Abstract
DRAL is a four and a half LIM domain protein identified because of its differential expression between normal human myoblasts and the malignant counterparts, rhabdomyosarcoma cells. In the current study, we demonstrate that transcription of the DRAL gene can be stimulated by p53, since transient expression of functional p53 in rhabdomyosarcoma cells as well as stimulation of endogenous p53 by ionizing radiation in wild-type cells enhances DRAL mRNA levels. In support of these observations, five potential p53 target sites could be identified in the promoter region of the human DRAL gene. To obtain insight into the possible functions of DRAL, ectopic expression experiments were performed. Interestingly, DRAL expression efficiently triggered apoptosis in three cell lines of different origin to the extent that no cells could be generated that stably overexpressed this protein. However, transient transfection experiments as well as immunofluorescence staining of the endogenous protein allowed for the localization of DRAL in different cellular compartments, namely cytoplasm, nucleus, focal contacts, as well as Z-discs and to a lesser extent the M-bands in cardiac myofibrils. These data suggest that downregulation of DRAL might be involved in tumor development. Furthermore, DRAL expression might be important for heart function.
Keywords: LIM domain protein, transcriptional regulation, p53, apoptosis, subcellular localization
Introduction
Development of cancer results from multiple genetic events occurring in a single cell and culminating in the inactivation and/or malfunction of several key proteins. Among these, the tumor suppressor gene p53 is the most frequently mutated gene in human cancer (Harris 1996; El-Deiry 1998). p53 plays a central role in cells by inducing or repressing transcription of a multitude of target genes. Transcriptional regulation of target genes by p53 requires the presence of two copies of the 10-bp motif 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ separated by 0 to 13 bp (El-Deiry et al. 1992). This tandem motif can be located either in the 5′ region, in introns, or the 3′ regions of the target genes. Although it has been suggested that there may be several hundred p53-inducible genes (Tokino et al. 1994), only about 50 have been identified and described to date. These can be subdivided into several specific groups according to their function, such as involvement in G1 arrest, DNA repair, G2/M arrest, and apoptosis. Among the target genes that participate in the apoptotic pathway (reviewed in Amundson et al. 1998) are proteins like BAX, IGF-BP3, FAS, DR5, PAG608, TRID, and the just recently described ei24, Noxa, and PERP (Buckbinder et al. 1995; Miyashita and Reed 1995; Owen-Schaub et al. 1995; Israeli et al. 1997; Wu et al. 1997; Sheikh et al. 1998, Sheikh et al. 1999; Attardi et al. 2000; Gu et al. 2000; Oda et al. 2000) all of which are induced, whereas Bcl-2, MAP4 (a microtubule stabilizing protein), and IGFI-R levels are repressed by p53 (Miyashita et al. 1994; Murphy et al. 1996; Prisco et al. 1997). However, the sole absence of one target gene is not sufficient to block the functions of p53. Recently, a number of additional p53-inducible genes involved in apoptosis or growth arrest have been identified by the SAGE (serial analysis of gene expression) technique (Polyak et al. 1997). Since the inducible genes exhibited variable kinetics of induction, p53 might act not only on multiple target genes, but also at different time points. Whereas most of the above mentioned target genes are ubiquitously expressed, some p53-inducible genes must act in a cell type–dependent manner, as shown for example for the muscle-specific phosphoglycerate mutase gene, which is induced by p53 specifically in muscle cells (Ruiz-Lozano et al. 1999).
Our previous studies focused on the identification of potential new tumor suppressor genes from a pediatric sarcoma model. For this purpose we performed a subtractive cDNA library screening between normal cultured myoblasts and the human embryonal rhabdomyosarcoma (RMS) cell line RD (Genini et al. 1996). 48 genes were identified as being downregulated in RD cells, 19 of which were coding for unknown genes. One of these genes was subsequently characterized as a novel LIM-domain protein and named DRAL (for downregulated in rhabdomyosarcoma LIM protein, now also called SLIM3, or FHL-2) (Genini et al. 1997).
The LIM domain is a cysteine-rich domain of ∼50 amino acids, which folds into two specialized zinc fingers and is involved primarily in protein–protein interactions (for reviews see Sanchez-Garcia and Rabbitts 1994; Schmeichel and Beckerle 1994; Dawid et al. 1998). DRAL was the first four and a half LIM domain (FHL) protein discovered. Now four additional proteins have been assigned to this subfamily, namely SLIM1 (FHL1) and SLIM2 (FHL3) both mainly expressed in skeletal muscle (Morgan et al. 1995; Greene et al. 1999), and ACT and FHL4 with highest expression in testis (Fimia et al. 1999; Morgan and Madgwick 1999). All these proteins range in size from 31 to 35 kD. Thus far, none of these FHL proteins have been implicated in cancer development and not much is known about their function. However, SLIM1 can interact with the mammalian homologue of the Drosophila transcription factor suppressor of hairless (Taniguchi et al. 1998) and ACT binds to and stimulates the cAMP-responsive element modulator (CREM; Fimia et al. 1999). Hence, proteins of the FHL subclass might be directly involved in modulation of transcription. This notion is supported by recent experiments demonstrating that DRAL can act as costimulatory factor for the androgen receptor (Muller et al. 2000). Hence, the available evidence suggests that these five LIM-only proteins might share similar functions, but are restricted to different tissues or developmental stages.
Here, DRAL was identified as a p53-responsive gene. Given the potential unique role of DRAL in tumor biology and to obtain insight into possible functions of this protein, we investigated the effects of ectopic DRAL expression and determined its intracellular localization in a range of cell types.
Materials and Methods
Cell Lines
All cell lines were grown in DME supplemented with 10% FBS (Life Technologies), except for primary myoblasts, which were cultured in F12 medium with 15% FBS; both media contained 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies). The nonmuscular cells were maintained in 5% CO2, muscle cells in 10% CO2 at 37°C. The human embryonal RMS cell line RD, NIH 3T3 mouse fibroblasts, and COS-1 African green monkey kidney cells were obtained from American Type Culture Collection. RD-tsp53 (RD cells expressing a temperature-sensitive p53 mutant, amino acid 135 Ala to Val) and RD-Neo cells (vector alone) were generated as described (De Giovanni et al. 1998). The p53+/+ and p53−/− mouse embryonal fibroblasts expressing either wild-type or mutant p53 are described elsewhere (Pruschy et al. 1999). Neonatal rat cardiomyocytes were isolated and maintained as described (Auerbach et al. 1999).
