Abstract
The pharmacological action of salicylate cannot be explained by its inhibition of cyclooxygenase (COX) activity. In this report, the effects of aspirin and sodium salicylate on COX-2 expressions in human umbilical vein endothelial cells and foreskin fibroblasts were evaluated. Aspirin and sodium salicylate at therapeutic concentrations equipotently blocked COX-2 mRNA and protein levels induced by interleukin-1β and phorbol 12-myristate 13-acetate. The suppressing effect was more pronounced in cultured cells deprived of fetal bovine serum for 24 h, suggesting that it may be cell cycle related. Salicylate inhibited nascent COX-2 transcript synthesis but had no effect on COX-2 mRNA stability. It inhibited COX-2 promoter activity in a concentration-dependent manner. In mice pretreated with aspirin (10 and 30 mg/kg), followed by challenge with lipopolysaccharide, COX-2 mRNA expression in peritoneal macrophages was markedly suppressed. These findings suggest that salicylate exerts its antiinflammatory action in part by suppressing COX-2 induction, thereby reducing the synthesis of proinflammatory prostaglandins.
Since the classic work of Vane (1), it has been widely accepted that the pharmacological action of nonsteroidal antiinflammatory drugs (NSAID) is mediated by inhibiting the activity of cyclooxygenase (COX), a key enzyme in biosynthesis of proinflammatory prostaglandins. Recent studies implicate COX-2 induction as a critical event in inflammation (2). This notion has been supported by effective suppression of inflammatory responses in experimental animals by selective COX-2 inhibitors (3, 4). Aspirin (acetylsalicylic acid) is a nonselective COX inhibitor (5, 6). Moreover, aspirin is rapidly deacetylated in blood to form salicylic acid. Salicylic acid has been a known NSAID for over a century, but its mechanism of action remains a pharmacological enigma. It has virtually no inhibitory activity against purified COX-1 or COX-2, although it inhibits prostaglandin synthesis in intact cells (7). To elucidate the mechanism by which salicylic acid exerts its antiinflammatory action, we evaluated the effects of aspirin and sodium salicylate on COX-2 expression in human umbilical vein endothelial cells (HUVEC) and human foreskin fibroblasts (HFF) induced by inflammatory mediators. We show here that aspirin and sodium salicylate at therapeutic concentrations suppress COX-2 gene transcription. When administered to mice pretreated with lipopolysaccharide, aspirin inhibited COX-2 mRNA levels in peritoneal macrophages.
MATERIALS AND METHODS
Materials.
Recombinant IL-1β, phorbol 12-myristate 13-acetate (PMA), sodium salicylate, aspirin, NS398, indomethacin, and lipopolysaccharide (LPS) were obtained from Sigma.
Cell Culture.
HUVECs were cultured as described (8) in medium 199 containing 20% FBS, 50 μg/ml endothelium cell growth factor, 10 units/ml heparin, 100 μg/ml streptomycin, and 100 units/ml penicillin. Only first and second passage cells were used. HFFs were obtained from American Type Culture Collection and were cultured according to a standard procedure of American Type Culture Collection. Unless otherwise indicated, experiments were carried out in HFF or HUVEC that had been cultured in media containing 0.5% FBS or 0.5% BSA for 24 h. The cells were washed and incubated in fresh medium in the presence or absence of aspirin or sodium salicylate at 37°C for 30 min before the addition of PMA or IL-1β.
Reverse Transcription (RT)–PCR Assays.
