Abstract
DNA polymerase θ (pol θ) is a nuclear A-family DNA polymerase encoded by the POLQ gene in vertebrate cells. The biochemical properties of pol θ and of Polq-defective mice have suggested that pol θ participates in DNA damage tolerance. For example, pol θ was previously found to be proficient not only in incorporation of a nucleotide opposite a thymine glycol or an abasic site, but also extends a polynucleotide chain efficiently from the base opposite the lesion. We carried out experiments to determine whether this ability to extend from non-standard termini is a more general property of the enzyme. Pol θ extended relatively efficiently from matched termini as well as termini with A:G, A:T, and A:C mismatches, with less descrimination than a well-studied A family DNA polymerase, exonuclease-free pol I from E. coli. Although pol θ was unable to, by itself, bypass a cyclobutane pyrimidine dimer or a (6–4) photoproduct, it could perform some extension from primers with bases placed across from these lesions. When pol θ was combined with DNA polymerase ι , an enzyme that can insert a base opposite a UV-induced (6–4) photoproduct, complete bypass of a (6–4) photoproduct was possible. These data show that in addition to its ability to insert nucleotides opposite some DNA lesions, pol θ is proficient at extension of unpaired termini. These results show the potential of pol θ to act as an extender after incorporation of nucleotides by other DNA polymerases, and aid in understanding the role of pol θ in somatic mutagenesis and genome instability.
Introduction
Insertion of nucleotides opposite DNA adducts is one strategy taken by cells to enhance their probability of survival following DNA damage. Enzymes with “translesion bypass” activity are found in all organisms, from bacteria to mammals [1–3]. Vertebrate genomes harbor at, current count, 14 distinct DNA polymerase genes, each encoding an enzyme specialized for functions that are being gradually unravelled.
A frequent problem for cells is that enzymes normally carrying out chromosome replication can be stalled by the presence of lesions in DNA. Specialized DNA polymerases can help rescue the situation by allowing synthesis over such lesions. This is beneficial for cell survival, though it is sometimes mutagenic. Major subjects that are being investigated include understanding which DNA polymerases are involved in bypass of given lesions, in which cell types, and at what time in the cell cycle. For example, human pol η (POLH) inserts bases efficiently across from UV radiation-induced cyclobutane dimers (CPD) in DNA, while human pol ι (POL ) inserts bases most efficiently across from another UV radiation-induced lesion, the (6–4) photoproduct [(6–4)PP] [4]. It further appears that more than one specialized DNA polymerase might be needed to mediate bypass for some DNA lesions. This necessitates understanding signalling and switching mechanisms for DNA polymerase selection [5,6]. Here, we report some properties of the enzyme pol θ that are relevant to the consideration of its function in cells, and for translesion DNA synthesis in general.
DNA polymerase θ is an A-family DNA polymerase encoded by the POLQ gene in vertebrate cells. There are several indications that pol θ normally participates in DNA damage tolerance. Purified pol θ protein can catalyze DNA synthesis opposite an abasic (AP) site in DNA or a thymine glycol [7]. Mice with disruption of the Polq gene have elevated spontaneous and radiation-induced frequencies of micronuclei [8,9], and viability of the mice is severely compromised by an additional mutation in Atm [9]. Polq-defective mice also have reduced frequencies of somatic hypermutation of immunoglobulin genes [10–12]. Pol θ has both the ability to bypass an AP site and an inherently low fidelity, properties consistent with participation in somatic hypermutation according to current models.
Not only is pol θ proficient in incorporation of a nucleotide opposite a thymine glycol or an AP site in DNA, but it can also extend a polynucleotide chain efficiently from a base opposite the lesion. It is important to know whether this extension ability is restricted only to AP sites and thymine glycol, or whether the ability of pol θ to extend from unpaired termini is a more general property of the enzyme. Consequently, we investigated the ability of pol θ to extend a mismatched primer, and to extend primers opposite UV-radiation induced lesions.
Materials and Methods
Enzyme purification and DNA Polymerase Assays
Recombinant full-length pol θ with a 6X-His tag at the N-terminus and a FLAG-tag at the C-terminus was purified from a sequence-verified expression vector clone using the procedure previously described [7]. The RefSeq accession number for POLQ is NP_955452. Recombinant pol ι was kindly provided by R. Woodgate and E. Frank [13]. Standard reaction mixtures were incubated at 37°C for 10 min and contained 150 fmol of 5’-32P-labeled primers annealed to 30-mer template DNA, 168 fmol of pol θ and 100 mM of each dNTP (unless otherwise noted) in pol θ buffer: 20 mM Tris-HCl (pH 8.75), 4% glycerol, 80 μg/ml BSA, 8 mM MgCl2 and 0.1 mM EDTA. E. coli exo free pol I (Klenow fragment, Kf) was purchased from Promega Corporation, Madison, Wisconsin (catalog number M2181) and the supplied pol I buffer was used for the assays (50 mM Tris-HCl [pH 7.2], 10 mM MgSO4, 0.1 mM dithiothreitol). For Figure 3, the buffer used for both pol θ and pol I was 20 mM Tris-HCl (pH 8.0), 4% glycerol, 80 μg/ml BSA, 8 mM MgCl2, 1 mM dithiothreitol and 0.1 mM EDTA.