Northern Blot Analysis
Total RNA was extracted from different cells by guanidinium-isothiocyanate lysis followed by centrifugation through a 5.7 M caesium chloride cushion. It was then separated on a 1% agarose gel in the presence of 2.2 M formaldehyde and transferred to Nytran nylon membranes (Schleicher & Schuell, Inc.) by capillary transfer. Alternatively, a commercially available human RNA Master Blot™ was used (CLONTECH Laboratories, Inc.). Equal loading of the blots was confirmed by hybridization with β-actin or ubiquitin, respectively. Probes (inserts of the clones A33-35; nucleotides 64–451 of the DRAL cDNA), a 505-bp EcoRI–KpnI DRAL fragment for hybridization of the mouse Northern blot, A33-89, A33-124, human EST clone 470149, and mouse EST clone 533961 (p21WAF1) were generated by random priming (Prime-a-gene; Promega) with α[32P]dATP (NEN Life Science Products) and used for hybridization at 68°C with QuickHyb Hybridization Solution (Stratagene) according to the manufacturer's instructions. The membranes were exposed to x-ray films (Eastman Kodak Co.) with intensifying screens at −70°C.
Ionizing Radiation (IR) Treatment
Primary human myoblasts, RD cells, and wild-type and mutant p53 expressing mouse fibroblasts were plated on 10 cm dishes 24 h before treatment, for each time point in duplicate. After exposure to 20 Gy of IR (from a 137Cs source) cells were fed with fresh medium and cultured until harvesting.
Cloning of the Human DRAL Promoter
A human P1 library was screened by PCR (Genome Systems, Inc.) using the DRAL-specific primers DRAL-FOR2 5′-ACCCGCAAGATGGAGTA-3′ and DRAL-REV3 5′-GCAGGGCACACAGAAATTCTG-3′ under the following cycling conditions: 15 s at 94°C, 30 s at 56°C, and 60 s at 72°C for 30 cycles. Southern blot analysis was carried out either using nucleotide (nt) 64 to nt 150 of the DRAL cDNA or a 2.5-kb PstI subfragment of the P1 clone as probes with QuickHyb solution according to the manufacturer's procedures (Stratagene). Probes were random primed as described above. Subcloning into pUC18 and sequencing was done according to standard procedures. Putative transcription factor binding sites in the promoter region were identified by computer programs (WebSignalScanProgram 4.05 [TFD database] and MatInspector 2.2).
Plasmid Constructions
Two constructs tagging the COOH- and NH2-terminal end of the DRAL coding region with the FLAG epitope DYKDDDDK (DRAL-CF and DRAL-NF, respectively) were constructed by PCR amplification from full-length human cDNA using the Expand™ High Fidelity PCR System (Roche) and the following primers, which were designed to encode a BamHI (5′ end) or Xba I (3′ end) restriction site and the FLAG epitope: for DRAL-CF (COOH-terminally FLAG-tagged DRAL): 5′-CGGGATCCGCCACCATGACTGAGCGCTTTGACTGC-3′ and 5′-GCTCTAGATCACTTGTCATCGTCGTCCTTGTAGTCGATGTCTTTCC- CACAGTC-3′; and for DRAL-NF (NH2-terminally FLAG-tagged DRAL): 5′-CGGGATCCGCCACCATGGACTACAAGGACGACGATGACAAGACTGAGCGCTTTGACTGC-3′ and 5′-GCTCTAGATCAGATGTCTTTCCCACA-3′. Amplified fragments were digested and ligated into pcDNA3 (Invitrogen). All constructs were verified by sequencing before use.
Transfection and Immunofluorescence
RD, NIH 3T3, and COS-1 cells were plated 1 d before transfection on 35-mm dishes (Falcon). For transfections 5 μg of DNA (DRAL, DRAL–CF, DRAL–NF, or pFLAG–CMV-2–BAP as control plasmid; Eastman Kodak Co.) and 5 μl lipofectamine (Life Technologies) for each dish were added to the cells in unsupplemented DME. Medium was changed 6–8 h after transfection. Cells were fixed with 3.7% formaldehyde, permeabilized with methanol, and then stained with the anti-FLAG primary monoclonal antibody M2 (Sigma-Aldrich; diluted 1:300), followed by a Cy3-conjugated goat anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories; diluted 1:200). DNA was visualized by Hoechst staining. Cells were viewed through a fluorescence microscope (Carl Zeiss, Inc.) and transfected cells were quantified by photographing three representative regions of each plate. Every picture showed 280 to 500 cells, the exact cell number was obtained by counting the Hoechst-stained nuclei. DRAL-positive cells were counted as apoptotic when they were round and displayed condensed nuclei. Three independent experiments were carried out.
Neonatal rat cardiomyocytes were transfected as described (Auerbach et al. 1999) and stained with the anti-FLAG antibody M2 (see above) and a polyclonal anti–chicken heart myosin binding protein C antibody (Bähler et al. 1985) followed by secondary antibodies, FITC-coupled anti–mouse (Cappel), and Cy3-coupled anti–rabbit (Jackson ImmunoResearch Laboratories). Endogenous DRAL was localized with the polyclonal anti–human DRAL antibody (Genini et al. 1997) as described. Sarcomeric α-actinin and myomesin were visualized using monoclonal antibodies (clone EA-53 [Sigma-Aldrich] and B4 [Grove et al. 1984]). Secondary antibodies were as described above.
AnnexinV staining was carried out as described previously (Bernasconi et al. 1996).
Caspase Activity
Cells were resuspended in lysis buffer (50 mM Pipes, 50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM PMSF, 1 mM DTT, 1 μg/ml chymostatin, leupeptin, aprotinin, and pepstatin each) and lysed by at least two freeze–thaw cycles. 150 μg of lysate was incubated with either caspase-3 inhibitor (DEVD-CHO; Bachem) and substrate (DEVD-pNA; Bachem) or substrate alone at 80 μM in 0.1 ml. Substrate cleavage was monitored at 405 nm and background activity subtracted for data presentation.
Confocal Microscopy
Cell lines were cultured in 35-mm plastic dishes (Falcon) or on fibronectin-coated glass coverslips in 24-well plates. Immunofluorescence labeling was done as described above, except that DNA was labeled using Pico Green (Molecular Probes, Inc.; diluted 1:200). Stainings were visualized with a confocal laser scanning microscope consisting of a Leica inverted microscope (DM IRB/E) equipped with an argon–krypton mixed gas laser. Image processing was done on a Silicon Graphics workstation using the software, Imaris (Bitplane AG; Messerli et al. 1993).
