Abstract
Previously, we reported that chromosomes contain a giant filamentous protein, which we identified as titin, a component of muscle sarcomeres. Here, we report the sequence of the entire titin gene in Drosophila melanogaster, D-Titin, and show that it encodes a two-megadalton protein with significant colinear homology to the NH2-terminal half of vertebrate titin. Mutations in D-Titin cause chromosome undercondensation, chromosome breakage, loss of diploidy, and premature sister chromatid separation. Additionally, D-Titin mutants have defects in myoblast fusion and muscle organization. The phenotypes of the D-Titin mutants suggest parallel roles for titin in both muscle and chromosome structure and elasticity, and provide new insight into chromosome structure.
Keywords: chromosome condensation, Drosophila melanogaster, myoblast fusion, sarcomere, titin
Introduction
Chromosomes are highly ordered, elastic structures that maintain their integrity throughout the physically strenuous process of cell division. Each human chromosome contains a single DNA molecule of 50–250 × 106 base pairs, which would extend from 1.7–8.5 cm if uncoiled (Alberts et al. 1994). Chromosomal DNA must be compacted to fit inside the cell nucleus, which measures only ∼5–20 μm. Chromosomes are further compacted by 5–10-fold during mitosis, reaching an overall 10,000-fold compaction, ensuring that tangles between sister chromatids or between neighboring nonhomologous chromosomes are eliminated and that chromosomes do not get trapped in the cleavage furrow during cytokinesis. Compaction is not random, since each mitotic chromosome within a given cell type has a fixed axial diameter and length, and each chromosome gives a reproducible banding pattern when stained with DNA dyes. Fluorescence in situ hybridization (FISH) experiments indicate that specific DNA probes localize to reproducible positions on the chromosomes (for review see Koshland and Strunnikov 1996).
Several models have been proposed for the structure of mitotic chromosomes, including the formation of a simple solenoid structure (Thoma et al. 1979), the formation of a cross-linked unorganized gel (McDowall et al. 1986), a hierarchical folding of chromatin (Sedat and Manuelidis 1978; Manuelidis 1990), and the attachment of chromatin loops to a central scaffold (Paulson and Laemmli 1977; Gasser et al. 1986; Boy de la Tour and Laemmli 1988). A new model for chromosome structure has been proposed based on elasticity measurements, specifically the longitudinal deformability and bending rigidity of whole chromosomes. This model proposes that the condensed mitotic chromosome is formed by one or a few thin rigid elastic axes, built of titin-like molecules, surrounded by a soft envelope of chromatin (Houchmandzadeh and Dimitrov 1999).
To fully understand mitotic chromosome structure, it is necessary to identify the protein components of the chromosome and to learn the role each protein plays in chromosome architecture. Several proteins required for chromosome condensation have been identified, including histones, topoisomerase II (topo II), and the structural maintenance of chromosomes (SMCs). The SMCs comprise the condensin and cohesin complexes, which mediate condensation and sister chromatid cohesion, respectively (for review see Koshland and Strunnikov 1996; Warburton and Earnshaw 1997; Hirano 1999; Strunnikov and Jessberger 1999).
We identified titin as a chromosomal protein in both human cells and Drosophila embryos (Machado et al. 1998). Titin, a protein more generally known for its structural and elastic roles in muscle, is the largest protein so far identified; single titin molecules span half a sarcomere, with their amino and carboxy termini anchored in the Z-disc and M-line, respectively (for review see Gregorio et al. 1999). Vertebrate titins are highly modular, comprised mostly of Ig-like and fibronectin type 3 (FN3)-like domains arranged in tandem (Trinick and Tskhovrebova 1999). Titins also contain an elastic region rich in proline (P), glutamic acid (E), valine (V), and lysine (K) residues, termed the PEVK domain, and a carboxy-terminal serine kinase domain. Titin transcripts are differentially spliced, particularly in regions encoding the tandem-Ig and PEVK domains, giving rise to many isoforms with different extensible properties (Labeit and Kolmerer 1995). Titin is responsible for the elasticity of striated muscle and is believed to function as a molecular scaffold specifying the correct assembly of myofibrils (for review see Gregorio et al. 1999; Trinick and Tskhovrebova 1999). The elastic properties of purified titin correspond well to the elastic properties of chromosomes from living cells and chromosomes assembled in vitro (Houchmandzadeh et al. 1997; Houchmandzadeh and Dimitrov 1999). Here, we show that mutations in the Drosophila titin gene, D-Titin, result in both muscle and chromosomal defects, suggesting that titin provides an elastic structural scaffold for both muscles and chromosomes.
Materials and Methods
Fly Stocks
Genetic abbreviations are used according to Lindsley and Zimm 1992. Five dre8 ethyl methanesulphonate (EMS)-induced alleles (Titin through Titin 5) and four dre8 γ-ray–generated alleles (Titin 6 through Titin 9) were provided by T. Sliter (University of Texas Southwestern, Dallas, TX; Sliter et al. 1989); the sallimus allele of Titin (D-Titin 14) was provided by J. Kennison (National Institutes of Health, Bethesda, MD); the P-element insertions l(3)4860 and l(3)6265 were provided by A. Spradling (Carnegie Institute, Baltimore, MD); the lethal P-element insertion l(3)j1D7 (Titin 10) was generated in the laboratory of H. Jan (University of California, San Francisco, San Francisco, CA) and was provided by the Bloomington Stock Center. The Df(3L)Aprt deficiencies were provided by J. Mason (NIEHS, Research Triangle Park, NC; Wang et al. 1994).
Excisional mutagenesis was carried out as described in Hamilton and Zinn 1994 with two P-element insertions in D-Titin: the l(3)j1D7/Titin10 insertion allele and the viable P-element insert, l(3)4860. We obtained both lethal (28 lines) and viable (16 lines) independent w − excision derivatives of l(3)j1D7/Titin10 and concluded that the P-element insertion was the cause of the loss-of-D-Titin function in the Titin10 stock. We obtained both lethal (eight lines) and viable (six lines) independent ry − and ry + excision derivatives of l(3)4860.
Molecular Biology
Plasmid, phage, and genomic DNA isolation, the labeling of radioactive probes and polymerase chain reactions were performed as described in Maniatis et al. 1989. Drosophila genomic DNA flanking the P-element inserts l(3)04860 and l(3)j1D7 was isolated by plasmid rescue as described in Hamilton and Zinn 1994. Quantitative Southern blots were used, in addition to polytene chromosome in situ of Df(3L)Aprt deficiencies (see below), to map the deficiency breakpoints within the 62B-C region. D-Titin clones were hybridized to Southern blots of EcoRI- and SalI-restricted genomic DNA from flies heterozygous to all of the Df(3L)Aprt deficiencies (data not shown). Southern blots were also used to map DNA polymorphisms associated with lesions in D-Titin. Since D-Titin alleles 1 through 9 were generated on the same parental third chromosome (Sliter et al. 1989), each allele serves as an internal control. Bands corresponding to the balancer chromosomes were identified by including on each gel a lane of DNA isolated from a Df(3L)Aprt143/Balancer stock that was digested with the appropriate restriction enzyme (either EcoRI, SalI, BamHI, or HindIII). GH05716, CK340, and CK55 are expressed sequence tag clones from The Berkeley Drosophila Genome Project (BDGP). DNA sequencing was performed by the DNA Analysis Core Facility at Johns Hopkins University. The DNA sequences linking the KZ cDNA with a ket cDNA (X72709) and the ket cDNA with the CK340 cDNA were obtained by PCR amplification using standard conditions, the PCR Supermix kit (Life Technologies) and the following pairs of primers, respectively: 5′-GGGGGAATTCCCAAGTAACTGCTGATC-3′ and 5′-GGGGCTCGAGCCTCAAAGTGCACAGC-3′; 5′- GGGGCTCGAGTCTAAGGTGCCGAATGC-3′ and 5′-ACATCAACGATCTGGGTG-3′. Sequence analyses were performed using the DNA Strider and Gene Finder program. Homology and motif searches were performed using the BLAST and Motif Programs at NCBI. The GenBank/EMBL/DDBJ accession numbers are as follows: D-Titin ORF AF241652; GH05716 cDNA; CK340 cDNA AF241648; CK55 cDNA AF241649; PR4860 AF241650; and PRj1D7 AF241651.
Polytene Chromosome In Situ Hybridization
Polytene chromosome in situs were done by hybridizing biotin-labeled D-Titin genomic and cDNA clones to fixed salivary gland polytene chromosomes from larvae heterozygous for a Df(3L)Aprt deficiency chromosome and a “wild-type” balancer chromosome, as described by Pardue 1994, omitting the RNase treatment and acetylation steps and using the Vectastain Kit (Vector Laboratories) for HRP signal detection. Every cDNA and genomic clone described as being part of the D-Titin gene were localized to the same genomic interval using the deficiency chromosomes.
Immunostaining of Embryos
Embryo fixation and staining were performed as described in Reuter et al. 1990. Primary antibodies were used as follows: rat polyclonal α-D-Titin-KZ (1:5,000; Machado et al. 1998), rat monoclonal α-KET3 (1:200; MAC155; Lakey et al. 1993), and rabbit α-MHC (1:500; Kiehart and Feghali 1986). Antibody-stained embryos were visualized and photographed using Nomarski optics on a Zeiss Axiophot microscope. Royal Gold ASA 100 print film was used for photography.