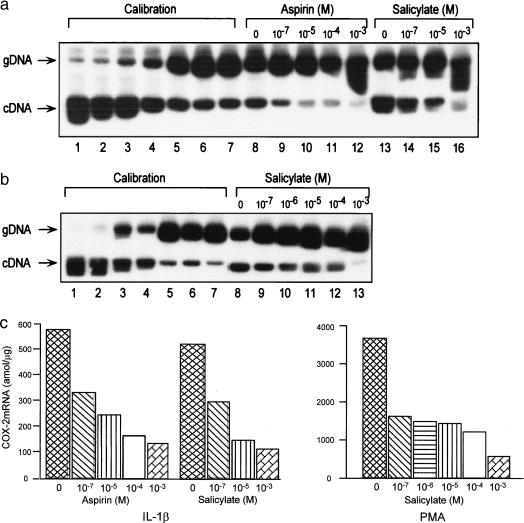
COX-2 mRNA concentrations were assayed by a quantitative RT-PCR procedure modified from a procedure previously described (9). In this modified assay, a calibration curve was constructed, and the mRNA levels in samples were derived from the calibration curve. As shown in Fig. 1 a and b, an increasing concentration of COX-2 genomic RNA (gRNA) fragment was competing against a fixed concentration of COX-2 mRNA in a RT-PCR reaction mixture as described (9). The amplified gDNA and cDNA were separated by a 6% polyacrylamide gel electrophoresis. The bands were excised, and radioactive counts were measured. Because there is a reciprocal competition between gDNA and cDNA, a reciprocal plot of log cDNA/gDNA radioactive count ratios vs. log cDNA concentrations was derived from a plot of log gDNA/cDNA vs. log gDNA concentrations. The COX-2 mRNA of unknown samples was determined by incubating a fixed quantity of gRNA with 0.5 μg of total cellular RNA. The gDNA and cDNA templates as well as primers for RT and PCR in this assay are designed to be specific for human COX-2 and not cross-detecting COX-1 messages. The gDNA fragment stretching from exon 6 to exon 7 containing a 119-bp intron matched the cDNA in sequence except for the intron, which is not present in cDNA. This results in a difference by 119 bp in size between gDNA and cDNA fragments, which can be easily distinguished on gel electrophoresis. These two fragments, on the other hand, have identical 5′ and 3′ sequences for PCR amplification: forward sequence, 5′-+659ACGGTGAAACTCTGGCT-3′ and reverse sequence, 5′-+921GGATGCTCCTGTTTAAG-3′. A 10-mer (5′-+927 CATTCAGGAT-3′) was used in reverse transcription of gRNA and mRNA.
Figure 1.
Effects of aspirin and sodium salicylate on induced HUVEC COX-2 mRNA levels measured by competitive PCR by using a gRNA generated from a human genomic COX-2 DNA fragment as the competitor in that assay. (a) A representative autoradiograph illustrating the calibration (lanes 1–7) using increasing concentrations of gRNA (lanes 1–7, 20–1280 amol in a 2-fold escalation) competing against a standard concentration (256 nmol/tube) of COX-2 mRNA. In lanes 8–16, each tube contained in a RT-PCR mixture 640 amol of gRNA and 0.5 μg of total RNA prepared from HUVEC stimulated with IL-1β (1 ng/ml, 4 h) in the absence of (lanes 8 and 13) or in the presence of aspirin (lanes 9–12) or sodium salicylate (lanes 14–16). gDNA and cDNA were clearly separated as indicated. (b) An autoradiograph illustrating the suppressing effect of sodium salicylate on PMA-induced COX-2 mRNA by competitive PCR. Lanes 1–7 serve as the calibration for quantifying COX-2 mRNA levels. Increasing concentrations of gRNA (lanes 1–7, 160–10240 amol per tube in a 2-fold escalation) were competing against a standard COX-2 mRNA concentration. Lanes 8–13 denote mRNA levels under PMA (100 nM, 4 h) stimulation in the absence (lane 8) or presence (lanes 9–13) of sodium salicylate. (c) COX-2 mRNA levels derived from the calibration curves of a and b.
The RT-PCR assay for human COX-2 mRNA failed to detect mouse macrophage COX-2 mRNA because of sequence divergence. A different pair of primers were designed to specifically detect mouse COX-2 mRNA. The sequences of these two primers are forward primer 5′-CAAGCAGTGGCAAGGCCTCCA-3′ and reverse sequence, 5′-GGCACTTGCATTGATGGTGGCT-3′.
Northern Blot Analysis.
RNA was isolated from cultured cells by using RNA-STAT 60 (Tel-Test, Friendswood, TX). A portion (25–30 μg) of RNA was fractionated on 1% agarose and was transferred to a positively charged nylon membrane. As a COX-2 probe, we used agarose gel-purified, full-length, 1.9-kilobase COX-2 cDNA (10) kindly provided by Timothy Hla (American Red Cross, Rockville, MD). Probe was labeled by using [α-32P]dCTP random labeling kit (Stratagene). Membranes were hybridized overnight in 5× standard saline citrate (SSC) containing 50% formamide, 0.02% SDS, 0.1% sarcosyl, and 2% blocking reagent (Boehringer Mannheim) at 55°C. Nonspecifically bound probe was washed during 45 min of high stringency washes, performed at 65°C in 0.1× SSC containing 0.1% SDS. Membranes were stripped at 95°C in 0.1× SSC containing 0.1% SDS for 15 min and were rehybridized to a digoxigenin-labeled glyceraldehyde-3-phosphate dehydrogenase RNA probe. Blots were quantitated by using the Bio Image system (MilliGen).