Results
Identification of DRAL as a p53-inducible Gene
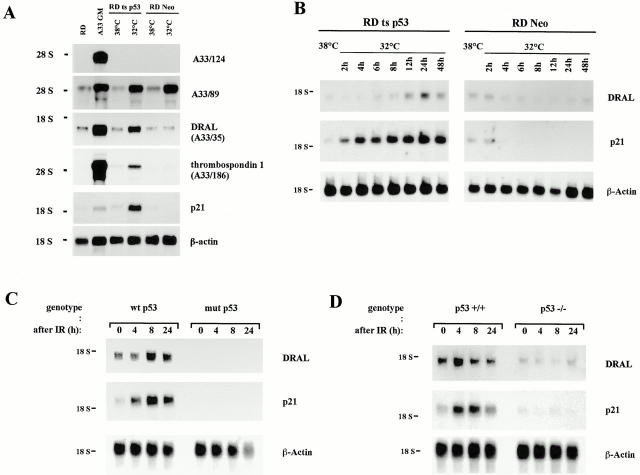
Previously, we applied a subtractive cloning procedure to identify molecular changes occurring during progression from normal myoblasts to RMS (Genini et al. 1997). From this experiment, a list of 48 different genes downregulated in the human RMS cell line RD was compiled, among them 19 unknown genes. Since RD cells have a mutation in the p53 tumor suppressor gene (Felix et al. 1992), we hypothesized that downregulation of some of these genes might be due to lack of p53. Indeed, analyzing the cDNAs obtained by the subtractive screen already identified one known p53 target gene, namely thrombospondin-1 (A33-186) (Dameron et al. 1994). Hence, taking advantage of a RD-tsp53 cell line, which stably expresses a temperature-sensitive p53 cDNA (p53Val137) that is inactive at 37°C and assumes wild-type conformation at 32°C, all unknown genes were screened for possible p53-dependent expression. As control, RD-Neo (vector alone) cells were used. As expected, by dropping the temperature to 32°C, expression of thrombospondin-1 was induced in the RD-tsp53 cells but not in the RD-Neo cells (Fig. 1 A). As a further control, enhanced mRNA levels of the p53 target gene p21WAF1 were detected in the same experimental series. Analyzing the expression of clones from the subtractive screen revealed that most clones tested from our library were not induced such as A33-124, or induced solely by dropping the culture temperature to 32°C like A33-89, suggesting that expression of the respective gene might be influenced by stress. In contrast, expression of the previously characterized cDNA coding for DRAL (A33/35) could be substantially stimulated in a p53-dependent manner (Fig. 1 A), analogous to the known p53 targets p21WAF1 and thrombospondin-1.
Figure 1.
(A) Screening for p53-inducible genes. 5 μg total RNA from the indicated human cell lines was loaded in each lane and hybridized with the labeled cDNAs A33/124, A33/89, A33/35, A33/186, p21WAF1, and β-actin. RD are RMS cells, A33 GM are primary myoblasts kept in growth medium, RD-tsp53 are RD cells expressing a temperature-sensitive p53 mutant, and RD-Neo are RD cells expressing the vector alone. The mutant p53 protein is inactive at 38°C, but dropping the temperature to 32°C leads to wild-type p53 protein functions. RNA was extracted one day after temperature shift. (B) Time course analyses of p53 target gene induction. Total RNA was extracted from RD-tsp53 or RD-Neo cells either grown at 38° or 32°C for the indicated time. 3 μg total RNA was loaded in each lane. The blot was hybridized with the labeled cDNAs coding for DRAL, p21WAF1, and β-actin. (C) Induction of DRAL expression after γ-irradiation. Normal human myoblasts (wtp53) and RD RMS cells (mutp53) were treated with 20 Gy, and afterwards fed with fresh medium. Total RNA was isolated at the time points indicated and subjected to Northern blot analysis (3 μg/lane). Inserts from the indicated cDNAs were isolated, labeled by random priming, and used for hybridization. The blot was also reprobed for β-actin as a control for equal loading and transfer. (D) Same as in C except that p53+/+ or p53−/− mouse fibroblasts were irradiated.
To confirm and characterize these initial findings in more detail, gene expression was analyzed in a time-dependent manner by extracting RNA at eight different time points after temperature shift (from 2 to 48 h). Northern blot analysis revealed an increase of DRAL mRNA levels specifically in the RD-tsp53 cell line and not in RD-neo cells (Fig. 1 B). The increase was highest after 24 h, similar to p21WAF1 used as control. Hence, induction of wild-type p53 in the RD cell line led to the selection of a potential new p53-inducible gene, DRAL, from the initially screened cDNAs.
To determine whether DRAL can also be induced upon activation of endogenous p53, its expression was studied both in primary human myoblasts (wild-type p53) compared with RD cells (mutant p53) and in p53+/+ compared with p53−/− mouse embryo fibroblasts (Fig. 1C and Fig. D). In untreated samples, the levels of both DRAL and p21 mRNA were strikingly lower in cells containing mutated or no p53 than in wild-type cells. After exposure to 20 Gy of IR increases in p21WAF1 as well as DRAL mRNA levels were observed in wild-type p53 but not in mutant p53 cells. Interestingly, the time course of activation after irradiation is similar for DRAL and p21, although slightly different for the two cell types examined. These data suggest that transcription of endogenous DRAL can be stimulated by irradiation in a p53-dependent manner.
Cloning and Partial Characterization of the DRAL Gene and Its 5′ Upstream Region
To gain some insight into the regulation of DRAL expression on the molecular level, the promoter region of the human DRAL gene was cloned. Screening of a human P1 library by PCR using DRAL-specific primers (amplifying nt 472 to nt 597 of the DRAL cDNA) resulted in one positive clone, P1:21280. To ensure that no recombination event occurred in the P1 clone, Southern blot analysis was performed with either human genomic DNA or the P1 clone digested with two different restriction enzymes and using a 2.5-kb PstI fragment from the P1 clone as a probe. Since both DNAs revealed the same hybridization pattern, any gross recombination in the relevant region of the P1 clone can be excluded (Fig. 2 B). The 7.8-kb HindIII fragment identified in this Southern blot analysis was then further analyzed in detail and its sequence determined completely (available from Genbank/EMBL/DDBJ under accession number AF211174). The fragment contained 1.5 kb of upstream sequence and exon 1 (63 bp) and exon 2 (51 bp), separated by a 2,276-bp intron of the DRAL gene (Fig. 2 A). The sequence of this intron is flanked by consensus splice donor and acceptor sites. In the upstream region, a TATA box is present 72-bp upstream of the start of the cDNA (which was isolated by RACE experiments). Therefore, it is likely that the start of transcription is located further upstream and the cDNA extends another 40–50 bp. Additionally, two GC rich sequences (GC-boxes) were identified at positions −132 and −166 (relative to the start of the cDNA). A search for putative sequence elements able to bind upstream regulatory transcription factors within the promoter, identified several sites including one for myocyte enhancer binding factor 2 (MEF-2) at position −890, five E-boxes at positions −480, −508, −708, −878, and −1274 as well as five binding sites for the homeobox protein NKX2.5 at positions −417, −636, −815, −868, and −1426 (only the first four sites for both are indicated in Fig. 2 A). The presence of multiple E-boxes might explain expression of DRAL in normal muscle tissue. Additionally, the five NKX2.5 sites are likely to contribute to the strong expression of DRAL in cardiac tissue (see below). Intriguingly, five putative p53-binding sites were identified within the 7.8-kb fragment and designated as p53 I to p53 V (Fig. 2 C). The sites p53 I and p53 II are located in the 5′ region whereas the sites p53 III, IV, and V are located in intron 2.
Figure 2.