Cytological Analysis of Mitotic Chromosomes
The cytology of mitotic chromosomes was investigated in larval neuroblasts. Squashed third instar larval brains stained with aceto-orcein were prepared as described in Gatti and Goldberg 1991, with incubation of the brains in colchicine and treatment in hypotonic solution. Untreated mitotic chromosomes were prepared for Hoechst staining as described in Gatti et al. 1994. Mitotic chromosome preparations were scored and photographed on a Zeiss Axiophot microscope, under phase contrast. Kodak PanFilm-ESTAR-AH or Kodak Ektachrome 400 were used for photography.
Results
D-Titin Encodes a Two-Megadalton Protein
In previous work, we showed that D-TITIN (a) has significant homology to vertebrate titins, (b) is expressed in all striated muscle, (c) localizes to chromosomes and to sarcomeres, and (d) migrates as a megadalton (MDa)-sized polypeptide on SDS-PAGE (Machado et al. 1998). We reported a partial sequence of the D-Titin gene encoding 1608 residues, corresponding to a small fraction of the entire open reading frame (ORF). We also mapped the gene to cytological region 62C1-2.
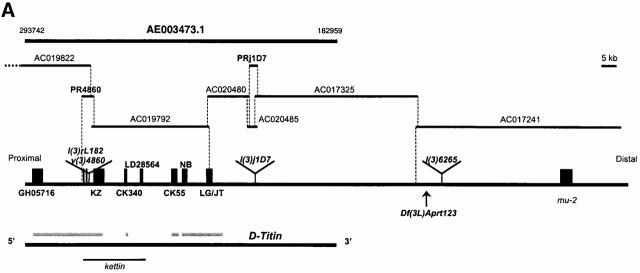
The Berkeley Drosophila Genome Project (BDGP) and Celera Genomics recently released sequence from the 62C region that allowed us to assemble a 290-kb contig that includes the entire D-Titin gene (Fig. 1 A). This large contig is composed of seven overlapping contigs that span the ∼110 kb of genomic DNA corresponding to the D-Titin gene, plus almost 100 kb of sequence both upstream and downstream of D-Titin. Our assembly of these contigs has been confirmed by Celera Genomics; D-Titin spans position 293742 through 182959 in Celera contig AE003473.1 (Table ). We determined the orientation of D-Titin on the chromosome; the 5′ end of D-Titin is closer to the centromere (proximal) and the 3′ end is closer to the telomere (distal). We mapped the proximal breakpoint of the deficiency Df(3L)Aprt123 to the most 3′ clone in the series (contig AC017241) (Fig. 1 A). Df(3L)Aprt123 genetically defines the boundary between the D-Titin gene (see below) and its nearest known distal neighbor, mutator2 (mu2) (Kasravi et al. 1999). Consistent with our mapping, the mu2 sequence is included in the AC017241 contig. We also independently obtained genomic DNA sequence linking several previously identified D-Titin fragments using a PCR-based cloning strategy (see Materials and Methods).
Figure 1.
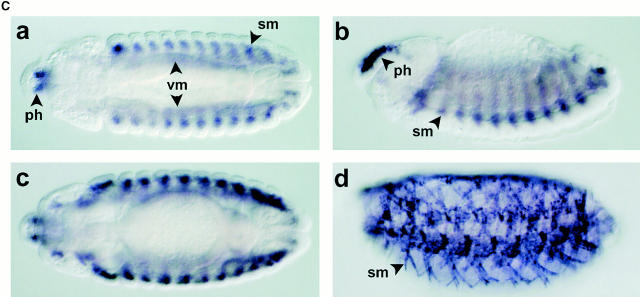
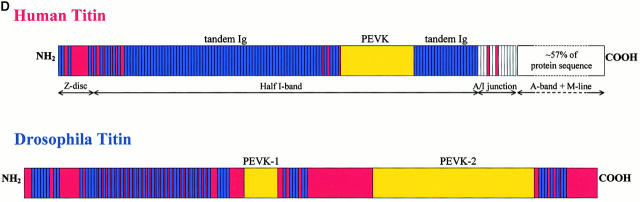
Molecular characterization of the D-Titin gene. (A) Molecular map of the region in 62C containing D-Titin. A series of seven overlapping contigs span the entire D-Titin gene and link previously isolated D-Titin clones (KZ, NB, LG, JT) and four new D-Titin cDNAs (GH05716, CK340, CK55, LD28564). D-Titin spans from nucleotide (nt) 293742 (5′) to nt 182959 (3′) in Celera contig AE003473.1. The ORF in the LG clone was completely contained within the large contig, however; LG is a chimeric genomic clone that also includes noncoding sequences from cytological region 62B. The kettin sequence (Hakeda et al. 2000; Kolmerer et al. 2000) is entirely included within the D-Titin gene. Three P-element insertions in D-Titin, l(3)rL182, v(3)04860, and l(3)j1D7, are indicated, as well as the flanking DNA, isolated by plasmid rescue (PR4860 and PRj1D7). l(3)6265 is a P-element insertion used to clone flanking genomic DNA and map the proximal breakpoint of Df(3L)Aprt123. Df(3L)Aprt123 deletes the neighboring distal gene (mu2) but does not affect D-Titin function. (B) Protein expression profiles detected by an antibody against D-TITIN (α-KZ) (a–f) and an antibody against KET (Lakey et al. 1993) (g–l). Identical expression is detected in all the somatic (sm), visceral (vm), and pharyngeal muscles (ph) and their precursors during embryogenesis with both antibodies. (a and g) Stage 11 embryos, ventral view. (b and h) Stage 13 embryos, ventral view. (c and i) Stage 16 embryos, ventral view. (d and j) Stage 11 embryos, lateral view. (e and k) Stage 13 embryos, lateral view. (f and l) Stage 16 embryos, lateral view. All embryos are oriented with anterior to the left. For embryos shown in lateral view, dorsal is up. (C) D-Titin transcripts detected by whole-mount in situ hybridization. Genomic and cDNA clones mapping throughout the D-Titin gene (see gray lines in A) detected identical expression patterns throughout embryogenesis in all striated muscles and their precursors. Shown in a–d are embryos hybridized with the JT cDNA, which encodes a portion of the PEVK-2 domain. (a) ventral view of stage 13 embryo; (b) lateral view of stage 14 embryo; (c) ventral view of early stage 15 embryo; (d) lateral view of stage 16 embryo. (D) Domain structure and sarcomeric layout of the Z-disc and I-band region of human elastic (soleus) titin and predicted alignment with the Drosophila TITIN protein. Immunoglobulin-like domains (blue), interdomain sequences (red), and the FN3 domains (white) are shown. The single elastic PEVK domain (yellow) of the human titin consists of 70% proline (P), glutamic acid (E), valine (V), and lysine (K). PEVK-1 (yellow) of D-TITIN (1,240 residues) consists of 58.5% P, E, V, and K; PEVK-2 (yellow) of D-TITIN (5,065 residues) consists of 52.4% P, E, V, and K.
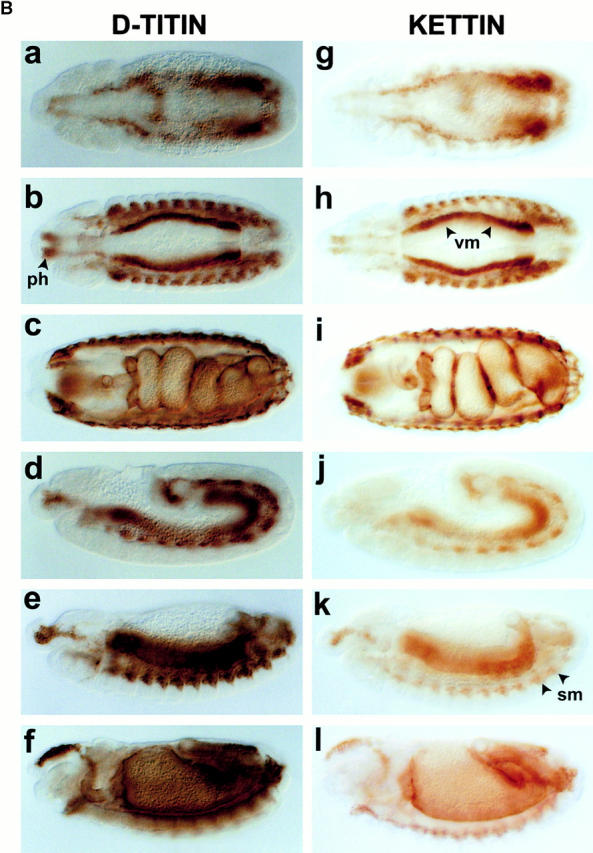
Table 1.
