177(200005)218:1<30::AID-DVDY4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Berger B., Kim P.S. LearnCoil-VMFcomputational evidence for coiled-coil–like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Boeke J.D., Fink G.R. Two genes required for cell fusion during yeast conjugationevidence for a pheromone-induced surface protein. Mol. Cell. Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F., Sollner T.H., Rothman J.E. SNAREpinsminimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Yuan Y.L., Fields S. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol. Cell. Biol. 1991;11:5910–5918. doi: 10.1128/mcb.11.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]