Partial genomic organization of the human DRAL gene. (A) The 7.8-kb HindIII fragment of the P1 clone 21280 contains the first two noncoding exons (exon 1: nt 1 to nt 63, exon 2: nt 64 to nt 114 of the DRAL cDNA). Letters correspond to the restriction sites: H, HindIII; P, PstI; S, SstI; E, EcoRI; and B, BamHI. Putative transcription factor binding sites in the promoter region are indicated. The five putative p53-binding sites within the 7.8-kb subclone are designated as sites p53 I–p53 V, sequences are listed separately in C. (B) Southern blot analysis using either human genomic DNA or the P1:21280 clone digested with HindIII (H) and PstI (P). The blot was hybridized with the 2.5-kb PstI fragment of the 7.8-kb subclone containing exon II. The sequence of the 7.8-kb HindIII fragment is available from Genbank/EMBL/DDBJ under accession number AF211174.
In summary, cloning and characterization of the DRAL promoter region identified five potential p53 target sites, further supporting the observed p53-dependent transcriptional activation of the DRAL gene.
Expression Pattern of DRAL in Normal Tissues
To obtain a broad view of DRAL expression in human tissues, we analyzed its mRNA expression pattern in 50 human adult and fetal tissues. The results confirmed previous observations (Genini et al. 1997) in that DRAL expression was highest in fetal and adult heart followed by ovary (Fig. 3). Equivalent levels of expression were further detected in placenta, uterus, mammary gland, and adrenal gland. In skeletal muscle, colon, bladder, prostate, stomach, trachea, testis, small intestine, thyroid gland, and kidney DRAL mRNA was still clearly detectable. No significant difference in DRAL expression was evident between fetal tissues and their adult counterparts (brain, heart, kidney, liver, spleen, thymus, and lung), suggesting that the average RNA levels are not altered during the developmental period analyzed. Furthermore, it appears that DRAL is only marginally expressed in the brain. The small signal observed for Escherichia coli DNA indicates the possibility that related sequences are present in E. coli. The strong expression of DRAL in heart is also reflected by examination of the origin of EST sequences present in the databases, since one fifth of the ESTs coding for DRAL originate from heart cDNA libraries. Apart from the predominant expression in human heart, lower levels of transcription are observed in a wide range of tissues.
Figure 3.
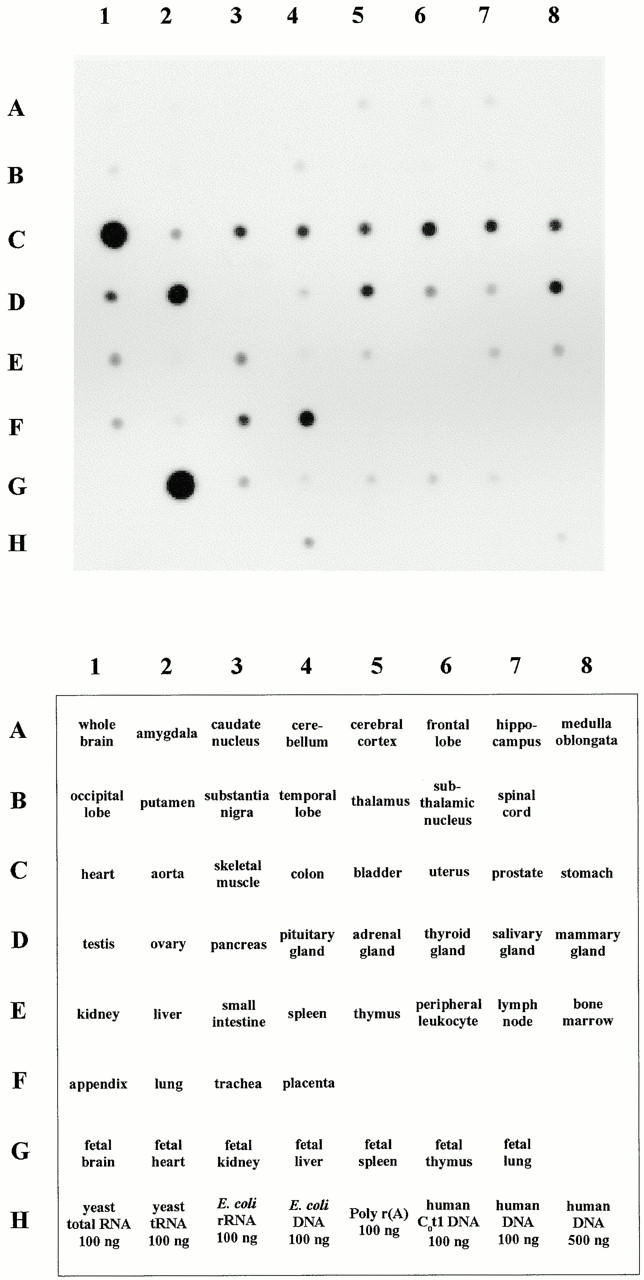
Expression pattern of DRAL in normal human fetal and adult tissues. A commercially available human RNA dot blot containing normalized quantities of mRNA per dot was hybridized with a labeled DRAL cDNA probe. The tissues used for mRNA extraction are indicated in the lower panel.
Ectopic Expression of DRAL Induces Apoptosis
To gain insight into possible functions of DRAL, an expression vector carrying the DRAL cDNA was transfected into human RMS (RD), transformed monkey kidney (COS-1), and normal mouse fibroblast (NIH 3T3) cells. After selection with neomycin only a small number of clones was obtained whereas empty vector readily generated several hundred colonies from a parallel transfection. Surprisingly, none of the clones stably expressed DRAL protein (data not shown). To analyze the possible reason for this phenomenon, transient transfection experiments were performed in the same cells. Since DRAL consists only of LIM domains, the previously raised polyclonal antibody might also interact with other LIM domain proteins. To eliminate this possibility, two plasmids were constructed producing epitope-tagged DRAL protein, namely DRAL-CF (COOH-terminally FLAG-tagged DRAL) and DRAL–NF (NH2-terminally FLAG-tagged DRAL). As control in these experiments, the pFLAG–CMV-2–BAP plasmid (FLAG-tagged bacterial alkaline phosphatase) was used. Expression of FLAG-tagged proteins was analyzed by indirect immunofluorescence staining together with Hoechst DNA staining at different time points after transfection. In all cells highest FLAG–DRAL expression was detected after 32 h. Interestingly, the number of FLAG–DRAL-positive cells rapidly declined afterwards. At the same time labeled remnants were observed in the culture, which were reminiscent of apoptotic bodies (Fig. 4). Neither decline in expressing cells nor cell remnants were observed with FLAG–BAP, indicating that the effects observed are specific for DRAL. In addition, no difference was observed between DRAL–CF and DRAL–NF constructs. To unambigously identify apoptotic cells, caspase-3 activity was measured in transfected cultures as well as simultaneous stainings performed with either pico green labeled DNA (Fig. 5) or annexin V. A quantitative summary of these results is shown in Fig. 6. A dramatic increase in the number of apoptotic cells (70–90% after 72 h depending on cell type) was paralleled by a rapid decline in the number of transfected cells in transfections with both FLAG–DRAL constructs. In contrast, expression of pFLAG–CMV-2–BAP resulted in a constant number of transfected cells over the observation period and no specific increase in apoptotic cells (Fig. 6 A). Annexin V stainings on COS-1 cells harvested 42 h after transfection were quantified with the fluorescence-activated cell sorter demonstrating an approximate threefold increase in positively stained cells. This value was obtained over the entire transiently transfected population (transfection efficiency 15–20%) (Fig. 6 B). To further confirm the induction of apoptosis, caspase-3 activity was measured 28 h after transfection (Fig. 6 C). Indeed, expression of DRAL resulted in at least twice as much activity as the controls (vector alone or unrelated protein).