D-Titin Gene and Protein Structure
| Exon # (nt) | Position in genomic sequence | cDNAs | Amino acids | Domains |
|---|---|---|---|---|
| 1 (1–1249) | AE003473.1 (293742–292494) | LP06352 | 1–416 | Ig 1 |
| LP06486 | Ig 2 (N) | |||
| GH05716 | ||||
| 2 (1250–1410) | AE003473.1 (292429–292269) | GH05716 | 417–470 | Ig 2 (C) Ig 3 (N) |
| 3 (1411–3202) | AE003473.1 (290753–288962) | GH05716 HL07373 GH09781 | 471–1067 | Ig 3 (C) Ig 4–7 |
| 4 (3203–3409) | AE003473.1 (287955–287749) | GH05716G H09781 | 1068–1136 | Ig 8 (N) |
| 5 (3410–3585) | AE003473.1 (287398–287223) | GH05716 | 1137–1195 | Ig 8 (C) |
| 6 (3586–5347) | AE003473.1 (286929–285168) | 1196–1781 | ||
| KZ exon 1 | AE003473.1 (273780–273–569) | KZ | ||
| KZ exon 2 | AE003473.1 (272635–272295) AE003473.1 (267011) AE003473.1 (266789) | KZ Insertion site of lethal P-element-—l(3)rL182 Insertion site of viable P-element—v(3)04860 | ||
| 7 (5348–6931) (=KZ exon 3) | AE003473.1 (265929–264246) | KZ | 1782–2310 | Ig 9–12 |
| 8 (6932—8260) | AE003473.1 (263987–262659) | KZ | 2311–2753 | Ig 13–16 |
| 9 (8261–11059) | AE003473.1 (262558–259760) | 2754–3686 | Ig 17–23 | |
| 10 (11060–16618) | AE003473.1 (259679–254121) | LP05016 LP03833 LD25812 CK340 | 3687–5539 | Ig 24–37 |
| 11 (16619–16906) | AE003473.1 (254055–253768) | 5540–5635 | Ig 38 (N) | |
| 12 (16907–17138) | AE003473.1 (253702–253471) | 5636–5713 | Ig 38 (C) | |
| 13 (17139–17310) | AE003473.1 (253374–253203) | 5714–5770 | Ig 39 (N) | |
| 14 (17311–17496) | AE003473.1 (253142–252957) | 5771–5832 | Ig 39 (C) | |
| 15 (17497–17846) | AE003473.1 (252163–251816) | 5833–5949 | ||
| 16 (17847–19158) | AE003473.1 (251754–250443) | LD28564 GM12455 | 5950–6386 | Ig 40–42 Ig 43 (N) |
| 17 (19159–24543) | AE003473.1 (246263–240879) | GH25930 CK00556 | 6387–8181 | Ig 43 (C) PEVK–1 Ig 44 |
| 18 (24544–25744) | AE003473.1 (240558–239395) | CK00556 CK55 | 8182–8581 | Ig 45–47 |
| 19 (25745–26805) | AE003473.1 (237157–236097) | NB | 8582–8935 | Ig 48–49 |
| 20 (26806–32470) | AE003473.1 (234931–229267) | 8936–10823 | ||
| 21 (32471–34180) | AE003473.1 (226335–224626) | JT | 10824–11393 | PEVK–2 |
| 22 (34181–41905) | AE003473.1 (223703–215979) | GM05288 GM05521 | 11394–13968 | PEVK–2 |
| 23 (41906–42619) | AE003473.1 (215460–214827) | 13969–14206 | PEVK–2 | |
| 24 (42620–47748) | AE003473.1 (214168–209036) | 14207–14903 | PEVK–2 | |
| AE003473.1 (209170) | Insertion site of lethal P-element—l(3)j1D7 | |||
| 25 (47749–48016) | AE003473.1 (208697–208430) | 14904–15916 | PEVK–2 | |
| 26 (48017–48736) | AE003473.1 (208087–207368) | 15917–16005 | ||
| 27 (48737–50316) | AE003473.1 (207288–205705) | 16006–16245 | Ig 50 Ig 51 (N) | |
| 28 (50317–50932) | AE003473.1 (205088–204473) | 16246–16772 | Ig 51 (C) Ig 52–54 Ig 55 (N) | |
| 29 (50933-51353) | AE003473.1 (200121-199697) | 16773–16977 | Ig 55 (C) Ig 56 | |
| 30 (51354-52611) | AE003473.1 (188467-184577) | 16978–17117 | ||
| 31 (52612-53709) | AE003473.1 (184055-182959) | 17118–17903 | ||
Column 1 indicates exons included in longest putative D-Titin transcript. KZ exons 1/2 correspond to the 5′ end of an alternative D-Titin splice form. Column 2 indicates the position of each exon relative to the Celera Contig in the region. Column 3 includes cDNAs that correspond to one or more D-Titin exon(s). Column 4 indicates the amino acids encoded by each exon. Column 5 indicates where the 56 immunoglobulin-like domains (Ig) and the two PEVK-rich regions are found in the protein. N, NH2-terminal; C, COOH-terminal. The insertion sites of both the viable and lethal P-element insertions are indicated.
To identify sequences corresponding to D-Titin within the large contig, we searched for ORFs that were both larger than 150 bp and flanked by consensus splice acceptor/donor sequences for Drosophila (Mount 1982; Mount et al. 1992). The conceptual translation of such sequences was used to search the GenBank/EMBL/DDBJ database, using the BLASTp program (Altschul et al. 1990). Through this analysis, we identified 31 exons that, when translated, showed high homology to all titin family members for which sequence is available (Table and Fig. 1 A). The total number of residues predicted to be encoded by the putative D-Titin exons is 17903, which would correspond to a 1.9–2-MDa polypeptide. This size is consistent with that revealed by immunoblots incubated with either of two antibodies against distinct D-TITIN domains: α-KZ, which was raised against a D-TITIN fragment spanning residues 1784–2411, and α-LG, which was raised against residues 10879–11042 (Machado et al. 1998). Our predicted D-Titin gene organization is largely consistent with that determined using the Gene Finder program (Smith et al. 1996). We identified 19 D-Titin cDNAs, 6 of which (GH05716, KZ, CK340, CK55, NB, and JT) were completely sequenced either in this or previous work (Machado et al. 1998); only 5′ and/or 3′ end-run sequence is available for the remaining 13 cDNAs. Several of the cDNAs spanned more than one exon, and corresponded to exons not predicted by the Gene Finder program but that were included in our predicted sequence.
A BLAST search of the entire D-Titin ORF revealed significant homology to human elastic and cardiac titin (E values = e−169 and 3e−93, respectively), to chicken and rabbit skeletal titin (E values = e−111 and 2e−79, respectively), and to the Caenorhabditus elegans UNC-89 protein (E value = e−132). D-TITIN contains some, but not all, of the structural domains found in human titin, including 56 immunoglobulin-like repeats and two large domains rich in P, E, V, and K residues (PEVK-1 and PEVK-2; Table and Fig. 1 D). The most NH2-terminal region of D-TITIN is predicted to align with the most NH2-terminal region of vertebrate titin, which localizes to the Z-disc and binds to α-actinin (Linke et al. 1997; Trombitas and Granzier 1997; Trombitas et al. 1997). The large COOH-terminal portion of D-TITIN is most homologous to the region of vertebrate titin that localizes to the I-band. This homology and the relative arrangement of the Ig and PEVK domains suggest that the COOH-terminal portion of D-TITIN localizes to the I-band. The characteristic domains found in the A-band and M-line regions of vertebrate titin, which include FN3 repeats flanking additional Ig domains and the serine kinase domain, were not found in D-TITIN.
A sequence recently reported for a Drosophila kettin (ket) gene (Hakeda et al. 2000; Kolmerer et al. 2000) is contained entirely within a 20-kb region of the D-Titin gene (Fig. 1 A). KET was originally discovered as a Z-disc “mini-titin” in adult Drosophila flight muscle, and was proposed to function as a scaffolding protein in the Z-disc (Lakey et al. 1993). Our data suggest that KET is either a proteolytic cleavage product of D-TITIN, or is encoded by an alternative splice form of D-Titin. Antibodies directed against KET and D-TITIN revealed identical expression patterns in embryos (Fig. 1 B). Since only the larger MDa-sized polypeptide is detected on immunoblots of embryonic extracts incubated with two D-TITIN antibodies, one directed to a region shared with KET and another to the PEVK-2 region (Machado et al. 1998), we conclude that D-TITIN is the major protein form expressed during embryogenesis. We detected D-Titin transcripts by whole-mount in situ hybridization using genomic and cDNA fragments corresponding to exons that map both upstream and downstream of ket. The expression patterns were identical to those detected with probes from the region of overlap between D-Titin and ket (Fig. 1 C; Machado et al. 1998). This result suggests that the entire 110-kb D-Titin gene is transcribed during embryogenesis. The processed D-Titin mRNA is predicted to be at least 54 kb, not including potential 5′ and 3′ UTR sequences. Therefore, it is not surprising that we have been unable to detect D-Titin transcripts by standard Northern analysis (data not shown).