Western Blot Analysis.
Cells were washed twice in ice-cold PBS, were resuspended in PBS, and were lysed by freezing. Protein concentration in lysates was determined by using BCA protein assay (Pierce). Lysates (30 μg of protein) were electrophoresed on 7.5% polyacrylamide gel and were electroblotted onto a nitrocellulose membrane. The COX-2 proteins on the membranes were detected by using a rabbit anti-human COX-2 polyclonal antibody specific for COOH-terminal insert of COX-2 and were visualized by using enhanced chemiluminescence system (Pierce).
Nuclear Run-Off Experiments.
Isolation of nuclei and synthesis of labeled RNA were performed according to a procedure previously described (11). Cell nuclei (107) were incubated in the presence of 0.25 mCi of [32P]GTP and other unlabeled nucleotides (1 mM) at 26°C in a volume of 200 μl for 25 min. Transcribed RNAs were isolated by using Ultraspec (Biotecx Lab, Houston), and equal amounts of the labeled RNA (6–8 × 106 cpm) were hybridized to denatured pSG5 containing COX-2 cDNAs (15 μg per blot) blotted onto nitrocelluose membranes by using a slot-blot apparatus (Schleicher & Schuell). Conditions for hybridization and washes were identical to those for Northern blotting.
COX-2 Promoter Activity.
The COX-2 promoter activity was determined by transient expression of a COX-2 5′-flanking region in HFF or HUVEC by a previously described method (12). In brief, a 5′-flanking DNA fragment from position −891 to +9 (−891/+9) of human COX-2 gene (13) was constructed into a promoterless luciferase expression vector, pGL3, and the vector was expressed in HFF or HUVEC by lipofection. The expressed luciferase activity was determined in a luminometer (Monolight 2010, Analytic Luminescence Laboratory, San Diego). In each experiment, pSV-luc, driven by an SV40 promoter, and pGL3 basic were included as positive and negative controls, respectively.
Analysis of COX Activity.
Cultured HFFs were treated with sodium salicylate or NS398 30 min before the addition of PMA for 6 h. The culture condition was identical to that described above. The media were collected, and prostaglandin E2 (PGE2) was measured by a highly specific enzyme-immunoassay. The COX activity was analyzed further by reverse-phase HPLC as described (7). In brief, cells were treated with [1-14C] arachidonic acid at 37°C for 10 min. The medium was collected, and eicosanoids were extracted with C-18 cartridge and were analyzed by HPLC.
Animal Experiments.
Male BALB/C mice were pretreated with aspirin via a gastric route and were injected with endotoxin through a tail vein as described (14). Mice were killed 12 h after treatment, and peritoneal macrophages were isolated. COX-2 mRNA was determined by a RT-PCR procedure using a pair of mouse COX-2 specific primers in the RT-PCR assay as described.
RESULTS
We initially carried out experiments to evaluate the optimal conditions for determining the effects of salicylate on COX-2 expressions in HUVEC and HFF. We found that HUVEC or HFF in media deprived of FBS for 24 h yielded more pronounced and consistent COX-2 suppressions than cells cultured in FBS (5–20%) (data not shown). Flow cytometric analysis of cells deprived of serum for 24 h revealed that a majority (89 ± 5%) of cells deprived of serum for 24 h were synchronized at G0/G1 phase of cell cycle. All of the experiments were performed on HFF or HUVEC cultured in medium containing 0.5% FBS or 0.5% BSA for 24 h as described in Materials and Methods. To evaluate the effects of aspirin and sodium salicylate on COX-2 mRNA levels, we pretreated cultured HUVEC with either compound at increasing concentrations for 30 min before maximally inducing COX-2 mRNA expression with 1 ng/ml IL-1β or 100 nM of PMA for 2 h. COX-2 mRNA levels were determined by a quantitative cPCR assay and Northern blot analysis. Fig. 1 shows representative results of concentration-related suppression of IL-1-induced COX-2 mRNA by aspirin and sodium salicylate (Fig. 1a) and PMA-induced COX-2 mRNA by sodium salicylate (Fig. 1b). PMA-induced COX-2 mRNA levels were similarly suppressed by aspirin (data not shown). Aspirin and sodium salicylate appeared to be equipotent in suppressing COX-2 mRNA induction by IL-1β and PMA. The inhibitory effect of aspirin and sodium salicylate was evident at a relatively low concentration, i.e., 10−7 M, and was more pronounced with increasing aspirin and sodium salicylate concentrations (Fig. 1c). COX-2 mRNA levels induced by IL-1β or PMA were reduced by ≈70% at 10−4 M of either salicylate compound (Fig. 1c). This concentration range (10−7 to 10−4 M) is consistent with the plasma salicylate concentrations in individuals taking therapeutic doses of aspirin or sodium salicylate. The PCR assay results were corroborated by Northern blot analysis. In both HUVEC and HFF, aspirin and sodium salicylate at 10−5 M markedly reduced the density of the COX-2 mRNA band (Fig. 2 a and b). Again, the potency of aspirin and sodium salicylate was comparable.