Figure 4.
Time course of DRAL expression in COS-1 cells. Cells were transfected with either DRAL-CF (A–C), DRAL-NF (D–F), or the control plasmid pFLAG–CMV-2–BAP (G–I). After 32, 48, and 72 h cells were fixed and stained with the monoclonal anti-FLAG antibody. Pictures were taken from representative areas. The scale bar line is valid for all the pictures and indicates 100 μm. Arrows point to antibody-labeled remnants, which were absent in the transfections with the control pFLAG–CMV-2–BAP construct.
Figure 5.
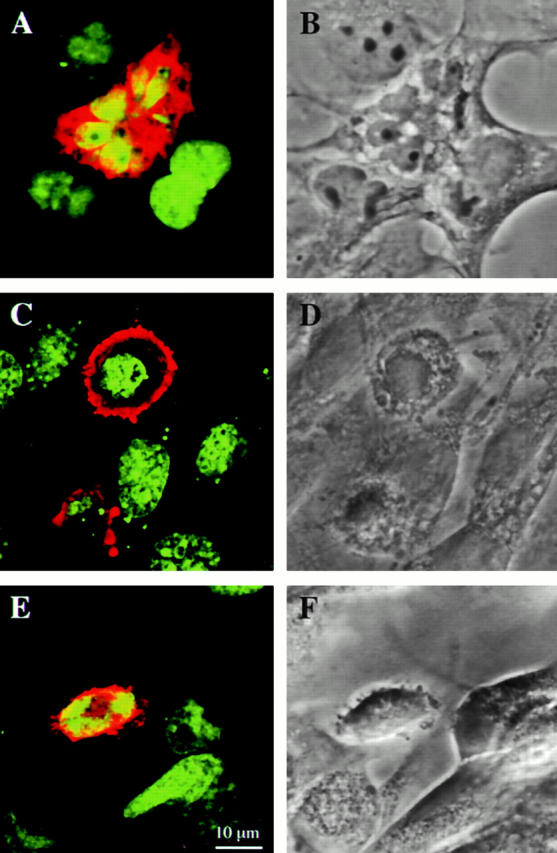
Induction of apoptosis by ectopic expression of DRAL. COS-1 (A and B), NIH 3T3 (C and D), and RD (E and F) cells were transiently transfected with FLAG-tagged DRAL. A, C, and E are composite confocal microscopy images of DRAL (red) and DNA (green) staining. B, D, and F represent the corresponding phase contrast picture. Bar: (A–F) 10 μm.
Figure 6.
Quantitative analysis of the events following ectopic expression of DRAL. (A) COS-1, RD, and NIH 3T3 cells were transfected with DRAL-CF (circle), DRAL-NF (square), or the control plasmid pFLAG–CMV-2–BAP (cross). After 24, 32, 48, and 72 h cells were fixed and stained for FLAG and Hoechst. Data are shown as mean values with standard error bars representing three independent transfection experiments. (% apoptotic cells) Refers to rounded FLAG-positive cells displaying nuclear condensation and fragmentation in percent of the total number of FLAG positive cells. In the right hand panels, transfected cells were counted as FLAG-positive cells and indicated as percentage of total cell number during time course. (B) COS-1 cells were stained with annexin V 42 h after transfection of the indicated DNA constructs. (C) COS-1 cell extracts were prepared 28 h after transfection with empty vector (black bars), pFLAG–CMV-2–BAP (hatched bars), DRAL-sense (punctated bars), DRAL-CF (grey bars), or DRAL-NF (white bars). OD was measured at 405 nm at 2, 4, 6, and 8 h after substrate addition. Three independent experiments were carried out.
We conclude from these experiments that ectopic expression of DRAL specifically induced apoptosis in all cell types analyzed, including human RMS cells. This notion is consistent with a possible role for DRAL as a potential tumor suppressor molecule.
Subcellular Localization of DRAL
To obtain some insight at potential protein–protein interactions that might be functionally relevant, the subcellular localization of DRAL was studied after transient transfection of FLAG–DRAL into the same cell types used before (RD, COS-1, and NIH 3T3). The previously reported predominant nuclear staining of DRAL (Genini et al. 1997) was confirmed in RD and NIH 3T3, to a lesser extent in COS-1 cells (Fig. 7). In addition, in some cells uniform staining was observed (Fig. 7D and Fig. G), sometimes even with exclusion of the nucleus (Fig. 7H and Fig. I). Interestingly, DRAL was also detected in the cellular periphery where spreading occurs, most likely resembling focal contact staining, in all three cell lines (Fig. 7, A–C). Indeed, this conclusion could be confirmed by colocalization of DRAL with vinculin (data not shown). No difference was detected in these experiments in regard to the position of the FLAG epitope on either side of DRAL.
Figure 7.
Subcellular localization of DRAL. NIH 3T3 (A and I), COS-1 (B, G, and H), and RD (C–F) cells were cultured on fibronectin coated cover slips or plastic dishes and transiently transfected with FLAG–DRAL. Cells were immunostained with anti-FLAG monoclonal antibody M2 and Cy3-conjugated goat anti–mouse polyclonal antibody and analyzed by confocal fluorescence microscopy. The arrows point to the focal contacts. Bars, 10 μm.
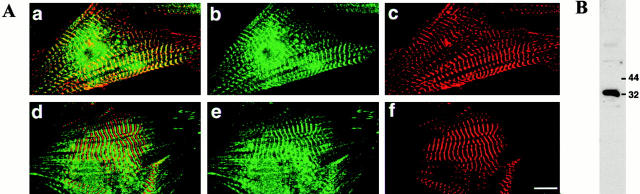
Since highest expression of DRAL was found in fetal and adult heart, we also wished to examine the subcellular localization of DRAL in cardiomyocytes. To this end neonatal rat cardiomyocytes were transiently transfected with either FLAG–DRAL (Fig. 8) or the FLAG–BAP plasmid (data not shown). Interestingly, in addition to the already described localization of DRAL in the nucleus and focal contacts, a distinct cross-striated pattern was observed. Double staining of the same cells with the myosin binding protein C and subsequent overlapping of the two staining patterns revealed that DRAL localizes specifically to the Z-discs and, to a lesser extent, to the M-band of myofibrils (Fig. 8C and Fig. F, small inset). In cardiomyocytes transfected with the FLAG–BAP plasmid only completely diffuse labeling could be observed, indicating that the localization is specific for DRAL (data not shown). To confirm these observations, immunofluorescence stainings of the endogenous DRAL in cardiomyocytes were performed as well (Fig. 9). Western blot of a cardiomyocyte extract with the anti-DRAL polyclonal antibody shows a major reactive species of about 32 kD, indicating that the antibody reacts specifically (Fig. 9 B). Immunofluorescence staining then confirmed localization of endogenous DRAL to the Z-discs and the M-band of cardiac myofibrils (Fig. 9 A).