Identification of D-Titin Mutations
D-Titin maps to 62C1-2, a region for which a saturation mutagenesis has been done (Sliter et al. 1989; Wang et al. 1994). Through a combination of genomic Southerns and in situ hybridization to polytene chromosomes, we localized D-Titin to a chromosomal interval that contains only a single known gene, formerly known as dre8 (Fig. 2). dre8 mutations disrupt D-Titin function. In genomic Southerns with internal D-Titin probes, we detected DNA polymorphisms in two of the dre8 γ-ray alleles that were not present in other dre8 alleles or in a dre6 allele that were generated on the same parental chromosome (data not shown). The lethal P-element, l(3)j1D7, inserted at position 210591 in AE003473.1 into an exon encoding part of the PEVK-2 domain of D-TITIN, in a region that is downstream of, and not included in, the putative ket gene (Fig. 1 A and Table ). We showed that the lethality and disruption of D-Titin/dre8 function in the l(3)j1D7 stock is due to the P-element insertion, rather than a second independent mutation on the same chromosome, by excising the P-element and restoring viability and D-Titin/dre8 function, i.e., the viable excisions complemented an EMS-induced allele of D-Titin (D-Titin2). Finally, we generated three new D-Titin/dre8 alleles by excisional mutagenesis of a viable P-element insertion, l(3)4860, that had inserted into an intron near the 5′-region of the D-Titin gene (Fig. 1 A). Because the l(3)4860 excision alleles were unstable, we did not rely solely on the characterization of these lines to determine D-Titin function. Despite their instability, these mutants had DNA polymorphisms in the 5′ D-Titin coding region that were detectable by genomic Southern blotting (data not shown) and produced phenotypes that were indistinguishable from those of animals carrying stable D-Titin mutations (see below).
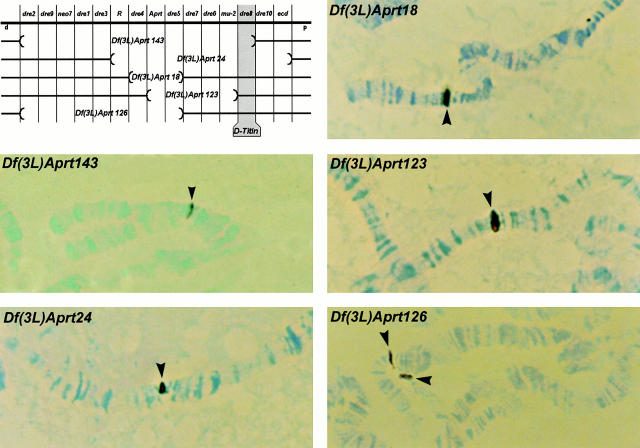
Figure 2.
Localization of D-Titin to a cytogenetic interval containing only a single known gene (dre8). The upper left panel is a map of cytological region 62B-C showing the known genes that have been mapped to the region and the deficiencies used to map D-Titin. The remaining panels show in situ hybridization to polytene chromosomes with biotinylated D-Titin genomic phage clone 5 (Machado et al. 1998). The polytene chromosomes are from larvae heterozygous for each deficiency. D-Titin is deleted in Df(3L)Aprt143 and Df(3L)Aprt24. D-Titin is not deleted in Df(3L)Aprt18, Df(3L)Aprt123, and Df(3L)Aprt126. Identical hybridization results were obtained using D-Titin clones that mapped throughout the D-Titin gene. (d, distal; p, proximal).
The colocalization of D-Titin DNA to the dre8 gene interval, together with the mapping of dre8 mutations and P-element insertions within the D-Titin coding region, indicate that dre8 mutations disrupt the D-Titin gene. This finding is consistent with results from Kolmerer et al. 2000, who showed that l(3)j1D7 fails to complement l(3)rL182, a P-element insertion at position 267011 in AE003473.1, which is within the kettin region (Hakeda et al. 2000; Kolmerer et al. 2000). l(3)rL182 also fails to complement the two dre8 alleles tested by Kolmerer et al. 2000. The finding that different D-Titin/dre8 mutations mapping over 56-kb apart (and mapping both inside and outside the ket contig) fail to complement indicates that the large ORF encoding 17903 residues corresponds to a single gene, D-Titin.
Loss of D-Titin Function Causes Embryonic Muscle Disorganization
We examined the muscles from all the D-Titin mutant alleles (Table II: D-Titin through D-Titin 13) by staining with two antisera: α-KZ, which is directed against an NH2-terminal region of D-TITIN and detects protein very early in myogenesis (Machado et al. 1998), and α-MHC, which is directed against muscle myosin heavy chain and detects protein at later stages (Kiehart and Feghali 1986). All except three of the D-Titin alleles expressed apparently normal levels of D-TITIN compared with their heterozygous siblings (Fig. 3); the three exceptions were the insertion allele l(3)j1D7/D-Titin 10, which showed reduced levels of protein, and two l(3)4860 excision alleles, D-Titin 12 and D-Titin 13, which were initially protein-null but several months later showed variable levels of expression. The levels of muscle myosin were normal in embryos homozygous for every D-Titin allele (data not shown). Consistent with previous studies showing nuclear D-TITIN throughout the cell cycle (Machado et al. 1998), the D-TITIN antisera detected protein in the cytoplasm and nuclei of muscle cells (Fig. 3), whereas MHC staining was excluded from nuclei (data not shown).
Figure 3.
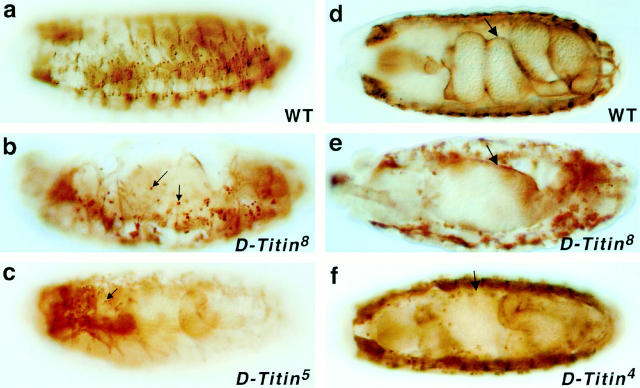
Muscle disorganization in D-Titin mutants. Myoblast fusion and gut defects in D-Titin mutants. Wild-type (a and d) and mutant (b, c, e, and f) embryos were stained with a D-TITIN antibody (α-KZ) (Machado et al. 1998). The mutations cause unfused myoblasts (b and c, arrows), disorganized muscle (b, c, e, and f), and gut morphogenesis defects (e and f, arrows). All embryos are oriented with anterior to the left. (a–c) are lateral views, dorsal is up. (d–f) are ventral views. (b and e) Titin8 homozygotes; (c) Titin5 homozygote; (f) Titin4 homozygote.
In stage 16 D-Titin mutant embryos, the muscles were very thin and spindly compared with WT muscles, and there were large populations of individual cells that stained for D-TITIN and muscle myosin that were not present in WT embryos (Fig. 3, a–c). Very similar phenotypes are observed in embryos mutant for myoblast city (mbc), blown fuse (blow), rolling stone (rost), and sticks and stones (sns); mutations affecting discrete steps in myoblast fusion (Paululat et al. 1995; Rushton et al. 1995; Doberstein et al. 1997, Erickson et al. 1997; Bour et al. 2000). Thus it appears that myoblast fusion is defective in D-Titin mutants. Indeed, transmission electron micrographs of stage 16 D-Titin mutant embryos revealed muscle cells with as few as one to three nuclei per cell whereas the corresponding muscle cells of WT embryos at a similar stage contained from 5 to 20 nuclei (data not shown). The other defect observed in the D-Titin mutants was a failure of the midgut constrictions to form (Fig. 3e and Fig. f). This phenotype is also observed in a fraction of embryos mutant for the mbc gene (Erickson et al. 1997).
D-Titin Mutations Severely Disrupt Chromosome Structure and Mitosis
The uniform distribution of titin along condensed mitotic chromosomes suggested a role for titin in chromosome structure and elasticity, similar to its proposed function in muscle (Machado et al. 1998). In Drosophila, almost all cell divisions that give rise to the embryo and larva occur in the first 2.5 h of development and depend on maternally supplied gene products. Consequently, many mitotic mutants die at the late larval–pupal transition, when pre-adult cells normally begin to replace larval tissues (Gatti and Baker 1989). Based on immunoblot analysis of 0–2 h total embryonic extracts, we know that D-Titin is supplied maternally (Machado et al. 1998). Thus, chromosome defects due to the loss of zygotic D-Titin will, most likely, be revealed only in late larval stages of development, specifically in the mitotically active tissues of the larva. Most mutations in D-Titin are early lethal, a likely result of the requirement for D-Titin in muscle development. However, three D-Titin alleles (titin 1, titin 8, and titin 11) survive until the larval–pupal transition (Sliter et al. 1989; Table ) allowing us to determine if D-Titin mutants have aberrant mitotic chromosome morphology and/or additional mitotic defects.
Table 2.