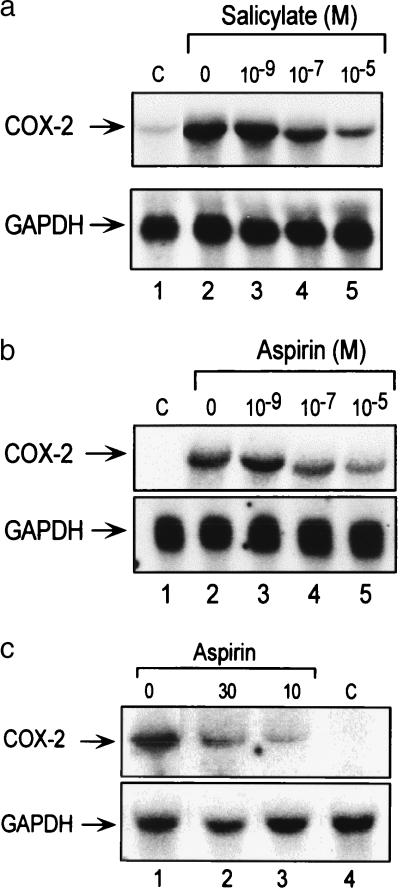
Figure 2.
Analysis of human COX-2 mRNA in cultured cells (a and b) by Northern blotting and murine COX-2 mRNA (c) by RT-PCR. a shows the effect of sodium salicylate at concentrations up to 10−5 M on HUVEC COX-2 mRNA induced by PMA. Concurrent glyceraldehyde 3-phosphate dehydroxygenase (GAPDH) mRNA blotting was included as the control. b shows the effect of aspirin on HFF COX-2 mRNA induced by PMA. c shows the in vivo effects of aspirin at dosages of 10 and 30 mg/kg on murine macrophage COX-2 mRNA expression induced by LPS. Lanes: 1–3, LPS-treated mice in the absence (lane 1) and presence (lanes 2 and 3) of aspirin; 4, normal mouse peritoneal macrophages. Glyceraldehyde 3-phosphate dehydroxygenase (GAPDH) mRNAs, which were included to serve as internal controls, are shown in the lower panel.
The in vivo effect of aspirin on COX-2 mRNA expression was evaluated further in a mouse model. Mice were given aspirin at 10 and 30 mg/kg body weight via a gastric route, followed by i.v. injection of LPS. These two doses of aspirin are estimated to produce a blood level of salicylate equivalent to that of human therapeutic doses of aspirin for treating inflammation. Peritoneal macrophage COX-2 mRNA was undetectable in untreated mice, was induced by LPS, and was suppressed by both doses of aspirin (Fig. 2c). Densitometric estimate of the extent of expression revealed that 10- and 30-mg aspirin dosages reduced the macrophage COX-2 mRNA level by 78 and 63%, respectively.