Figure 8.
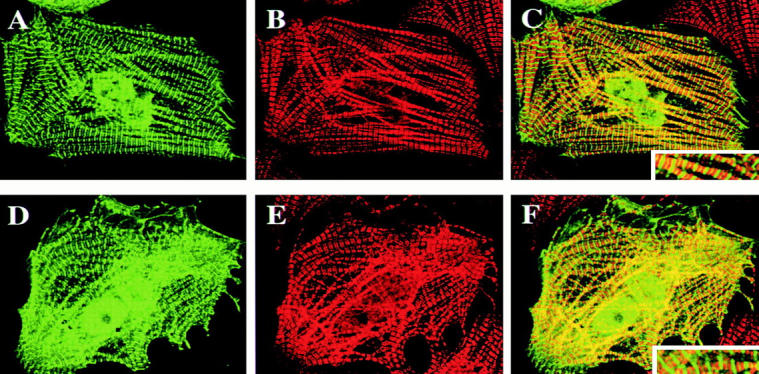
Subcellular localization of DRAL in cardiomyocytes. Neonatal rat cardiomyocytes were transiently transfected with FLAG–DRAL (DRAL-NF in A–C, DRAL-CF in D–F) and immunostained with anti-FLAG monoclonal antibody (A and D) or anti-myosin binding protein C antibody (B and E). C and F represent superposition of A and B or D and E, respectively. The insert shows an enlargement of myofibrils. Analysis was carried out with a confocal fluorescence microscope using Imaris software.
Figure 9.
Localization of endogenous DRAL in neonatal rat cardiomyocytes. (A) Double immunofluorescence staining with a polyclonal anti-DRAL antibody (green in a, b, d, and e) together with monoclonal antibodies recognizing sarcomeric α-actinin (red in a and c) or myomesin (red in d and f) reveals a signal primarily in the region of the Z-disc (overlap with α-actinin seen in yellow in a and alternating staining with myomesin in d). Striations can also be detected occasionally in the M-band. Bar, 10μm. (B) Western blot analysis of protein extract detects a major band at 32 kD.
Hence, DRAL could be unambigously localized in the nucleus, the focal contacts and the Z-discs as well as the M-band of heart myofibrils. Presumably, DRAL will have a different protein interaction partner at each of these locations.
Discussion
This study identifies a member of the FHL subfamily of LIM-only proteins, DRAL, as a novel target gene for the tumor suppressor protein p53. Intriguingly, DRAL protein was capable of efficiently triggering apoptosis in a wide range of cell types upon ectopic expression.
We provide three different experimental criteria suggesting that DRAL might be a direct transcriptional target of p53. First, induction of wild-type p53 in RMS cells through a temperature-sensitive p53 allele specifically increased transcription of endogenous DRAL, in a manner comparable to known p53 target genes like p21WAF1 and thrombospondin-1. Second, exposure of primary myoblasts and fibroblasts to γ-irradiation induced an increase in DRAL mRNA that paralleled the increase in p21WAF1 mRNA only in cells with wild-type p53. Because of different probes and exposure times, the mRNA levels of p21 and DRAL are not directly comparable. However, cells with mutated or no p53 consistently showed a striking reduction in the expression from both genes whereby basal levels of DRAL were higher in wild-type p53 cells than p21WAF1. This could be a possible explanation for the lower induction of DRAL mRNA after irradiation. Also, the basal expression level of DRAL was higher in fibroblasts than in myoblasts, possibly accounting for the more moderate induction observed in this cell type. Finally, analysis of the genomic structure of DRAL in the promoter region revealed five potential consensus p53-binding sites. Interestingly, the DRAL gene could be identified on recently published draft sequences of the human genome project (GenBank/EMBL/DDBJ accession numbers AC069576 and AC012360) indicating that the entire gene consists of seven exons.
In Northern blot experiments, DRAL expression was predominantly found in fetal and adult heart. The five putative NKX2.5-binding sites identified in the promoter region are in concurrence with this expression pattern. In fact the homeobox gene Nkx2.5 represents the earliest known marker of the cardiac lineage in vertebrates and its expression is maintained throughout the developing and adult heart (Schwartz and Olson 1999). Additionally, a MEF-2–binding site was found in the DRAL promoter. MEF-2 factors are known coregulators for myogenic basic helix-loop-helix proteins and play a pivotal role in determination and differentiation of skeletal and cardiac muscle cells (Black and Olson 1998). Finally, five E-boxes, which can be found in the control regions of many genes specifically expressed in skeletal muscle, are likely to contribute to expression of DRAL in skeletal muscle.
Since transcription of DRAL can be induced by wild-type p53, the next question was if DRAL would participate in any of the known functions of p53-like growth arrest or apoptosis. To test this notion, ectopic expression of DRAL was achieved in a range of cell types including human RMS cells. Interestingly, this resulted in efficient induction of apoptosis in all cell types analyzed, which was confirmed on the molecular level by annexin V staining and caspase-3 activity measurements. Indeed, we were unable to generate stable cell lines expressing DRAL. Even RMS cells ectopically expressing Bcl-2 (to protect cells from apoptosis) failed to produce DRAL expressing clones (results not shown). Therefore, DRAL might participate in the apoptotic response following activation of p53. Very recently, another LIM-only protein was described to induce apoptosis in myoblasts upon ectopic expression, namely the paxillin homologue Hic-5 (Hu et al. 1999), which is an LIM-only protein containing four LIM domains. Hic-5 is also known to bind to cell adhesion kinase β, which can induce apoptosis in fibroblasts (Xiong and Parsons 1997) and probably other cell types. Remarkably, like DRAL, Hic-5 levels are downregulated in transformed cells compared with normal cells (Shibanuma et al. 1994). Since a number of other LIM proteins are downregulated in transformed cells, such as Ril (Kiess et al. 1995) and Zyxin (Schenker and Trueb 1998), DRAL might not be the only LIM domain protein to play a suppressive role in tumor development.