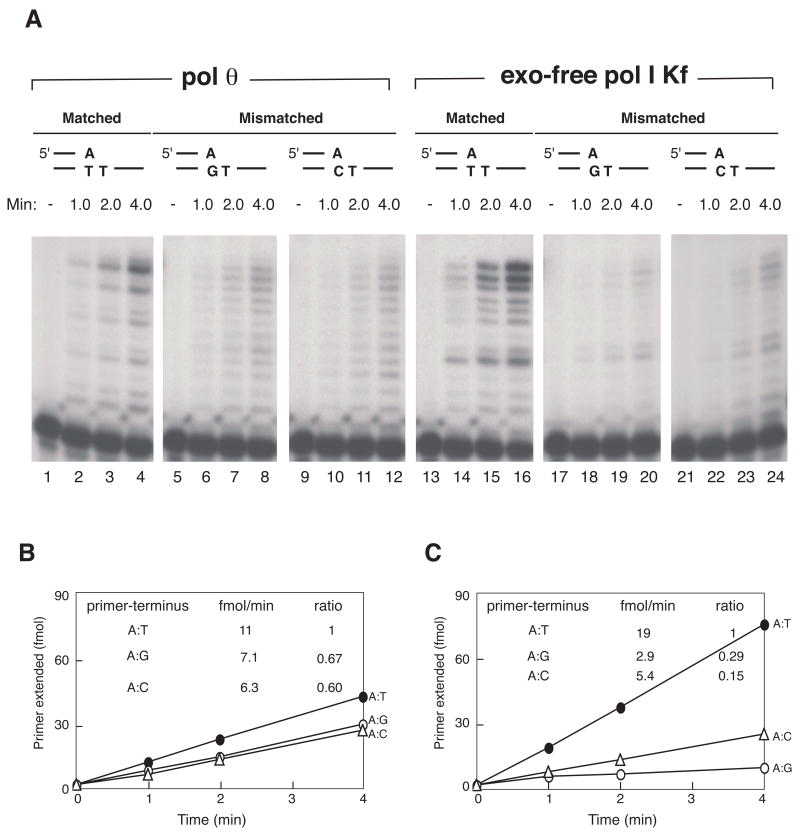
Figure 3. Low discrimination by pol θ for mismatch extension compared to exo-free pol I.
Extension reactions from matched or mismatched primers (same as used in Figs. 1 and 2) were tested under identical conditions at pH 8.0 (see Materials and Methods). (A). POLQ (168 fmol) was incubated with A:T matched (lanes 2–4), A:G mismatched (lanes 6–8) or A:C mismatched (lanes 10–12) primer-termini. Exo-free pol I Kf (9 x 10−4 Unit) was incubated with the same substrates, A:T matched (lanes 14–16), A:G mismatch (lanes 16–20) and A:C mismatch (lanes 22–24). Control experiments without enzymes are shown in lane 1, 5, 9, 13, 17, and 21. Products from part A were quantified for (B), pol θ and (C), exo-free pol I Kf. A: G mismatch (open circle), A:C mismatch (open triangle) and A:T matched (closed circle).
Oligonucleotide Substrates
Primers were purchased from Bio-Synthesis or Sigma Genosys and 5’-labeled by using polynucleotide kinase and γ-32P-[dATP]. Oligonucleotides containing a CPD and a thymine-thymine (6–4) PP were as described [7,14] and were kindly provided by Shigenori Iwai, Osaka University.