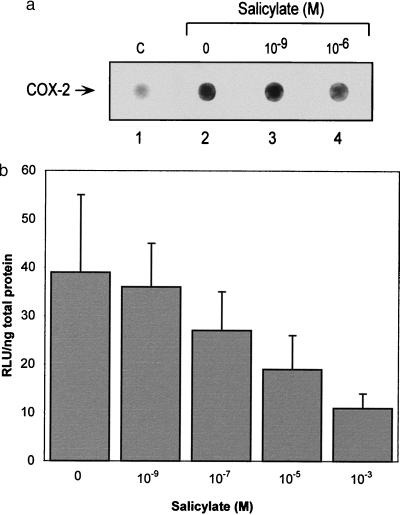
To determine whether reduction of COX-2 mRNA levels by salicylate may be mediated by blocking the PMA-induced COX-2 transcription, we measured nascent COX-2 transcript synthesis by nuclear run-off analysis. Sodium salicylate at 10−6 M significantly reduced the nascent transcript synthesis (Fig. 3a). Sodium salicylate had no effect on COX-2 mRNA degradation (data not shown). These results are consistent with suppression of COX-2 transcription. COX-2 promoter activity effected by salicylate in HUVEC and HFF was determined. It has been shown that the −891 to +9 region of COX-2 gene confers PMA and IL-1 inducible promoter activity (13). We constructed this fragment into a promoterless luciferase expression vector and expressed it in HUVEC or HFF. A basal promoter activity was detected in both types of cells. PMA increased the promoter activity by ≈2.5-fold over the basal activity (Fig. 3b). In this region, there is a consensus NF-κB site at −213 to −222 and another site at −438 to −447 (13). Site-directed mutation of either or both sites in this region did not dampen the induction of COX-2 by PMA (data not shown). Sodium salicylate suppressed the PMA-induced COX-2 promoter activity in a concentration-dependent manner (Fig. 3b). When the basal activity was subtracted, salicylate reduced the COX-2 promoter activity induced by PMA by ≈40% at 10−5 M and ≈63% at 10−3 M. 5′-deletion mutant analysis revealed −192 to +9 to be the minimal promoter fragment that responded to salicylate suppression.
Figure 3.
Effect of salicylate on COX-2 gene transcription. (a) Nuclear run-off experiments showing concentration-dependent suppression of PMA-induced COX-2 transcripts. The circles show the dot blot of COX-2 transcripts. C denotes non-PMA-induced basal control. (b) Effect of sodium salicylate on COX-2 promoter activity transiently expressed in HFF. Each bar represents mean of ±SD of five experiments. The data are statistically significant when analyzed by ANOVA (P < 0.05).
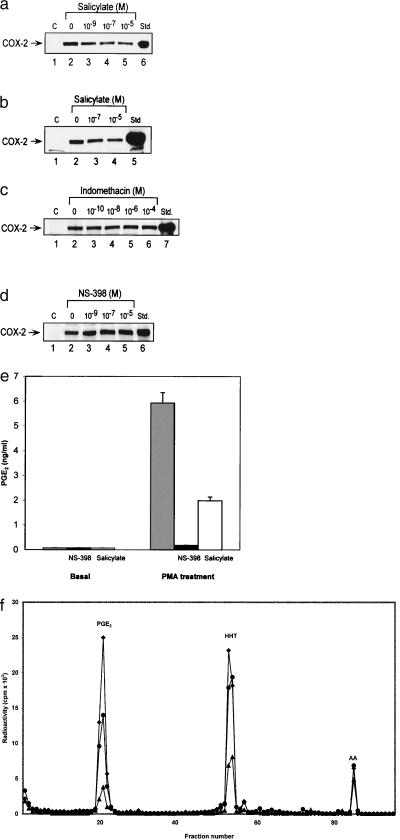
Sodium salicylate suppressed HUVEC and HFF COX-2 protein levels induced by PMA in a concentration-dependent manner (Fig. 4 a and b). Aspirin exhibited a comparable inhibitory effect as sodium salicylate (data not shown). By contrast, neither indomethacin nor NS-398 had a significant effect on COX-2 protein induction by PMA (Fig. 4 c and d). COX-2 activity was highly elevated by PMA treatment, as evidenced by a high level of PGE2 synthesis in HFF, which was blocked by a COX-2 selective inhibitor, NS398 (Fig. 4e). Sodium salicylate at 10−5 M inhibited PGE2 synthesis induced by PMA (Fig. 4e) and prostanoid synthesis in response to exogenous arachidonate in HFF treated with PMA (Fig. 4f). Suppressing effect of salicylate was concentration-dependent. The IC50 value was estimated to be ≈5 × 10−6 M. This value is consistent with that for COX-2 mRNA and protein inhibition. These results indicate that salicylate at therapeutic concentrations inhibits COX-2 activity via suppression of COX-2 protein expression.
Figure 4.
