Abstract
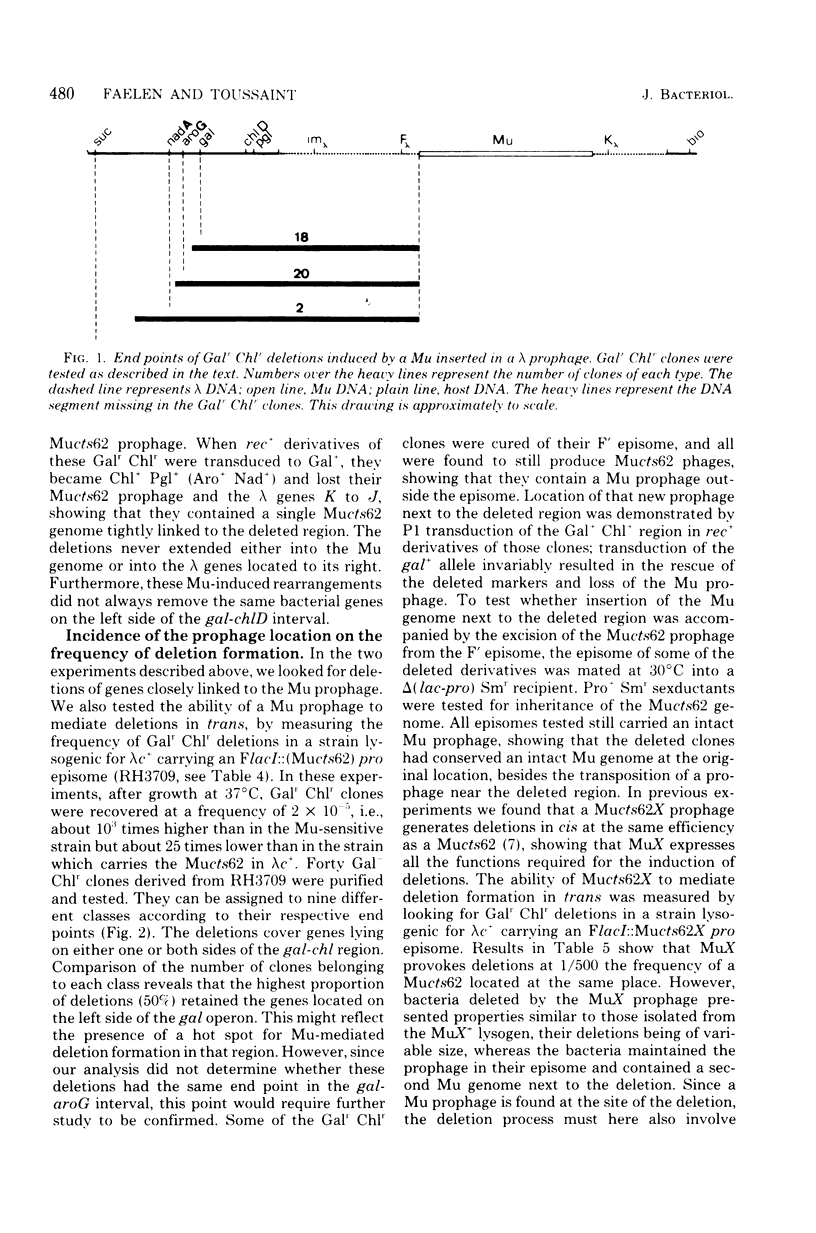
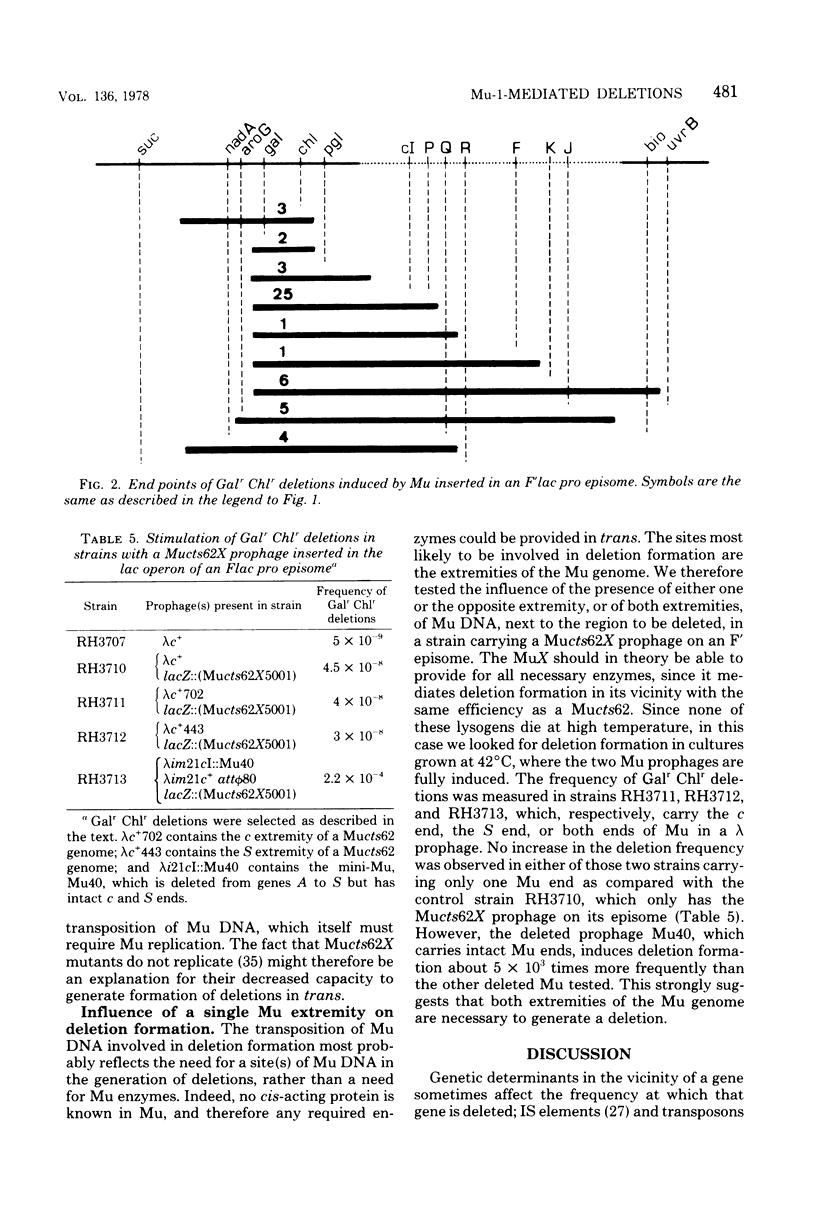
Deletion of bacterial DNA fragments is stimulated in induced Mucts62 lysogens. The host genes located proximally to the prophage are more frequently lost than those which are unlinked to the Mu genome. Genes located on either side of a Mu genome are deleted in the same manner. Like the other Mu-induced rearrangements, this process is recA independent and requires the participation of Mu DNA, as indicated by the fact that a phage genome always replaces the deleted genes. Data are presented which strongly suggest that both ends of the Mu genome are involved in deletion formation.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of Lambda Lysogenicity during Bacterial Recombination in Escherichia Coli K12. Genetics. 1954 Jul;39(4):429–439. doi: 10.1093/genetics/39.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I. Bacteriophage mu as a transposition element. Annu Rev Genet. 1976;10:389–412. doi: 10.1146/annurev.ge.10.120176.002133. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I. Reversal of mutator phage Mu integration. J Mol Biol. 1975 Jul 25;96(1):87–99. doi: 10.1016/0022-2836(75)90183-7. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Taylor A. L. Influence of insertions on packaging of host sequences covalently linked to bacteriophage Mu DNA. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4399–4403. doi: 10.1073/pnas.72.11.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón T., Faelen M., De Wilde M., Bollen A., Thomas R. Expression of ribosomal protein genes in Escherichia coli. Mol Gen Genet. 1975;137(2):125–129. doi: 10.1007/BF00341678. [DOI] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A. Bacteriophage Mu-1: a tool to transpose and to localize bacterial genes. J Mol Biol. 1976 Jul 5;104(3):525–539. doi: 10.1016/0022-2836(76)90118-2. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., Couturier M. Mu-1 promoted integration of a -gal phage in the chromosome of E. coli. Mol Gen Genet. 1971;113(4):367–370. doi: 10.1007/BF00272338. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., De Lafonteyne J. Model for the enchancement of lambde-gal integration into partially induced Mu-1 lysogens. J Bacteriol. 1975 Mar;121(3):873–882. doi: 10.1128/jb.121.3.873-882.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLANSDORFF N. TOPOGRAPHY OF COTRANSDUCIBLE ARGININE MUTATIONS IN ESCHERICHIA COLI K-12. Genetics. 1965 Feb;51:167–179. doi: 10.1093/genetics/51.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- KAISER A. D. A genetic study of the temperate coliphage. Virology. 1955 Nov;1(4):424–443. doi: 10.1016/0042-6822(55)90036-2. [DOI] [PubMed] [Google Scholar]
- KAISER A. D. Mutations in a temperate bacteriophage affecting its ability to lysogenize Escherichia coli. Virology. 1957 Feb;3(1):42–61. doi: 10.1016/0042-6822(57)90022-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kupor S. R., Fraenkel D. G. 6-phosphogluconolactonase mutants of Escherichia coli and a maltose blue gene. J Bacteriol. 1969 Dec;100(3):1296–1301. doi: 10.1128/jb.100.3.1296-1301.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOEB T. Isolation of a bacteriophage specific for the F plus and Hfr mating types of Escherichia coli K-12. Science. 1960 Mar 25;131(3404):932–933. doi: 10.1126/science.131.3404.932. [DOI] [PubMed] [Google Scholar]
- Lederberg E M, Lederberg J. Genetic Studies of Lysogenicity in Escherichia Coli. Genetics. 1953 Jan;38(1):51–64. doi: 10.1093/genetics/38.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUSHIRO A. Specialized transduction of tryptophan markers in Escherichia coli K12 by bacteriophage phi-80. Virology. 1963 Apr;19:475–482. doi: 10.1016/0042-6822(63)90041-2. [DOI] [PubMed] [Google Scholar]