Results
Pol θ extends mismatched primer-termini
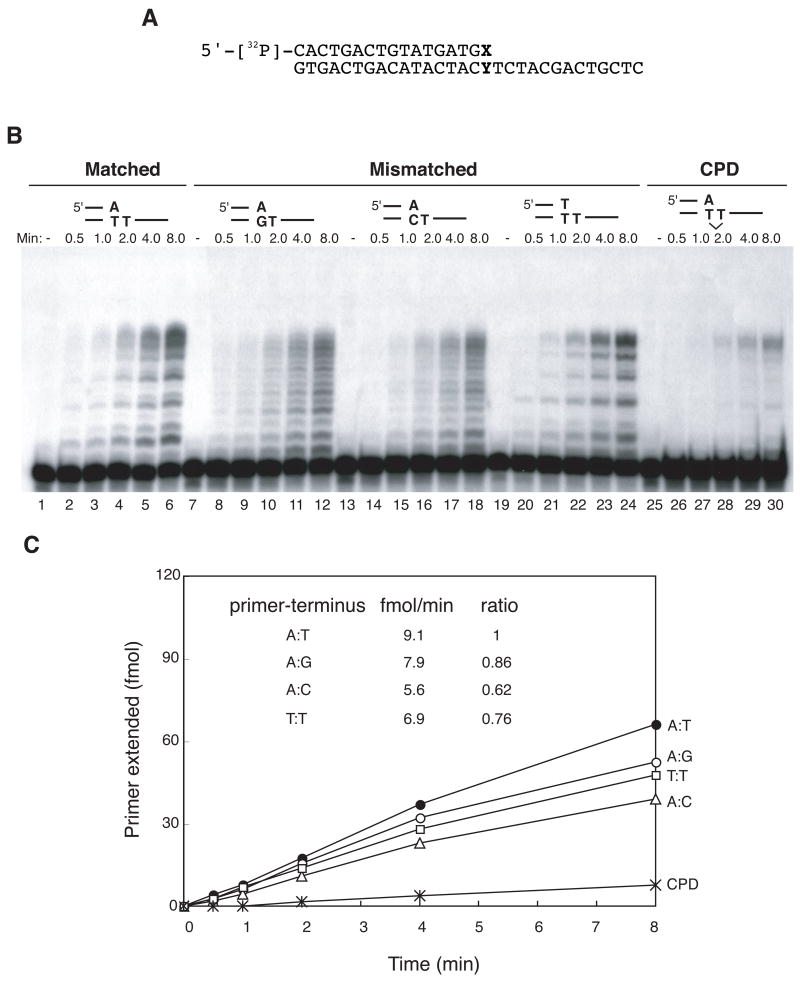
Extension from primer termini having a 3’-mismatch was tested with a 30-mer oligonucleotide template annealed to a 17-mer primer (Fig. 1A). On a matched primer-template (Fig. 1B, lane 1–6), pol θ utilized 9.1 fmol primer/min (Fig. 1C). On primer-termini having A:G or A:C mismatches, pol θ extended relatively efficiently (Fig. 1B, lanes 7–12, 13–18), with ratios of 0.86 and 0.62, respectively, compared to matched template (Fig. 1C). In both these cases, the unpaired base at the primer terminus was A, and the second template base was T. In this instance, primer-template realignment is a conceivable mechanism for extension past a mismatch. Such synthesis can occur with pol κ , for example [15], with the result that the final product length is one nucleotide shorter. No evidence was seen previously for primer/template slippage during translesion synthesis by pol θ [7] and no evidence for this was seen here with mismatched vs. matched primers (Fig. 1B). When the template and primer terminus were also both T to obviate the possibility of pairing-related slippage, the rate of primer-template utilization was 0.76, comparable to the other mismatches tested (Fig. 1B, lane 19–24).
Figure 1. Pol θ efficiently extends mismatched primer termini and weakly from an A opposite the first T of a CPD.
A) Substrate used for the assay. A 5’-32P-labeled 17-mer was annealed to 30-mer DNA. The primer terminus is denoted by X and the template base opposite the primer terminus by Y.
B) Extension from primers having a mismatch. Pol θ (168 fmol) was incubated in pol θ buffer for the indicated times with the following substrates, A:T matched (lanes 2–6), A:G mismatch (lanes 8–12), A:C mismatch (lanes 14–18) and T:T mismatch (lanes 20–24). Extension from an A opposite the first T of a CPD was also tested (lanes 24–30). Control experiments without pol θ are shown in lane 1, 7, 13, 19, and 25. Reaction products were separated by electrophoresis on a 20% polyacrylamide DNA sequencing gel and an autoradiograph is shown.
C) Quantification of the extension reactions shown in B). Extended products were quantified with a Fuji PhosphoImager and Image Analyzer software. A: G mismatch (open circle), A:C mismatch (open triangle), T:T mismatch (open square) and A:T match (closed circle). Rates of primer-terminus utilization (from data for the first 4 min of incubation) are shown in the inset, with the ratio calculated relative to the matched terminus.