(a–d) Western blot analysis of COX-2 proteins in cultured HFF. (a and b) The effect of sodium salicylate on PMA-induced COX-2 proteins with (a) and without (b) FBS in the medium. (c and d) The effects of indomethacin (c) and NS398 (d) on COX-2 proteins in HFF. Std., purified COX-2 proteins as standards. (e–f) COX-2 activity in cultured HFF. (e) The effect of sodium salicylate (10−5 M) on basal and PMA-induced PGE2 levels. Each bar represents mean ± SD of four determinations. (f) The effect of sodium salicylate (10−5 M) on eicosanoid production in response to [1-14C] arachidonate treatment in HFF stimulated with PMA. The data are representative of two experiments with similar results. ♦ denotes PMA plus vehicle, ● denotes PMA plus sodium salicylate, and ▴ denotes PMA plus NS398.
DISCUSSION
A major finding of this report is that sodium salicylate and aspirin at pharmacological concentrations inhibit COX-2 transcription induced by PMA, IL-1β, and LPS. A close correspondence of concentration-dependent inhibition of COX-2 mRNA and protein levels by sodium salicylate and aspirin is consistent with the interpretation that inhibition of COX-2 induction is elicited by salicylic acid. This finding explains why salicylate, although inactive in blocking COX-1 or COX-2 activity, is capable of inhibiting prostaglandin biosynthesis in intact cells and inflammatory exudates (7, 15). By suppressing COX-2 induction, salicylate eliminates the robust PGE2 synthesis induced by inflammatory mediators. Furthermore, our finding provides a plausible explanation for the reported suppression of excretion rates of PGE2 and PGE1 by sodium salicylate and aspirin in healthy human volunteers (16). Thus, salicylate controls synthesis of proinflammatory prostaglandins by a mechanism independent of a direct inhibition of COX activity. By suppressing COX-2 induction, it confers a dynamic control of productions of prostaglandins, notably PGE2 at the inflammatory sites.
The classic work of Vane has established inhibition of prostaglandin synthesis as the paradigm for the action of NSAIDS (1, 17). As COX-2 has emerged as a major culprit responsible for synthesis of proinflammatory prostaglandins, the pharmacological action of most NSAIDS has been explained by inhibiting COX-2 activity. The action of salicylate, however, remains paradoxical. It has comparable antiinflammatory properties as aspirin (2), but, unlike aspirin, it does not inhibit COX-2 activity when added to a purified enzyme (6, 18). By demonstrating COX-2 gene suppression, our finding adds a new dimension to Vane’s paradigm regarding the actions of aspirin and salicylate.
How salicylate at pharmacological concentrations suppresses the transcriptional activation of COX-2 induced by different stimuli is unclear at the present time. Recent studies have shown that salicylate inhibits NF-κB- and AP-1-mediated gene transcription (19, 20). However, these effects of salicylate were observed only at suprapharmacological concentrations (>5 mM); no effect was noted at pharmacological concentrations. Franz and O’Neill (21) have pointed out that salicylate at suprapharmacological concentrations inhibit kinase activities nonspecifically and that the observed inhibition of NF-κB may simply be attributable to nonspecific and toxic properties of high salicylate concentrations. Results from Mitchell et al. (22) and the present study cast doubts about NF-κB inactivation as a major mechanism for COX-2 suppression. However, while this manuscript was under review, Yin et al. (23) reported a selective inhibition of IκB kinase β by aspirin and salicylate at pharmacological concentrations. IκB kinase β and α phosphorylate IκB, thereby causing its degradation, resulting in NF-κB translocation and activation of genes involved in inflammation. Hence, aspirin and salicylate may exert their antiinflammatory actions by suppressing the expression of NF-κB-activated inflammatory genes. It is important to reexamine whether the suppressive effect of salicylate on COX-2 induction is mediated by inhibiting IκB kinase β. Another possible mechanism for the salicylate action may be mediated by peroxisome proliferator-activated receptors. It has been suggested that NSAIDs, notably indomethacin, may exert their antiinflammatory activities through peroxisome proliferator-activated receptor activation (24–26). However, salicylate does not bind and activate either form of peroxisome proliferator-activated receptors (24). It is unlikely that the action of salicylate on COX-2 is mediated by the peroxisome proliferator-activated receptor pathway. Because salicylate is capable of suppressing COX-2 expressions induced by diverse stimuli whose activation of COX-2 transcription involves distinct regulatory elements and transactivators besides NF-κB (27–29), it is possible that salicylate suppresses COX-2 induction by altering a common signaling pathway that reduces the transcriptional activation of COX-2. Further work is needed to define the mechanism and determine whether it is distinct from its action on IκB kinase β.