Complementation Tests Among dre8/Titin Alleles and Local Deficiencies
| Df (3L)Aprt 143 | Df (3L)Aprt 123 | Df (3L)Aprt 126 | Titin1(dre8e4) | Titin2(dre8e22) | Titin3(dre8e80) | Titin4(dre8e83) | Titin5(dre8e90) | Titin6(dre8g27) | Titin7(dre8g32) | Titin8(dre8g65) | Titin9(dre8g72) | Titin10(l(3)j1D7) | Titin11(110ry-) | Titin12(136ry-) | Titin13(136ry+) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Titin1 (dre8e4) | 139:0 | 80:34 | 100:54 | 283:0 | ||||||||||||
| Titin2 (dre8e22) | 117:0 | 90:23 | 101:48 | 165:0 | 150:0 | |||||||||||
| Titin3 (dre8e80) | 136:0 | 89:35 | 102:30 | 173:0 | 71:0 | 154:0 | ||||||||||
| Titin4 (dre8e83) | 92:0 | 81:48 | 96:30 | 109:1 | 84:0 | 111:26 | 89:0 | |||||||||
| Titin5 (dre8e90) | 98:0 | 69:33 | 67:25 | 107:0 | 80:0 | 135:0 | 120:1 | 45:0 | ||||||||
| Titin6 (dre8g27) | 121:0 | 46:32 | 108:38 | 131:0 | 151:0 | 117:0 | 42:0 | 117:0 | 115:0 | |||||||
| Titin7 (dre8g32) | 82:0 | 100:44 | 113:41 | 21:0 | 83:0 | 38:0 | 77:0 | 71:0 | 74:0 | 40:0 | ||||||
| Titin8 (dre8g65) | 96:0 | 70:31 | 80:19 | 29:0 | 96:0 | 46:0 | 89:0 | 52:0 | 93:32 | 39:0 | 26:0 | |||||
| Titin9 (dre8g72) | 84:0 | 81:33 | 115:55 | 52:3 | 125:0 | 135:0 | 48:0 | 155:0 | 92:0 | 46:20 | 64:0 | 80:1 | ||||
| Titin10 (l(3)j1D7) | 476:0 | 153:101 | 70:16 | 251:0 | 87:0 | 73:0 | 139:22 | 82:0 | 107:0 | 97:0 | 98:0 | 96:0 | 267:0 | |||
| Titin11 (110ry−) | 159:0 105:33 | -- 40:28 | -- 104:64 | -- 45:26 | 42:0 -- | -- 99:14 | 80:0 -- | 31:0 -- | 74:0 60:5 | -- 63:32 | -- 39:12 | 162:0 71:57 | 140:0 75:18 | 203:0 49:16 | ||
| Titin12 (136ry−) | 219:0 51:4 | -- 60:30 | --- 46:20 | -- 72:24 | -- 64:18 | -- -- | -- -- | -- -- | -- 87:20 | -- 45:36 | -- -- | 101:0 93:18 | 149:0 33:10 | 92:0 53:22 | 164:0 124:3 | |
| Titin13 (136ry+) | 94:0 41:14 | -- 29:19 | -- 106:48 | -- 109:30 | -- -- | -- -- | -- -- | -- -- | -- -- | -- 61:30 | -- -- | 53:0 86:22 | 59:0 43:22 | 86:0 89:11 | 88:0 55:28 | 82:0 66:47 |
| Titin14 (sls) | 97:0 | 88:48 | 63:20 | 124:0 | 92:0 | 119:0 | 125:0 | 187:0 | 111:0 | 42:0 | 95:0 | 92:0 | 116:0 | -- | -- | -- |
Titin through Titin 5 are EMS-induced alleles. Titin 6 through Titin 9 are γ-ray–induced alleles. Titin 10 is a lethal P-element insertion allele. Titin 11 through Titin 13 are unstable excision alleles of dre8/D-Titin derived from the viable P-element insertion l(3)4860, which are inserted into an intron of the D-Titin gene. Previous allele names are indicated in parentheses below the Titin allele designation. The excision alleles initially failed to complement two Titin alleles and Df(3L)Aprt143, a deficiency that removes Titin DNA (top numbers in rows 11–13). Complementation tests done several months later revealed that these unstable Titin alleles complemented all other tested Titin alleles and Df(3L)Aprt143 (bottom numbers in rows 11–13). The symbol “--” indicates crosses that were not done either when the excision mutants were first generated or several months later. Titin 14 is an EMS-induced allele of a gene also known as sallimus (sls), which was first identified as a dominant suppressor of Polycomb mutations (Kennison and Tamkun 1988).
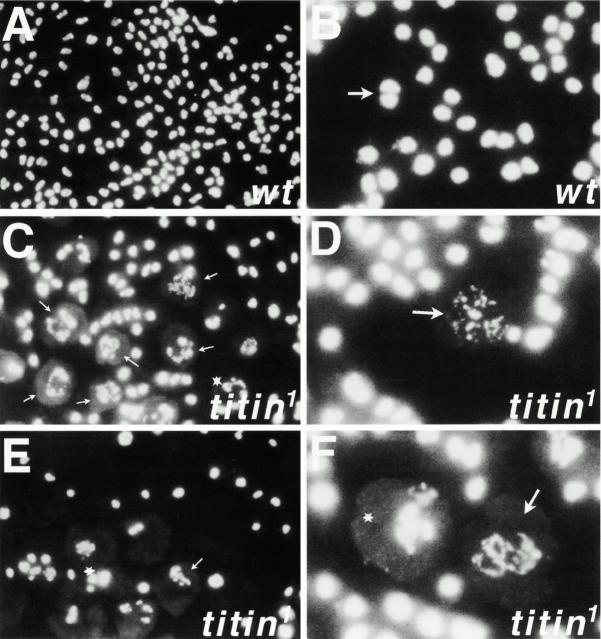
We first compared preparations of untreated chromosomes from titin 1 homozygous larvae to those of wild-type larvae. Hoechts staining of wild-type neuroblasts revealed large fields of uniformly sized nuclei, with the vast majority of cells in interphase (Fig. 4A and Fig. B). In contrast, there was considerable nuclear size variability in the titin 1 mutants, with several cells in mitosis (Fig. 4, A–F). Most of the mitotic nuclei were polyploid and showed both chromosome fragmentation (Fig. 4C and Fig. D) and/or irregular condensation (Fig. 4C, Fig. E, and Fig. F).
Figure 4.
Mitotic phenotypes in D-Titin mutants. Mitotic figures of wild-type and D-Titin mutants from untreated brain squashes of third instar larvae. Brains were stained with Hoechst. (A) Low magnification view of wild-type neuroblasts showing a field of uniformly-sized cells in interphase. (B) Higher magnification of wild-type neuroblasts showing a field of interphase cells. Arrow indicates a cell in anaphase. (C) Low magnification view of titin neuroblasts. Note large polyploid nuclei (white star) and large polyploid nuclei with variably condensed chromatin and chromosome breaks (small white arrows). (D) Higher magnification of a titin neuroblast showing variably condensed chromatin in addition to chromosome breakage. (E and F) Low and high magnification views of titin neuroblasts with polyploid nuclei with highly condensed chromatin (stars) and polyploid nuclei with variable chromosome condensation (arrows). Images were viewed and photographed under 100× objective, 10× eyepiece.
To determine the primary defect in the titin mutants, we examined aceto-orcein squashes of third instar larval brains from larvae homozygous for each of the late larval titin mutations, and from larvae heterozygous for two combinations of mutations (titin 1/titin 8 and titin 1/titin 11). Brains were incubated in colchicine, to increase the number of cells in metaphase, and treated with hypotonic solution, to better visualize chromosome morphology and number. The predominant mitotic defect observed in neuroblasts of all three D-Titin alleles and all heteroallelic combinations was chromosome undercondensation: ∼30% of mutant cells showed very poorly condensed chromosomes, whereas only 7.8% of wild-type cells had poorly condensed chromosomes (Fig. 5B, Fig. C, and Fig. E and Table ). The defect in chromosome condensation was not restricted to euchromatin, since several examples of undercondensation of only centromeric heterochromatin were also observed (Fig. 5 D). Chromosome undercondensation was often accompanied by additional mitotic defects, including premature sister chromatid separation, chromosome loss, polyploidy, and chromosome breakage (Fig. 5, E–H and Table ). We also examined neuroblasts of larvae heterozygous for the late lethal EMS mutation (Titin) and the P-element induced embryonic lethal allele (Titin 10) to determine if the early lethal mutations also affected chromosome structure and behavior. Indeed, chromosomes from titin 1/titin 10 larval neuroblasts showed the same range of defects observed in the titin homozygotes, although slightly fewer cells were affected (Fig. 5 C and Table ). Neuroblasts from titin 1/+ larvae showed a minor increase in chromosome defects compared with those from wild-type larvae, although the defects seen were not nearly as severe as those in any of the mutant combinations (Table ). The increase in mitotic defects in these heterozygotes versus wild type suggests a mild haploinsufficiency for D-Titin function on chromosomes.
Figure 5.