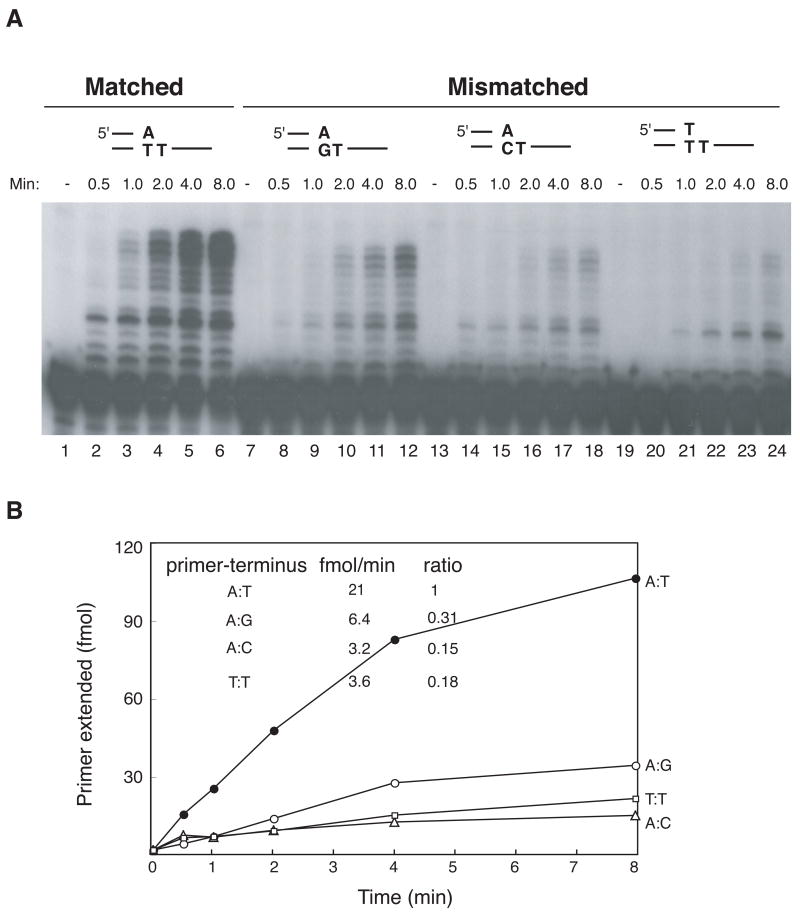
Parallel experiments were done with exo-free pol I. Exo-free pol I is recombinant E. coli pol I Klenow fragment lacking 5’ to 3’ exonuclease, and carrying an additional mutation conferring 3’ to 5’ exonuclease deficiency. Exo-free pol I was chosen for comparison wtih pol θ because both are A-family DNA polymerases (pol I is the founding member) and neither protein has exonuclease activities. The Klenow fragment of pol I is able to extend mismatches to some extent in some sequence contexts [16]. In the present experiments, exo-free pol I discriminated considerably better between paired and mispaired termini than did pol θ (Fig. 2A and B). Exo-free pol I utilized mismatched primer-templates at a 3–4 fold lower relative ratio compared to a matched primer-template (ratios of 0.31, 0.18, and 0.15 for A:G, T:T, and A:C mismatches).
Figure 2. Discrimination against extension of mismatched termini by exo-free Klenow fragment of E. coli pol I.
A) Extension from primers having a mismatch. Exo-free Klenow fragment of E. coli pol I (9 x 10−4 Unit) was incubated in pol I buffer with the same substrates as in Fig. 1: A:T match (lanes 2–6), A:G mismatch (lanes 8–12), A:C mismatch (lanes 14–18) and T:T mismatch (lanes 20–24). Control reaction mixtures without DNA polymerase are in lanes 1, 7, 13, and 19.
B) Quantification of the extension reactions. A:G mismatch (open circle), A:C mismatch (open triangle), T:T mismatch (open square) and A:T match (closed circle). Rates of primer-terminus utilization (from data for the first 4 min of incubation) are shown in the inset, with the ratio calculated relative to the matched terminus.
The experiments in Figs. 1 and 2 used optimized reaction conditions for each enzyme. As DNA conformations can change with variations in buffer pH and cation concentrations, extension of matches and mismatches was compared with the two enzymes under exactly the same buffer conditions (Fig. 3). A very similar relative discrimination between matched and mismatched termini was seen, with the primer extension ratios indicated in Fig. 3B and C.
Extension from primer-termini opposite a (6–4) photoproduct
As pol θ efficiently extends from a primer-terminus opposite an AP site or a thymine glycol, extension was tested for UV radiation-induced photoproducts, which can be bypassed by some other specialized DNA polymerases. Pol θ cannot insert a nucleotide opposite the first base of a cyclobutane pyrimidine dimer (CPD) [7]. However, it was able to perform some extension of a primer with an A residue placed opposite the first T of a CPD (Fig. 1B, lane 25–30), with 5% of the primer utilized (Fig. 1C).