Our finding may explain the epidemiological observations that aspirin use at relatively low doses reduces human colon cancer (30). COX-2 overexpression in colon epithelial and cancer cells has been linked to a reduced apoptosis (31) and increased metastatic potential and colon cancer growth (32). The beneficial effect of aspirin observed in the epidemiological studies was attributed to inhibition of COX-2 activity, but this was considered to be unlikely because the concentration of aspirin was too low to have a significant inhibition of COX-2 activity. Our results suggest that the anti-colon cancer effect of aspirin could be contributed by suppression of COX-2 expression by salicylate.
O’Sullivan et al. reported suppression of COX-2 expression in rabbit alveolar macrophages induced with LPS by dexamethasone but not by aspirin (33). Mitchell et al. reported inhibition of COX-2 activity by salicylic acid but did not observe suppression of COX-2 mRNA or protein levels by IL-1β in a human pulmonary epithelial cell line A549 (22). Anteby et al. (34), on the other hand, showed that aspirin inhibited COX-2 promoter activities in cultured human primary trophoblasts. The reasons for the discrepancy are unclear, but one potential reason attributes it to different experimental conditions. In our experiments with cultured HFF and HUVEC, we found that the COX-2 suppressing effect of salicylate is more pronounced and consistent in synchronized cells after 24 h of serum deprivation, as contrasted to the reported results (22, 23) from experiments performed in asynchronized cells without serum deprivation. It is speculated that salicylates suppress COX-2 transcription at the early- to mid-G1 phase of the cell cycle. This intriguing hypothesis is being tested in our laboratory.
The inhibitory effect of salicylate on COX-2 expression appears selective and not shared by indomethacin, a nonselective but preferential COX-1 inhibitor, or NS-398, a selective COX-2 inhibitor. The structure–function relationship of salicylate activity remains unclear. It would be important to evaluate the effects of other NSAIDS, selective COX-2 inhibitors, and salicylate analogs and derivatives to understand the structural basis for COX-2 gene suppression. This information will be valuable for designing new drugs to treat diseases caused by COX-2 overexpression.
Acknowledgments
We thank Dr. Karsten Schrör for comments, Dr. Derek Gilroy for performing cell cycle analysis, and Scott Biederman for technical assistance. This work is supported in part by Grants NS-23327 and HL-50675 to K.K.W. from the National Institutes of Health.
ABBREVIATIONS
- COX-2
cyclooxygenase 2
- HUVEC
human umbilical vein endothelial cells
- HFF
human foreskin fibroblasts
- PMA
phorbol 12-myristate 13-acetate
- NSAID
nonsteroidal antiinflammatory drugs: LPS, lipopolysaccharide
- RT
reverse transcription
- gRNA
genomic RNA
- PGE2
prostaglandin E2
References
- 1.Vane J R. Nat New Biol. 1971;231:232–235. doi: 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- 2.Vane J R, Mitchell J A, Appleton I, Tomlinson A, Bishop-Bailey D, Croxtall J, Willoughby D A. Proc Natl Acad Sci USA. 1994;91:2046–2050. doi: 10.1073/pnas.91.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Perkins W, Lee L, Isakson P. Proc Natl Acad Sci USA. 1994;91:12013–12017. doi: 10.1073/pnas.91.25.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan C C, Black C, Boyce S, Brideau C, Ford-Hutchinson A W, Gordon R, Guay D, Hill R, Li C-S, Mancini J, et al. J Pharmacol Exp Ther. 1995;274:1531–1537. [PubMed] [Google Scholar]
- 5.Meade E A, Smith W L, DeWitt D L. J Biol Chem. 1993;268:6610–6614. [PubMed] [Google Scholar]