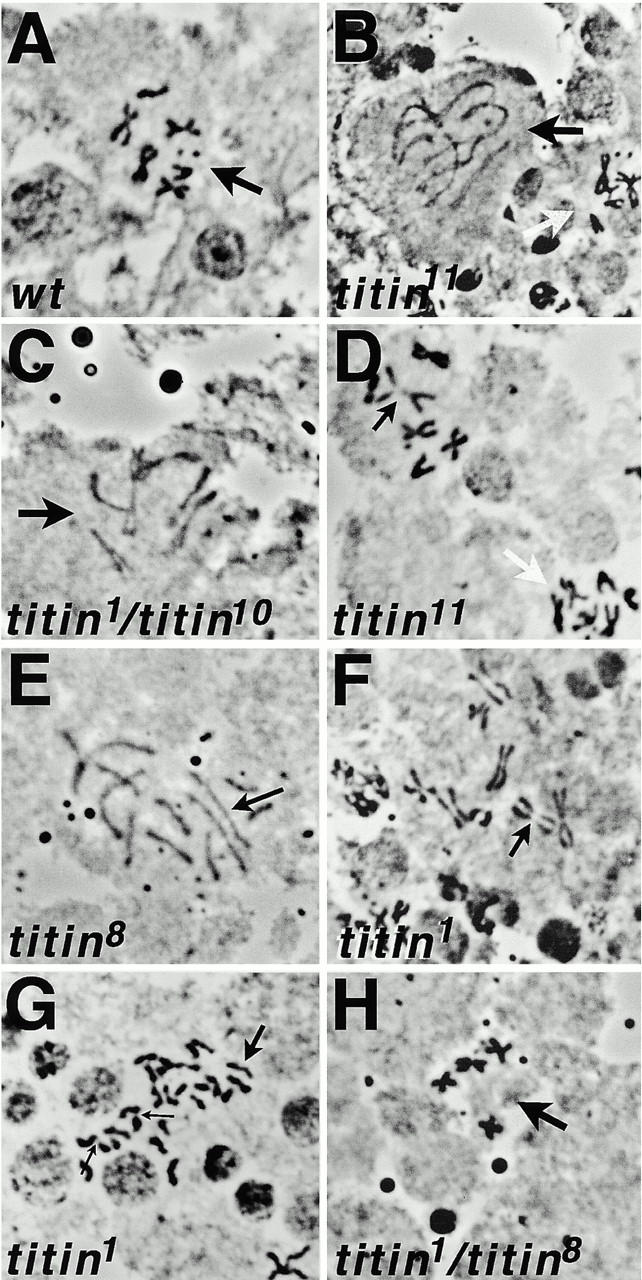
Chromosomal defects in D-Titin mutants. Brains were treated with colchicine/hypotonic shock and stained with orcein. (A) Wild-type neuroblast showing well spread, condensed, diploid chromosomes characteristic of those treated with colchicine and hypotonic shock. (B) Two neuroblasts from a titin 11 homozygous larva showing one cell with severe chromosome undercondensation (black arrow) and an adjacent cell with relatively normal levels of chromosome condensation (white arrow). (C) A neuroblast from a titin 1/titin 10 larva showing chromosome undercondensation. (D) Two neuroblasts from a titin 11 homozygous larva showing one cell with undercondensation of the centromeric heterochromatin (black arrow) and another cell with relatively normal chromosomes. (E) A neuroblast from a titin 8 homozygous larva showing chromosome decondensation and premature sister chromatid separation. The black arrow indicates one of the large autosomes where the two sister chromosomes have completely separated. (F) A neuroblast from a titin homozygous larva showing premature sister chromatid separation and decondensation of the centromeric heterochromatin (black arrow). (G) A neuroblast from a titin homozygous larva showing polyploidy, premature sister chromatid separation (for example the chromosomes indicated with the large black arrow) and decondensation of centromeric heterochromatin (small black arrows). (H) A neuroblast from a titin 1/titin 8 larva showing normal levels of condensation but missing the two sex chrosomes. Images were viewed and photographed under 100× objective, 10× eyepiece.
Table 3.
Quantification of the Mitotic Phenotypes in D-Titin Late Larval Lethals
| Phenotype | D-Titin allelic combinations | |||||||
|---|---|---|---|---|---|---|---|---|
| + | Titin1 | Titin1 | Titin1 | Titin1 | Titin1 | Titin8 | Titin11* | |
| + | + | Titin1 | Titin8 | Titin10 | Titin11* | Titin8 | Titin11* | |
| Normal | 87.4 | 72.7 | 35.1 | 45.8 | 57.8 | 30.0 | 44.3 | 36.4 |
| Undercondensation (total): | 7.8 | 13.3 | 33.7 | 30.0 | 23.7 | 32.7 | 28.8 | 31.8 |
| Undercondensation only | 6.3 | 12.1 | 24.4 | 23.6 | 22.5 | 23.5 | 24.3 | 24.2 |
| +Premature separation | 0.3 | 0.4 | 1.8 | 3.4 | 0.2 | 2.5 | 1.8 | 1.5 |
| +Polyploidy | 0.0 | 0.0 | 1.6 | 0.4 | 0.0 | 0.0 | 0.0 | 2.1 |
| +Aneuploidy | 0.6 | 0.0 | 2.2 | 0.6 | 0.4 | 2.5 | 1.1 | 1.3 |
| +Centromere | 0.6 | 0.8 | 3.7 | 2.0 | 0.6 | 4.2 | 1.6 | 3.7 |
| Premature separation (total) | 2.6 | 1.7 | 7.4 | 13.4 | 3.3 | 11.7 | 12.0 | 8.9 |
| Premature separation only | 2.6 | 1.7 | 4.8 | 12.8 | 3.3 | 5.0 | 9.6 | 6.4 |
| +Aneuploidy | 0.0 | 0.0 | 2.6 | 0.6 | 0.0 | 6.7 | 2.4 | 2.5 |
| Polyploidy only | 0.0 | 0.5 | 6.1 | 2.4 | 4.4 | 8.4 | 4.2 | 4.0 |
| Aneuploidy only | 0.3 | 8.4 | 10.4 | 4.9 | 8.2 | 5.9 | 6.3 | 14.4 |
| Breakage | 0.0 | 0.0 | 2.2 | 3.1 | 0.4 | 4.2 | 2.3 | 0.8 |
| Number of nuclei scored | 1520 | 553 | 1480 | 709 | 512 | 930 | 615 | 1600 |
Chromosomes were examined in orcein-stained brain squashes of third instar larvae. The number of nuclei scored per genotype corresponds to a total of five brains. All brains were incubated in colchicine and treated in a hypotonic solution for better visualization of chromosome abnormalities.
Polytene chromosomes from the salivary glands of the titin homozygotes were indistinguishable from wild type (data not shown), suggesting that either D-TITIN is not required in these “interphase-equivalent” chromosomes or that this allele has sufficient residual D-Titin activity to support normal chromosome function during interphase. Overall, the defects observed in D-Titin mutants strongly suggest roles for D-Titin in mitotic chromosome condensation, chromosome integrity and sister chromatid cohesion.
Discussion
D-Titin Encodes an MDa Protein with Structural Features Found in Vertebrate Titins
We identified D-TITIN as a protein associated with mitotic chromosomes, and cloned portions of the corresponding gene, D-Titin. Here, we report the sequence of the entire D-Titin gene, which links all of the previously identified D-Titin genomic and cDNA clones (Machado et al. 1998), as well as more recently isolated cDNAs. From our analysis, D-Titin begins with exons encoding ORFs most homologous to the NH2-terminal region of vertebrate titin, specifically sequence encoding the Z1-Z3 Ig domains, and ends ∼110-kb downstream with sequences most homologous to the COOH-terminus of the I-band region of vertebrate titin. The D-Titin gene is predicted to encode a 17,903-residue polypeptide accounting for a 1.9- to 2-MDa protein, which is consistent with the MDa-sized polypeptide detected on immunoblots using D-TITIN antisera (Machado et al. 1998).
D-TITIN is only 54% the size of human elastic (soleus) titin, its most homologous vertebrate counterpart, and D-TITIN is most homologous to the NH2-terminal half of the human protein (Fig. 1 D). Moreover, D-TITIN contains structural features (tandem or near-tandem Ig-like domains and PEVK-rich regions) that are found in the Z-disc and I-band of human titin, but does not contain the characteristic domains of A-band and M-line titin. Thus, we predict that D-TITIN will span the sarcomeric Z-disc and I-band, but not the A-band nor the M-line. We further predict that either there is a second molecule in Drosophila that encodes the true homologue of vertebrate titin, or that two (or more) titin-related proteins do the work of the one vertebrate protein. A TBLASTN search of the Drosophila genome, which is essentially complete in the euchromatic regions, with small fragments of human elastic titin (1,000–2,000 residues) revealed that the most homologous ORF to the first 15,000 residues was D-Titin. The most homologous ORF to the remaining 18,449 residues of human elastic titin was projectin. PROJECTIN is the homologue of C. elegans TWITCHIN, another so-called “mini-titin” that binds to myosin filaments in body wall muscles. TWITCHIN has the Ig/FN3 repeats and a serine kinase domain near the COOH terminus, but does not have a PEVK domain (Benian et al. 1989, Benian et al. 1996). Mutations in twitchin (unc-22) cannot develop or sustain muscle contractions, and can be suppressed by mutations in the myosin heavy chain gene (Waterston et al. 1980; Moerman et al. 1988). These phenotypes support a role for TWITCHIN in sarcomere function and provide evidence for a direct physical association between TWITCHIN and MYOSIN in the A-band. Mutations in projectin (bent mutations) cause early embryonic lethality but have apparently normal muscle contractions (Fyrberg et al. 1992; Ayme-Southgate et al. 1995). However, these mutations may not be null for projectin function. Based on the above findings, we favor a model in which two (or more) proteins in flies (D-TITIN and PROJECTIN) and worms (a D-TITIN homologue and TWITCHIN) combine to do the work of the single vertebrate titin protein. D-TITIN would function as a protein equivalent to the Z-disc/I-band region of vertebrate titin, whereas PROJECTIN and TWITCHIN would function as proteins equivalent to the A-band/M-line region of vertebrate titin.