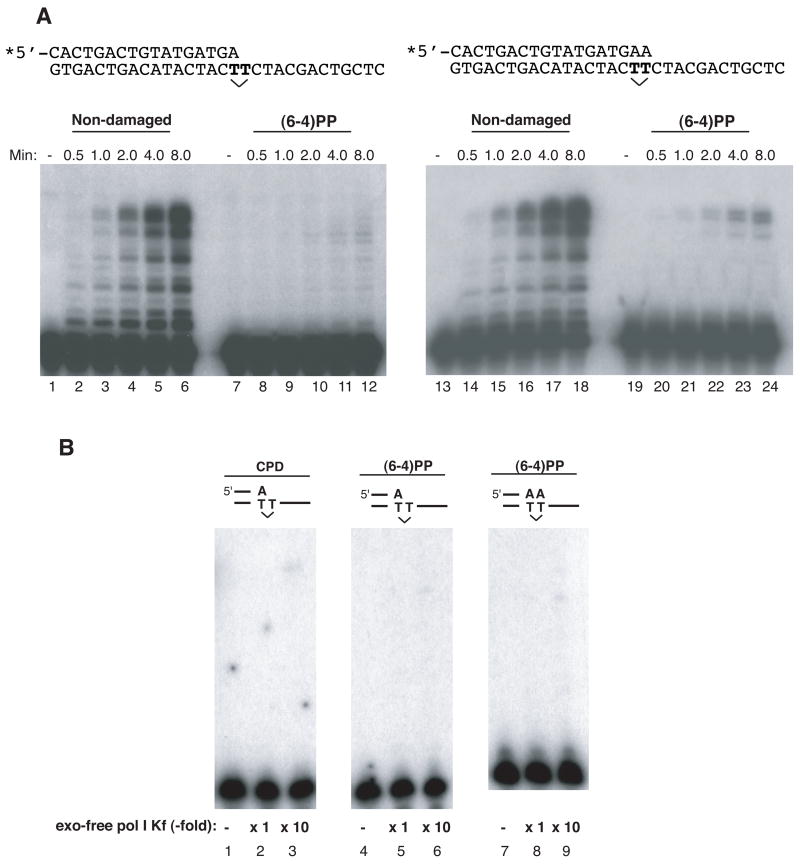
The possibility was tested that pol θ may act at a (6–4)PP in a DNA template (Fig. 4). Very weak extension (1% of primer utilized) was seen when the primer had its 3’ end opposite the first template T (3’ T) of a (6–4)PP (Fig. 4A, lanes 7–12; compare with nondamaged template in lanes 1–6). When the primer had its 3’end opposite the second template T of the (6–4)PP lesion, more extension to the end of the template was possible (Fig. 4A, lanes 19–24). In neither case were extension reactions stimulated by PCNA (data not shown).
Figure 4. Pol θ extends from an A opposite the second T of a (6–4) photoproduct.
(A) A 17-mer (left, lanes 1–12) or an 18-mer (right, lanes 13–24) of the DNA sequence shown was labeled with 32P at the 5’ end (denoted by the asterisk) and annealed to template 30-mer DNA containing either a (6–4)PP at a TT sequence, or normal TT bases at the same sequence (undamaged). Extension from an A opposite a T of an undamaged template (lanes 2–6) or the first T of a (6–4)PP (lanes 8–12); extension from an A opposite the T of an undamaged template (lanes 14–18) or the second T of a (6–4)PP (lanes 20–24). A time course assay was performed and control experiments without pol θ are shown in lanes 1, 7, 13, and 19.
(B) exo-free Klenow fragment of E. coli pol I does not extend from an A opposite a (6–4)PP or a CPD. Extension was tested with 9 x 10−3 Unit (x 1) and 9 x 10−2 Unit (x 10) of enzyme at 37°C for 10 min. Extension reactions from an A opposite the first T of a CPD (lanes 2 and 3), opposite the first T of a (6–4)PP (lanes 5 and 6) and opposite the second T of a (6–4)PP (lanes 8 and 9) were examined. Reaction mixtures with no enzyme are shown in lanes 1, 4 and 7.
In contrast, exo-free pol I could not extend from primers placed opposite a (6–4)PP or a CPD (Fig. 4B), even with 10–100 fold higher enzyme concentrations than used in Figs. 2 and 3.