- 6.Mitchell J A, Akarasereenont P, Thiemermann C, Flower R J, Vane J R. Proc Natl Acad Sci USA. 1993;90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu K K, Sanduja R, Tsai A-L, Ferhanoglu B, Loose-Mitchell D S. Proc Natl Acad Sci USA. 1991;88:2384–2387. doi: 10.1073/pnas.88.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X-M, Tang J-L, Chen X, Wang L-H, Wu K K. J Biol Chem. 1997;272:6943–6950. doi: 10.1074/jbc.272.11.6943. [DOI] [PubMed] [Google Scholar]
- 9.Xu X-M, Tang J-L, Hajibeigi A, Loose-Mitchell D S, Wu K K. Am J Physiol. 1996;270:C259–C264. doi: 10.1152/ajpcell.1996.270.1.C259. [DOI] [PubMed] [Google Scholar]
- 10.Hla T, Neilson K. Proc Natl Acad Sci USA. 1992;89:7384–7388. doi: 10.1073/pnas.89.16.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zembowicz A, Tang J-L, Wu K K. J Biol Chem. 1995;270:17006–17010. doi: 10.1074/jbc.270.28.17006. [DOI] [PubMed] [Google Scholar]
- 12.Tang J-L, Zembowicz A, Xu X-M, Wu K K. Biochem Biophys Res Commun. 1995;213:673–680. doi: 10.1006/bbrc.1995.2184. [DOI] [PubMed] [Google Scholar]
- 13.Tazawa R, Xu X M, Wu K K, Wang L H. Biochem Biophys Res Commun. 1994;203:190–199. doi: 10.1006/bbrc.1994.2167. [DOI] [PubMed] [Google Scholar]
- 14.Masferrer J L, Zweifel B S, Seibert K, Needleman P. J Clin Invest. 1990;86:1375–1379. doi: 10.1172/JCI114850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittle B J R, Higgs G A, Eakins K E, Moncada S, Vane J R. Nature (London) 1980;248:271–273. doi: 10.1038/284271a0. [DOI] [PubMed] [Google Scholar]
- 16.Hamberg M. Biochem Biophys Res Commun. 1972;49:720–726. doi: 10.1016/0006-291x(72)90470-6. [DOI] [PubMed] [Google Scholar]
- 17.Vane J R, Botting R M. Postgrad Med J. 1991;66:S2–S17. [PubMed] [Google Scholar]
- 18.Cromlish W A, Kennedy B P. Biochem Pharmacol. 1996;52:1777–1785. doi: 10.1016/s0006-2952(96)00599-0. [DOI] [PubMed] [Google Scholar]
- 19.Kopp E, Ghosh S. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 20.Dong Z, Huang C, Brown R E, Ma W-Y. J Biol Chem. 1997;272:9962–9970. doi: 10.1074/jbc.272.15.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franz B, O’Neill E A. Science. 1995;270:2017–2018. doi: 10.1126/science.270.5244.2017. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell J A, Saunders M, Barnes P J, Newton R, Belvisi M G. Mol Pharmacol. 1997;51:907–912. doi: 10.1124/mol.51.6.907. [DOI] [PubMed] [Google Scholar]
- 23.Yin M J, Yamamoto Y, Gaynor R B. Nature (London) 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann J M, Lenhard J M, Oliver B B, Ringold G M, Kliewer S A. J Biol Chem. 1997;272:3406–3410. doi: 10.1074/jbc.272.6.3406. [DOI] [PubMed] [Google Scholar]
- 25.Jiang C, Ting A T, Seed B. Nature (London) 1998;391:82–85. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 26.Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra I P, Delerive P, Fadel A, Chinetti G, Fruchart J-C, et al. Nature (London) 1998;393:790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- 27.Inoue H, Yokoyama C, Hara S, Tone Y, Tanabe T. J Biol Chem. 1995;270:24965–24971. doi: 10.1074/jbc.270.42.24965. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. J Biol Chem. 1995;270:31315–31320. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 29.Xie W, Herschman H R. J Biol Chem. 1996;271:31742–31748. doi: 10.1074/jbc.271.49.31742. [DOI] [PubMed] [Google Scholar]
- 30.Thun M J, Namboodiri M M, Heath C W. N Engl J Med. 1991;325:1593–1596. doi: 10.1056/NEJM199112053252301. [DOI] [PubMed] [Google Scholar]
- 31.Tsujii M, DuBois R N. Cell. 1995;83:493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- 32.Tsujii M, Kawano S, DuBois R N. Proc Natl Acad Sci USA. 1997;94:3336–3340. doi: 10.1073/pnas.94.7.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Sullivan M G, Huggins E M, McCall C E. Biochem Biophys Res Commun. 1993;191:1294–1300. doi: 10.1006/bbrc.1993.1358. [DOI] [PubMed] [Google Scholar]
- 34.Anteby E Y, Johnson R D, Huang X, Nelson D M, Sadovsky Y. J Clin Endocrinol Metab. 1997;82:2289–2293. doi: 10.1210/jcem.82.7.4040. [DOI] [PubMed] [Google Scholar]