Kettin Is Encoded by the D-Titin Gene
The sequence of the proposed Drosophila ket gene is contained entirely within a 20-kb region mapping near the 5′ end of D-Titin (Fig. 1 A; Hakeda et al. 2000; Kolmerer et al. 2000). Zeugmatin, like KET, was first identified as a “smaller” 600–800-kD Z-disc associated protein in chicken cardiac muscle (Maher et al. 1985). However, recent studies suggest that the zeugmatin polypeptide is an NH2-terminal cleavage product of chicken titin (Turnacioglu et al. 1996, Turnacioglu et al. 1997a,Turnacioglu et al. 1997b; Ayoob et al. 2000). KET may derive from an alternative splice form of D-Titin, since embryonic transcripts consistent with the size of KET have been reported (Hakeda et al. 2000). However, the smaller kettin mRNA does not appear to be translated during embryogenesis since a polypeptide of the corresponding size (500–700 kD) was not detected in immunoblots of embryonic extracts incubated with antibodies recognizing a shared domain of KETTIN and D-TITIN (Machado et al. 1998). These same immunoblots did detect the MDa-sized D-TITIN, as well as smaller polypeptides below 200 kD, which are likely to be degradation products of full-length D-TITIN (Machado et al. 1998). This finding suggests that only the larger MDa form of D-TITIN functions during embryogenesis. Thus, mutations reported to disrupt ket must be considered as mutations that disrupt D-Titin, and at least during embryogenesis, are likely to produce defects as a result of lesions in the D-TITIN protein.
D-TITIN Is Required for Myoblast Fusion and Sarcomere Structure
Two prominent embryonic muscle phenotypes were common to all of D-Titin mutants: failure of myoblasts to fuse with existing myofibers and defects in the embryonic gut (Fig. 3). The myoblast fusion defects were similar to those caused by loss-of-function mutations in mbc, blow, rost, or sns (Paululat et al. 1995; Rushton et al. 1995; Doberstein et al. 1997; Bour et al. 2000), by the muscle-specific expression of a constitutively active form of Drac1 (Luo et al. 1994) and by mutations in mef2, which encodes a transcription factor that regulates the expression of several muscle-specific structural protein genes (Bour et al. 2000). Each of these genes affects a discrete step in the fusion of myoblasts with either muscle pioneers (also known as founder myoblast cells) or existing myotubes (Doberstein et al. 1997). As with mbc (Erickson et al. 1997), D-Titin may not be directly involved in fusion but may instead be essential for a cytoskeletal rearrangement step required for fusion. For example, D-Titin may be required in the elongation step, which occurs after cell-cell recognition and adherence but before the formation of the prefusion complex (Doberstein et al. 1997).
Transmission electron microscopy (TEM) analyses reveal that the Z-discs of the body wall muscles fail to organize during late embryogenesis in ket mutants, whereas Z-discs have nearly completely formed in wild-type embryos (Hakeda et al. 2000). At later stages, the ket mutant embryos have completely disorganized sarcomeres with apparently randomly-arrayed myofilaments. Because the ket mutations also disrupt the D-Titin ORF, and because only the MDa form of D-TITIN (and not KET) is detected in embryos, the defects observed in embryos or in early larvae are more likely due to the absence of the larger form of the protein, D-TITIN. The ket/D-Titin mutant phenotypes are consistent with long-standing models that titin serves not only to provide elasticity to the sarcomere, but that it may also serve as the template for sarcomere organization and assembly. Thus, the muscle phenotypes described for the ket mutations are absolutely consistent with those expected for mutations in D-Titin.
Titin Is Required for Chromosome Structure and Mitosis
Based on our finding that titin localizes to chromosomes, and the proposed functions of titin in muscle, we proposed that titin has two functions on chromosomes: to organize higher-order chromosome structure and to provide elasticity (Machado et al. 1998). The chromosomal and mitotic defects seen in actively dividing cells of D-Titin mutants fully support this proposal. Even mild D-Titin mutations that allow survival to late developmental stages cause severe chromosomal and mitotic defects, affecting chromosome condensation and interfering with several other aspects of mitosis (Fig. 4 and Table ).
In muscle, titin is thought to act as a molecular scaffold for sarcomere assembly by specifying the precise position of its ligands within each half-sarcomere; this model is supported by the defects in D-Titin and ket mutants (Hakeda et al. 2000). The chromosome undercondensation phenotype of the D-Titin mutants suggests that titin has a critical role in establishing or maintaining higher-order chromosome compaction. Analogous to its role in muscle, titin may localize to chromosomes and provide a template for the correct binding and assembly of other proteins involved in chromosome condensation. These binding partners might include known proteins, such as the SMC proteins of the condensin complex, or new unidentified partners. The defects in sister chromatid cohesion in D-Titin mutants suggest that titin may also organize and/or stabilize cohesin complexes, which maintain sister chromatid cohesion before the metaphase–anaphase transition (for review see Biggins and Murray 1999).
Titin also functions as a molecular spring that confers elasticity and maintains the structural integrity of the contracting myofibrils (Gregorio et al. 1999). The chromosome breakage phenotype in D-Titin mutants suggests that titin may provide similar elasticity to chromosomes to prevent their breakage during metaphase and anaphase. Mitotic chromosomes behave as highly extensible, elastic objects that maintain their integrity throughout the physically strenuous process of cell division (Claussen et al. 1994; Houchmandzadeh et al. 1997). Indeed, the elastic properties of in vitro assembled mitotic chromosomes are consistent with those described for titin, and have led to a model for chromosome structure based on one or several thin elastic axes, consisting of elastic titin-like molecules, surrounded by a soft envelope of chromatin (Houchmandzadeh and Dimitrov 1999). Based on the localization of titin to both chromosomes and sarcomeres and the defects in D-Titin mutants, our results support a model in which titin serves parallel functions in the organization and elasticity of each of these two very different macromolecular assemblies, a remarkable feat of evolution.
Acknowledgments
We thank D. Barrick, P. Bradley, Y. Gruenbaum, K. Wilson, and the anonymous reviewers for their critical comments on the manuscript. We thank A. Cardoso for assistance with the figures. We are grateful to J. Kennison, J. Mason, T. Sliter, A. Spradling, and the Bloomington Stock Center for providing fly stocks. We thank J. Kennison for suggesting that sallimus is a mutation in D-Titin. We thank D. Kiehart for the antimuscle myosin antiserum and B. Bullard for the MAC155 rat mAb. We also thank the BDGP for D-Titin cDNAs GH05716, CK340, and CK55.
This work was supported by an Institutional Research Grant from the Johns Hopkins University School of Medicine and by a research grant from the March of Dimes, FY99-295. C. Machado was supported in part by a postdoctoral fellowship from the Fundação para a Ciência e a Tecnologia (FCT, Portugal).
Note Added in Proof. In September of 2000, a paper by Zhang et al. (Zhang, Y., D. Featherstone, W. Davis, E. Rushton, and K. Broadie. 2000. J. Cell. Sci. 113:3103–3115) reported similar defects in myoblast fusion with mutations in D. Titin.
Footnotes
Abbreviations used in this paper: FISH, fluorescence in situ hybridization; FN3, fibronectin type 3; MDa, megadalton; nt, nucleotide; ORF, open reading frame; SMC, structural maintenance of chromosome; topo II, topoisomerase II.