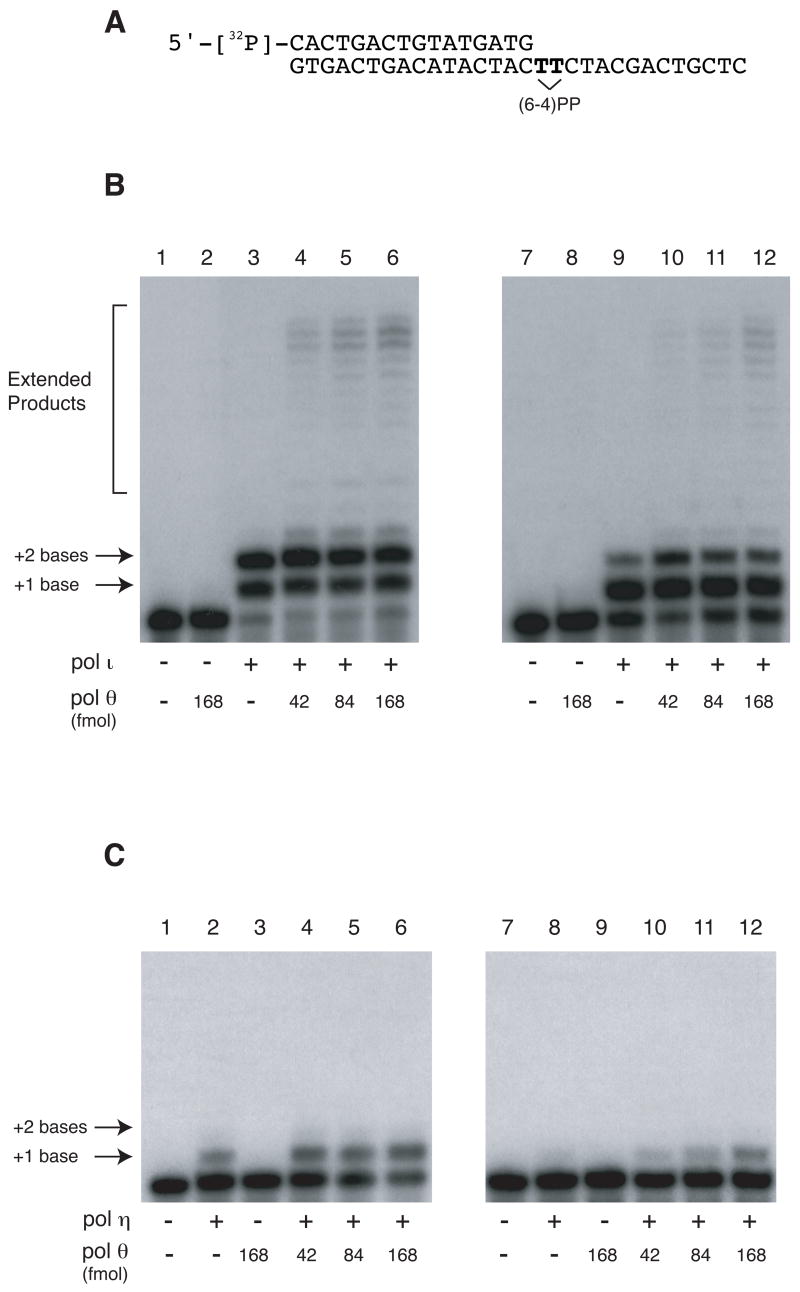
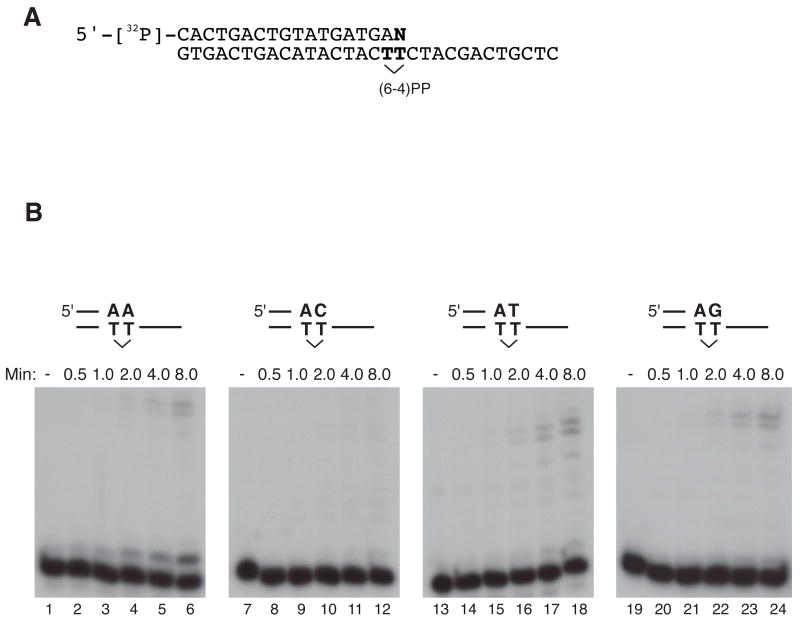
Complete bypass of a (6–4)PP has only been demonstrated in a mixed system with human pol ι , which can insert nucleotides opposite a (6–4)PP, and yeast pol ζ , which can extend the products of insertion [17]. We tested whether a combination of two enzymes encoded by the human genome, pol θ and pol ι , could perform a complete translesion synthesis reaction opposite a (6–4)PP (Fig. 5A). Pol ι alone inserted two bases opposite a (6–4)PP but did not extend further (Fig. 5B, lane 3), as described previously [18]. Pol θ by itself could not insert a base opposite a (6–4)PP (Fig. 5B, lane 2), as reported [7]. However, a combination of the two enzymes mediated incorporation opposite the photoproduct and extension past it (Fig. 5B, lane 4–6), with 15% of the primer extended with 168 fmol pol θ (lane 6). When the amount of pol ι was reduced tenfold, pol ι principally inserted a base opposite only the first T of most (6–4)PP templates (Fig. 5B, lane 9), and there was correspondingly less total extension by pol θ (Fig. 5B, lanes 10–12) with 8% of the primer extended with 168 fmol pol θ (lane 6). This is consistent with the observation that pol θ extends much better from a base opposite the second T of a (6–4)PP than from a base opposite the first (Fig. 4A). Addition of purified PCNA did not stimulate the extension reaction (data not shown). Pol ι has low fidelity when inserting opposite the second template base T of a (6–4)PP [19]. In some DNA sequences, any of the four nucleotides can be inserted [19]. The extension from each of these primer-termini opposite the second template T of a (6–4)PP lesion was examined. Pol θ extended all of these primer termini, with A the best extended and C the least well extended (Fig. 6).