To begin to investigate the mechanisms by which ectopic expression of DRAL can induce apoptosis, its intracellular localization was determined. Staining of RD-tsp53 cells with the polyclonal DRAL antibody at the permissive temperature provided the first evidence for localization of DRAL at focal contacts (data not shown). Later, DRAL was found in focal contacts, the cytoplasm, nucleus as well as the Z-discs and the M-band of cardiac myofibrils. These localizations were observed both by staining of the endogenous DRAL protein or staining of an ectopically expressed tagged version of the protein. As a further confirmation, a recently carried out yeast two-hybrid screen using DRAL as a bait identified several transcription factors, myofibrilar proteins and focal contact proteins as potential interaction partners (data not shown). These potential interactions are currently being analyzed in more detail.
Focal contacts are known to establish a transmembrane linkage between the actin cytoskeleton and the extracellular matrix and are implicated in a number of signaling pathways. Some of the proteins at these specialized sites (e.g., focal adhesion kinase, paxillin) serve as scaffolding molecules and can act as signaling centers by providing docking sites for other proteins (Burridge and Chrzanowska-Wodnicka 1996). Intriguingly, several of these scaffolding molecules are characterized LIM domain proteins, such as Hic-5, paxillin, PINCH, and zyxin (Salgia et al. 1995; Beckerle 1997; Hagmann et al. 1998; Tu et al. 1999). Hence, DRAL might fulfil a similar function at this site.
On the other hand, nuclear localization of DRAL might also be responsible or contribute to its pro-apoptotic function. Interestingly, dual localization in focal contacts and the nucleus is known for a number of other protein, e.g., zyxin, Hic-5, and β-catenin, a multi-functional protein activated by the Wnt signaling pathway (Miller et al. 1999). Whether DRAL localization could be influenced by external signals in a manner analogous to β-catenin is currently not known.
The Z-disc localization of DRAL is shared with a second LIM domain protein, MLP (Arber et al. 1997), whose expression seems to be very important for the structural organization of myofibrils, since mice deficient in this protein show a disrupted cytoarchitecture leading to dilated cardiomyopathy and finally heart failure. A possible function of DRAL in this respect will have to be investigated. MLP also shares nuclear expression with DRAL, but only MLP is associated with the actin filaments. Hence, the two proteins might have some overlapping, but not completely redundant functions.
Although the knowledge of the function of FHL proteins is still very limited, they may act either as scaffolding molecules to link several proteins and thus activating distinct signaling pathways or as modulators of transcription by complexing transcription factors (Chan et al. 2000; Muller et al. 2000) or both. The possible role of DRAL as a p53-dependent class II tumor suppressor molecule might involve one of these mechanisms.
Acknowledgments
We are very grateful to Dr. Sacchi (Molecular Oncogenesis Laboratory, Regina Elena Cancer Institute, Rome, Italy) for kindly providing RD-tsp53 and RD-Neo cells, and to Dr. M. Pruschy for his help with the γ-irradiation experiments including the p53+/+, p53−/− mouse fibroblasts. We thank Prof. C. W. Heizmann and Prof. F. E. Würgler for their support, Prof. J.C. Perriard (Institute of Cell Biology, ETH Zürich, Zürich, Switzerland) for making the confocal microscope available for these studies and Dr. A. Lauber-Biason for critical reading of the manuscript.
This work was sponsored by grants from the Krebsforschung Schweiz, the Krebsliga of the Kt. Zug, and the Swiss National Science Foundation (31-46886.96 and 31-56869.99).
Footnotes
Abbreviations used in this paper: BAP, bacterial alkaline phosphatase; DRAL, down-regulated in rhabdomyosarcoma LIM protein; DRAL-CF, COOH-terminally FLAG-tagged DRAL; DRAL-NF, NH2-terminally FLAG-tagged DRAL; FHL, four and a half LIM domain; IR, ionizing radiation; MEF-2, myocyte enhancer binding factor 2; RMS, rhabdomyosarcoma.
References
- Amundson S.A., Myers T.G., Fornace A.J. Roles for p53 in growth arrest and apoptosisputting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–3299. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- Arber S., Hunter J.J., Ross J., Jr., Hongo M., Sansig G., Borg J., Perriard J.C., Chien K.R., Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Attardi L.D., Reczek E.E., Cosmas C., Demicco E.G., McCurrach M.E., Lowe S.W., Jacks T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- Auerbach D., Bantle S., Keller S., Hinderling V., Leu M., Ehler E., Perriard J.C. Different domains of the M-band protein myomesin are involved in myosin binding and M-band targeting. Mol. Biol. Cell. 1999;10:1297–1308. doi: 10.1091/mbc.10.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler M., Moser H., Eppenberger H.M., Wallimann T. Heart C-protein is transiently expressed during skeletal muscle development in the embryo, but persists in cultured myogenic cells. Dev. Biol. 1985;112:345–352. doi: 10.1016/0012-1606(85)90405-1. [DOI] [PubMed] [Google Scholar]
- Beckerle M.C. Zyxinzinc fingers at sites of cell adhesion. Bioessays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Bernasconi M., Remppis A., Fredericks W.J., Rauscher F.J., 3rd, Schafer B.W. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc. Natl. Acad. Sci. USA. 1996;93:13164–13169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.L., Olson E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Buckbinder L., Talbott R., Velasco-Miguel S., Takenaka I., Faha B., Seizinger B.R., Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Burridge K., Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- Chan K.K., Tsui S.K., Ngai S.M., Lee S.M., Kotaka M., Waye M.M., Lee C.Y., Fung K.P. Protein-protein interaction of FHL2, a LIM domain protein preferentially expressed in human heart, with hCDC47. J. Cell. Biochem. 2000;76:499–508. [PubMed] [Google Scholar]
- Dameron K.M., Volpert O.V., Tainsky M.A., Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- Dawid I.B., Breen J.J., Toyama R. LIM domainsmultiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- De Giovanni C., Nanni P., Sacchi A., Soddu S., Manni I., G.D'Orazi S., Bulfone-Paus T.Pohl, Landuzzi L., Nicoletti G. Wild-type p53-mediated down-modulation of interleukin 15 and interleukin 15 receptors in human rhabdomyosarcoma cells. Br. J. Cancer. 1998;78:1541–1546. doi: 10.1038/bjc.1998.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry W.S. The p53 pathway and cancer therapy. Cancer J. 1998;11:229–235. [Google Scholar]
- El-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Felix C.A., Kappel C.C., Mitsudomi T., Nau M.M., Tsokos M.T., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J. Frequency and diversity of p53 mutations in childhood rhabdomysarcoma. Cancer Res. 1992;52:2243–2247. [PubMed] [Google Scholar]