References
- Alberts B., Bray D., Lewis J., Raff M., Roberts K., Watson J.D. Molecular Biology of the Cell 1994. Garland Publishing, Inc; New York: pp. 1294 pp [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ayme-Southgate A., Southgate R., Saide J., Benian G.M., Pardue M.L. Both synchronous and asynchronous muscle isoforms of projectin (the Drosophila bent locus product) contain functional kinase domains. J. Cell Biol. 1995;128:393–403. doi: 10.1083/jcb.128.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoob J.C., Turnacioglu K.K., Mittal B., Sanger J.M., Sanger J.W. Targeting of cardiac muscle titin fragments to the Z-bands and dense bodies of living muscle and non-muscle cells. Cell. Motil. Cytoskeleton. 2000;45:67–82. doi: 10.1002/(SICI)1097-0169(200001)45:1<67::AID-CM7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Benian G.M., Tang X., Tilney T.L. Twitchin and related giant Ig super family members of C. elegans and other invertebrates. Adv. Biophys. 1996;33:183–197. [PubMed] [Google Scholar]
- Benian G.M., Kiff J.E., Neckelmann N., Moerman D.G., Waterston R.H. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans . Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A.W. Sister chromatid cohesion in mitosis. Curr. Opin. Genet. Dev. 1999;9:230–236. doi: 10.1016/s0959-437x(99)80034-3. [DOI] [PubMed] [Google Scholar]
- Bour B.A., Chakravarti M., West J.M., Abmayr S.M. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Boy de la Tour E., Laemmli U. The metaphase scaffold is helically foldedsister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Claussen U., Mazur A., Rubtsov N. Chromosomes are highly elastic and can be stretched. Cytogenet. Cell Genet. 1994;66:120–125. doi: 10.1159/000133681. [DOI] [PubMed] [Google Scholar]
- Doberstein S.K., Fetter R.D., Mehta A.Y., Goodman C.S. Genetic analysis of myoblast fusionblown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson M.R.S., Galletta B.J., Abmayr S.M. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure and cytoskeleton organization. J. Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg C.C., Labeit S., Bullard B., Leonard B., Fyrberg E. Drosophila projectinrelatedness to titin and twitchin and correlation with lethal(4)102 CDa and bent-dominant mutants. Proc. R. Lond. B. Biol. Sci. 1992;249:33–40. doi: 10.1098/rspb.1992.0080. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U.K. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Gatti M., Baker B.S. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gatti M., Goldberg M. Mutations affecting cell division in Drosophila . Methods Cell Biol. 1991;35:543–586. doi: 10.1016/s0091-679x(08)60587-7. [DOI] [PubMed] [Google Scholar]
- Gatti M., Bonaccorsi S., Pimpinelli S. Looking at Drosophila mitotic chromosomes. In Drosophila melanogasterPractical Uses in Cell and Molecular Biology 1994. Academic Press, ; San Diego, CA: pp. 372–391 [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Granzier H., Sorimachi H., Labeit S. Muscle assemblya titanic achievement. Curr. Opin. Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- Hakeda S., Endo S., Saigo K. Requirements of Kettin, a giant muscle protein highly conserved in overall structure in evolution, for normal muscle function, viability and flight activity of Drosophila . J. Cell Biol. 2000;148:101–114. doi: 10.1083/jcb.148.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, B.A., and K. Zinn. 1994. From clone to mutant gene. In Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Academic Press, New York. 81–94. [DOI] [PubMed]
- Hirano T. SMC-mediated chromosome mehanicsa conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B., Dimitrov S. Elasticity measurements show the existence of thin rigid cores inside mitotic chromosomes. J. Cell Biol. 1999;145:215–223. doi: 10.1083/jcb.145.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B., Marko J.F., Chatenay D., Libchaber A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 1997;139:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasravi A., Walter M.F., Brand S., Mason J.M., Biessmann H. Molecular cloning and tissue-specific expression of the mutator2 gene (mu2) in Drosophila melanogaster. Genetics. 1999;152:1025–1035. doi: 10.1093/genetics/152.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J.A., Tamkun J.W. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila . Proc. Natl. Acad. Sci. USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D.P., Feghali R. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmerer B., Clayton J., Benes V., Allen T., Ferguson C., Leonard K., Weber U., Knekt M., Ansorge W., Labeit S., Bullard B. Sequence and expression of the kettin gene in Drosophila melanogaster and Caenorhabditis elegans . J. Mol. Biol. 2000;296:435–448. doi: 10.1006/jmbi.1999.3461. [DOI] [PubMed] [Google Scholar]
- Koshland D., Strunnikov A. Mitotic chromosome condensation. Ann. Rev. Cell Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Labeit S., Kolmerer B. Titins, giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Lakey A., Labeit S., Gautel M., Ferguson J., Barlow D.P., Leonard K., Bullard B. Kettin, a large modular protein in the Z-disc of insect muscles. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:2863–2871. doi: 10.1002/j.1460-2075.1993.tb05948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D.L., Zimm G.G. The genome of Drosophila melanogaster. Academic Press, ; San Diego, CA: 1992. [Google Scholar]
- Linke W.A., Ivemeyer M., Labeit S., Hinssen H., Ruegg J.C., Gautel M. Actin-titin interaction in cardiac myofibrilsprobing a physiological role. Biophy. J. 1997;73:905–919. doi: 10.1016/S0006-3495(97)78123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Liao Y.J., Jan L.Y., Jan Y.N. Distinct morphogenetic funcitons of similar small GTPasesDrosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Machado C., Sunkel C.E., Andrew D.J. Human autoantibodies reveal titin as a chromosomal protein. J. Cell Biol. 1998;141:321–333. doi: 10.1083/jcb.141.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher P.A., Cox G.F., Singer S.J. Zeugmatina new high molecular weight protein associated with Z lines in adult and early embryonic striated muscle. J. Cell Biol. 1985;101:1871–1883. doi: 10.1083/jcb.101.5.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular CloningA Laboratory Manual. Cold Spring Harbor Laboratory Press, ; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Manuelidis L. A view of interphase chromosome. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- McDowall A.W., Smith J.M., Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO (Eur. Mol. Biol. Organ.) J. 1986;5:1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman G.D., Benian G.M., Barstead R.J., Schrieffer L.A., Waterston R.H. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans . Genes & Dev. 1988;2:93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- Mount S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S.M., Burks C., Hertz G., Stormo G.D., White O., Fields C. Splicing signals in Drosophilaintron size, information content and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M.-L. Looking at polytene chromosomes. In Drosophila melanogasterPractical Uses in Cell and Molecular Biolog 1994. y; Academic Press: 333–351. pp. San Diego, CA [Google Scholar]
- Paulson J.R., Laemmli U.K. The structure of histone-depleted chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Paululat A., Burchard S., Renkawitz-Pohl R. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila . Development. 1995;121:2611–2620. doi: 10.1242/dev.121.8.2611. [DOI] [PubMed] [Google Scholar]
- Reuter R., Panganiban G.E.F., Hoffmann F.M., Scott M.P. Homeotic genes regulate the spatial expression of putative growth factors in the visceral mesoderm of Drosophila embryos. Development. 1990;110:1031–1040. doi: 10.1242/dev.110.4.1031. [DOI] [PubMed] [Google Scholar]
- Rushton E., Drysdale R., Abmayr S.M., Michelson A.M., Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sedat J., Manuelidis L. A direct approach to the structure of eukaryote chromosomes. Cold Spring Harbor Symp. Quant. Biol. 1978;42:331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Sliter T.J., Henrich V.C., Tucker R.L., Gilbert L.I. The genetics of the Dras3-Roughened-ecdysoneless chromosomal region (62B3-4 to 62D3-4) in Drosophila melanogasteranalysis of recessive lethal mutations. Genetics. 1989;123:327–336. doi: 10.1093/genetics/123.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.F., Wiese B.A., Wojzynski M.K., Davison D.B., Worely K.C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V., Jessberger R. Structural maintenance of chromosomes (SMC) proteinsconserved molecular properties for multiple biological functions. Eur. J. Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructure of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick J., Tskhovrebova L. Titina molecular control freak. Trends Cell Biol. 1999;9:377–380. doi: 10.1016/s0962-8924(99)01641-4. [DOI] [PubMed] [Google Scholar]
- Trombitas K., Granzier H. Actin removal from cardiac myocytes shows that near Z line titin attaches to actin while under tension. Am. J. Physiol. 1997;273:662–670. doi: 10.1152/ajpcell.1997.273.2.C662. [DOI] [PubMed] [Google Scholar]
- Trombitas K., Greaser M.L., Pollack G.H. Interaction between titin and thin filaments in intact cardiac muscle. J. Muscle Res. Cell Motil. 1997;18:345–351. doi: 10.1023/a:1018626210300. [DOI] [PubMed] [Google Scholar]
- Turnacioglu K.K., Mittal B., Sanger J.M., Sanger J.W. Partial characterization of zeugmatin indicates that it is part of the Z-band region of titin. Cell Motil.Cytoskel. 1996;34:108–121. doi: 10.1002/(SICI)1097-0169(1996)34:2<108::AID-CM3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Turnacioglu K.K., Mittal B., Dabiri G.A., Sanger J.M., Sanger J.W. An N-terminal fragment of titin coupled to green fluorescent protein localizes to the Z-bands in living muscle cellsoverexpression leads to myofibril disassembly Mol. Biol. Cell 8 1997. 705 717a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnacioglu K.K., Mittal B., Dabiri G.A., Sanger J.M., Sanger J.W. Zeugmatin is part of the Z-band targeting region of titin Cell Struct. Func. 22 1997. 73 82b [DOI] [PubMed] [Google Scholar]
- Wang M., Champion L.E., Biessmann H., Mason J.M. Mapping a mutator, mu2, which increases the frequency of terminal deletions in Drosophila melanogaster. Mol. Gen. Genet. 1994;245:598–607. doi: 10.1007/BF00282222. [DOI] [PubMed] [Google Scholar]
- Warburton P.E., Earnshaw W.C. Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. BioEssays. 1997;19:97–99. doi: 10.1002/bies.950190203. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Thomson J.N., Brenner S. Mutants with altered muscle structure in Caenorhabditis elegans . Dev. Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]