Figure 5. Pol θ catalyzes translesion synthesis past a (6–4) photoproduct together with pol ι but not pol η.
(A) Substrate used for the assay. 5’-32P-labeled 16-mer was annealed to 30-mer DNA containing a (6–4)PP, denoted by the bridged TT. (B) The indicated amounts of pol θ were incubated with 630 fmol of pol ι (lanes 3–6) or 63 fmol of pol ι (lanes 9–12) for 10 min at 37°C. 150 fmol of primer-template was used in each reaction mixture. (C) The indicated amounts of pol θ were incubated with 156 fmol of pol η (lanes 2, 4–6) or 15.6 fmol of pol η (lanes 8, 10–12) under the same conditions as in part A.
Figure 6. Extension of pol θ from primer terminus opposite the second T of a (6–4) photoproduct.
(A) Substrate used for the assay. A 5′-32P-labeled 18-mer was annealed to 30-mer DNA containing a (6–4)PP, denoted by the bridged TT. The 3′ terminal base opposite the second T of the (6–4)PP (denoted by N) was varied. (B) Extension from the primer terminus opposite the second T of a 6–4 PP lesion template at 37°C for 10 min in pol θ buffer. Extension from A opposite the second T of a 6–4 PP (lanes 2–6), extension from C (lanes 8–12), extension from a T (lanes 14–18) and extension from a G (lanes 20–24). Control experiments without enzymes are shown in lane 1, 7, 13 and 19.
A combination of pol η and pol θ was also tested. Pol η alone can insert a base opposite the first T of a fraction of the (6–4)PP (Fig. 5C, lane 2) [19–21]. Adding pol θ (Fig. 5C, lanes 4–6) did not result in detectable extension (about 1% of the products opposite the first T are extended, Fig. 4B). With a ten-fold lower amount of pol η , insertion opposite the first T of a (6–4)PP was enhanced slightly by pol θ (Fig. 5C, lanes 10–12), though there is no evidence that this was catalytically mediated by pol θ itself. However, unlike the combination of pol ι and pol θ , a combination of pol η and pol θ could not bypass a (6–4)PP.
Discussion
The unusual helicase-polymerase domain structure of pol θ distinguishes it from other mammalian DNA polymerases. Although similar to Mus308, a homolog in Drosophila with this domain structure, the function of pol θ appears to be biologically different [7]. Polq mutant mice have an elevated level of spontaneous and radiation-induced micronuclei in reticulocytes [8,9]. POLQ is highly expressed in hematopoietic cells and tissues, including the spleen and germinal centres [9,22,23]. A knockout of POLQ in the chicken DT40 B-cell line shows hypersensitivity to H2O2, indicating that pol θ is involved in tolerance of damage caused by reactive oxygen species. CH12 mouse B lymphoma cells with a knockdown of pol θ DNA polymerase activity showed elevated sensitivity to UV irradiation, mitomycin C, cisplatin, etoposide, γ-irradiation and MMS [24]. It is important to explore the biochemical capabilities of the pol θ enzyme in order to fully understand the detailed function of pol θ in maintaining genome stability.
The data reported here show that in addition to its ability to insert nucleotides opposite some DNA lesions, pol θ is proficient at extension of unpaired termini, including termini arising from incorporation opposite certain lesions. Pol θ is unusual in this respect compared to other DNA polymerases, and it is worth briefly considering the implications for function of the enzyme.