- McEntee K., Estein W. Isolation and characterization of specialized transducing bacteriophages for the recA gene of Escherichia coli. Virology. 1977 Mar;77(1):306–318. doi: 10.1016/0042-6822(77)90427-5. [DOI] [PubMed] [Google Scholar]
- Spudich J. A., Horn V., Yanofsky C. On the production of deletions in the chromosome of Escherichia coli. J Mol Biol. 1970 Oct 14;53(1):49–67. doi: 10.1016/0022-2836(70)90045-8. [DOI] [PubMed] [Google Scholar]
- Starlinger P., Saedler H. IS-elements in microorganisms. Curr Top Microbiol Immunol. 1976;75:111–152. doi: 10.1007/978-3-642-66530-1_4. [DOI] [PubMed] [Google Scholar]
- Szpirer J., Thomas R., Radding C. M. Hybrids of bacteriophages lambda and phi 80: a study of nonvegetative functions. Virology. 1969 Apr;37(4):585–596. doi: 10.1016/0042-6822(69)90276-1. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Leurs C., Dambly C., Parmentier D., Lambert L., Brachet P., Lefebvre N., Mousset S., Porcheret J., Szpirer J. Isolation and characterization of new sus (amber) mutants of bacteriophage lambda. Mutat Res. 1967 Nov-Dec;4(6):735–741. doi: 10.1016/0027-5107(67)90082-6. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M. Connecting two unrelated DNA sequences with a Mu dimer. Nat New Biol. 1973 Mar 7;242(114):1–4. doi: 10.1038/newbio242001a0. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Gruijthuijsen M. Chromosome mobilization and integration of F-factors in the chromosome of RecA strains of E. coli under the influence of bacteriophage Mu-1. Mol Gen Genet. 1972;118(2):173–183. doi: 10.1007/BF00267086. [DOI] [PubMed] [Google Scholar]