- Fimia G.M., De Cesare D., Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- Genini M., Schwalbe P., Scholl F.A., Schäfer B.W. Isolation of genes differentially expressed in human primary myoblasts and embryonal rhabdomyosarcoma. Int. J. Cancer. 1996;66:571–577. doi: 10.1002/(SICI)1097-0215(19960516)66:4<571::AID-IJC24>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Genini M., Schwalbe P., Scholl F.A., Remppis A., Mattei M.-G., Schäfer B.W. Substractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol. 1997;16:433–442. doi: 10.1089/dna.1997.16.433. [DOI] [PubMed] [Google Scholar]
- Greene W.K., Baker E., Rabbitts T.H., Kees U.R. Genomic structure, tissue expression and chromosomal location of the LIM-only gene, SLIM1. Gene. 1999;232:203–207. doi: 10.1016/s0378-1119(99)00125-0. [DOI] [PubMed] [Google Scholar]
- Grove B.K., Kurer V., Lehner C., Doetschman T.C., Perriard J.C., Eppenberger H.M. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J. Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Flemington C., Chittenden T., Zambetti G.P. ei24, a p53 response gene involved in growth suppression and apoptosis. Mol. Cell Biol. 2000;20:233–241. doi: 10.1128/mcb.20.1.233-241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann J., Grob M., Welman A., van Willigen G., Burger M.M. Recruitment of the LIM protein hic-5 to focal contacts of human platelets. J. Cell Sci. 1998;111:2181–2188. doi: 10.1242/jcs.111.15.2181. [DOI] [PubMed] [Google Scholar]
- Harris C.C. Structure and function of the p53 tumor suppressor geneclues for rational cancer therapeutic strategies. J. Natl. Cancer Inst. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- Hu Y., Cascone P.J., Cheng L., Sun D., Nambu J.R., Schwartz L.M. Lepidopteran DALP, and its mammalian ortholog HIC-5, function as negative regulators of muscle differentiation. Proc. Natl. Acad. Sci. USA. 1999;96:10218–10223. doi: 10.1073/pnas.96.18.10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli D., Tessler E., Haupt Y., Elkeles A., Wilder S., Amson R., Telerman A., Oren M. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiess M., Scharm B., Aguzzi A., Hajnal A., Klemenz R., Schwarte-Waldhoff I., Schafer R. Expression of ril, a novel LIM domain gene, is down-regulated in Hras-transformed cells and restored in phenotypic revertants. Oncogene. 1995;10:61–68. [PubMed] [Google Scholar]
- Messerli J.M., Van der Voort H.T.M., Rungger-Brändle E., Perriard J.C. Three-dimensional visualization of multi-channel voxel data. Cytometry. 1993;14:725–735. doi: 10.1002/cyto.990140705. [DOI] [PubMed] [Google Scholar]
- Miller J.R., Hocking A.M., Brown J.D., Moon R.T. Mechanism and function of signal transduction by the Wnt/β-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- Miyashita T., Reed J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Miyashita T., Harigai M., Hanada M., Reed J.C. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
- Morgan M.J., Madgwick A.J. The fourth member of the FHL family of LIM proteins is expressed exclusively in the testis. Biochem. Biophys. Res. Commun. 1999;255:251–255. doi: 10.1006/bbrc.1999.0180. [DOI] [PubMed] [Google Scholar]
- Morgan M.J., Madgwick A.J., Charleston B., Pell J.M., Loughna P.T. The developmental regulation of a novel muscle LIM-protein. Biochem. Biophys. Res. Commun. 1995;212:840–846. doi: 10.1006/bbrc.1995.2045. [DOI] [PubMed] [Google Scholar]
- Muller J.M., Isele U., Metzger E., Rempel A., Moser M., Pscherer A., Breyer T., Holubarsch C., Buettner R., Schule R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Hinman A., Levine A.J. Wild-type p53 negatively regulates the expression of a microtubule- associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Owen-Schaub L.B., Zhang W., Cusack J.C., Angelo L.S., Santee S.M., Fujiwara T., Roth J.A., Deisseroth A.B., Zhang W.W., Kruzel E., Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol. Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Xia Y., Zweier J.L., Kinzler K.W., Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Prisco M., Hongo A., Rizzo M.G., Sacchi A., Baserga R. The insulin-like growth factor I receptor as a physiologically relevant target of p53 in apoptosis caused by interleukin-3 withdrawal. Mol. Cell Biol. 1997;17:1084–1092. doi: 10.1128/mcb.17.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruschy M., Resch H., Shi Y.Q., Aalame N., Glanzmann C., Bodis S. Ceramide triggers p53-dependent apoptosis in genetically defined fibrosarcoma tumour cells. Br. J. Cancer. 1999;80:693–698. doi: 10.1038/sj.bjc.6690411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lozano P., Hixon M.L., Wagner M.W., Flores A.I., Ikawa S., Baldwin A.S., Jr., Chien K.R., Gualberto A. p53 is a transcriptional activator of the muscle-specific phosphoglycerate mutase gene and contributes in vivo to the control of its cardiac expression. Cell Growth Differ. 1999;10:295–306. [PubMed] [Google Scholar]
- Salgia R., Li J.L., Lo S.H., Brunkhorst B., Kansas G.S., Sobhany E.S., Sun Y., Pisick E., Hallek M., Ernst T. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL. J. Biol. Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia I., Rabbitts T.H. The LIM domaina new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Schenker T., Trueb B. Down-regulated proteins of mesenchymal tumor cells. Exp. Cell Res. 1998;239:161–168. doi: 10.1006/excr.1997.3896. [DOI] [PubMed] [Google Scholar]
- Schmeichel K.L., Beckerle M.C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schwartz R.J., Olson E.N. Building the heart piece by piecemodularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- Sheikh M.S., Burns T.F., Huang Y., Wu G.S., Amundson S., Brooks K.S., Fornace A.J., Jr., El-Deiry W.S. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- Sheikh M.S., Huang Y., Fernandez-Salas E.A., El-Deiry W.S., Friess H., Amundson S., Yin J., Meltzer S. J., Holbrook N.J., Fornace A.J., Jr. The antiapoptotic decoy receptor TRID/TRAIL-R3 is a p53-regulated DNA damage-inducible gene that is overexpressed in primary tumors of the gastrointestinal tract. Oncogene. 1999;18:4153–4159. doi: 10.1038/sj.onc.1202763. [DOI] [PubMed] [Google Scholar]
- Shibanuma M., Mashimo J., Kuroki T., Nose K. Characterization of the TGF beta 1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J. Biol. Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokino T., Thiagalingam S., El-Deiry W.S., Waldman T., Kinzler K.W., Vogelstein B. p53 tagged sites from human genomic DNA. Hum. Mol. Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- Tu Y., Li F., Goicoechea S., Wu C. The LIM-only protein PINCH directly interacts with integrin-linked kinase and Is recruited to integrin-rich sites in spreading cells. Mol. Cell Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.S., Burns T.F., McDonald E.R., 3rd, Jiang W., Meng R., Krantz I.D., Kao G., Gan D.D., Zhou J.Y., Muschel R. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- Xiong W., Parsons J.T. Induction of apoptosis after expression of PYK2, a tyrosine kinase structurally related to focal adhesion kinase. J. Cell Biol. 1997;139:529–539. doi: 10.1083/jcb.139.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]