Somatic hypermutation
Several studies suggest that pol θ is involved in somatic hypermutation, a process to generate antibodies [10–12]. Recent models for somatic hypermutation consider two major pathways. One is mutagenesis at G:C base pairs, potentially involving translesion synthesis reactions opposite AP sites created by excision of a uracil residue arising by deamination of cytosine. A second pathway of mutagenesis at A:T base pairs may involve low-fidelity DNA synthesis by one or more specialized DNA polymerases. Both A:T mutagenesis and G:C mutagenesis are reported to be reduced by ~20% in Polq-deficient mice [12]. Polh deficiency results in ~80% reduction in mutagenesis at A:T base pairs. A combination of Polq and Polh deficiency does not result in further reduction of somatic hypermutation, suggesting that pol η and pol θ may work in the same pathway of somatic hypermutation [25]. Possibly, pol θ participates in the extension from a primer having a mismatch generated by pol η.
Mismatch extension and translesion DNA synthesis
In order to complete translesion synthesis reactions opposite some lesions, a concerted action is needed between any DNA polymerase that inserts a base and a DNA polymerase that extends the incorporation past a lesion. A (6–4)PP represents a particular challenge among common environmentally-induced DNA lesions. It substantially distorts DNA and no single DNA polymerases in human cells can complete the translesion synthesis reaction alone. Both of the Y-family enzymes pol ι and pol η are involved in bypass and associated mutagenesis by UV radiation [26–29]. Pol ι is the only bypass DNA polymerase in human cells that correctly inserts an A opposite the first T of (6–4)PP; insertion opposite the second T of a (6–4)PP is error-prone [19]. Pol η preferentially inserts a G opposite the first T of (6–4)PP lesion with lower efficiency [19–21].
Pol θ both inserts a base opposite an AP site and efficiently extends from primer-termini opposite AP sites and thymine glycol adducts [7]. This strong extender activity of pol θ prompted a test of extension from mispaired termini and bases opposite UV photoproducts. The DNA polymerases in human cells catalyzing extension reactions from primer termini opposite a (6–4)PP have not been identified. Because (6–4)PP are avidly repaired by nucleotide excision repair, DNA replication forks may rarely encounter a (6–4)PP in normal cells. However, a combination of human pol ι and yeast Saccharomyces cerevisiae pol ζ can complete translesion DNA synthesis opposite a (6–4)PP in vitro [17]. It is not yet known whether human pol ζ can also participate in such a reaction. Thus, pol θ is the first human bypass DNA polymerase known to catalyze an extension reaction from primer termini opposite a (6–4)PP.
Acknowledgments
We are grateful to Roger Woodgate and Ekaterina Frank (NICHD, NIH, USA) for providing purified human DNA polymerase ι , Shigenori Iwai (Osaka University, Japan) for providing the oligonucleotide containing a (6–4)PP, and members of our laboratory for discussion. This work was supported by NIH grant CA101980 to RDW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 2.Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 4.Vaisman A, Lehmann AR, Woodgate R. DNA polymerasesη and ι. Adv Protein Chem. 2004;69:205–228. doi: 10.1016/S0065-3233(04)69007-3. [DOI] [PubMed] [Google Scholar]
- 5.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase ss contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci U S A. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J. Absence of DNA polymerase θ results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Tissier A, McDonald JP, Frank EG, Woodgate pol R. ι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 14.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase χ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 16.Minnick DT, Bebenek K, Osheroff WP, Turner RM, Jr, Astatke M, Liu L, Kunkel TA, Joyce CM. Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment) J Biol Chem. 1999;274:3067–3075. doi: 10.1074/jbc.274.5.3067. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 18.Vaisman A, Frank EG, McDonald JP, Tissier A, Woodgate R. pol ι-dependent lesion bypass in vitro. Mutat Res. 2002;510:9–22. doi: 10.1016/s0027-5107(02)00248-8. [DOI] [PubMed] [Google Scholar]
- 19.Vaisman A, Frank EG, Iwai S, Ohashi E, Ohmori H, Hanaoka F, Woodgate R. Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase ι. DNA Repair (Amst) 2003;2:991–1006. doi: 10.1016/s1568-7864(03)00094-6. [DOI] [PubMed] [Google Scholar]
- 20.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase ζ in the bypass of a (6–4) TT photoproduct. Mol Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki M, Marini F, Wood RD. POLQ (Pol θ) a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, O-Wang J. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 24.Ukai A, Maruyama T, Mochizuki S, Ouchida R, Masuda K, Kawamura K, Tagawa M, Kinoshita K, Sakamoto A, Tokuhisa T, O-Wang J. Role of DNA polymerase θ in tolerance of endogenous and exogenous DNA damage in mouse B cells. Genes Cells. 2006;11:111–121. doi: 10.1111/j.1365-2443.2006.00922.x. [DOI] [PubMed] [Google Scholar]
- 25.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, O-Wang J. DNA polymerases η and θ function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007 Apr 20;282 doi: 10.1074/jbc.M611849200. accession no. 17449470. [DOI] [PubMed] [Google Scholar]
- 26.Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- 27.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, Masutani C, Hanaoka F. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol Cell Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci U S A. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]