Abstract
Exposure to inflammatory agents or cytokines causes the suppression of cytochrome P450 (CYP) enzyme activities and expression in liver and primary hepatocyte cultures. We showed previously that phenobarbital-induced CYP2B protein is down-regulated in primary cultures of rat hepatocytes following exposure to bacterial endotoxin (LPS) in a nitric oxide (NO)-dependent manner. In the present study, we found that CYP2B proteins in primary rat hepatocyte cultures were suppressed more than 60% after 6h treatment with interleukin-1β (IL-1). This effect was NO-dependent, and treatment of cells with the NO-donors (Z)-1-[2-(2-aminoethyl)-N-(2-ammonioethyl)amino] diazen-1-ium-1,2-diolate (NOC-18), S-nitrosoglutathione (GSNO), and S-nitroso, N-acetylpenicillamine (SNAP) also suppressed CYP2B proteins. However, the down-regulation by IL-1 was insensitive to inhibition of cGMP-dependent protein kinases. The down-regulation by IL-1 or NO donors was abolished by treatments with the proteasome inhibitors MG132 and lactacystin that did not affect NO production. The calpain inhibitor E64-d or the lysosomal protease inhibitors NH4Cl and chloroquine did not attenuate the down-regulation of CYP2B by IL-1. Treatment of HeLa cells expressing c-myc-tagged CYP2B1 with NOC-18 down-regulated its expression and enhanced its ubiquitination. Treatment of rat liver microsomes with GSNO caused S-nitrosylation of CYP2B protein, and enhanced the ubiquitination pattern of CYP2B compared to unmodified CYP2B in an in vitro ubiquitination assay. These data are consistent with the hypothesis that NO-dependent CYP2B ubiquitination and proteasomal degradation are dependent on protein modification by reactive nitrogen species.
Cytochrome P450s (CYP2) are products of multigene families and many CYPs are involved in biosynthesis and catabolism of physiologically important active molecules such as steroid hormones, sterol and fatty acids (1). One third of them are involved in clearance of xenobiotic substances, most of which are expressed in the liver. Inflammation and infection cause the down-regulation of enzyme activity and expression of many CYPs leading to decreased drug clearance, elevation of plasma drug levels, and drug toxicity (2). Inflammation and infection can decrease the metabolic clearances of CYP substrates by 20 to 70% (3).
Substantial evidence exists to suggest a role of nitric oxide in the regulation of CYP enzyme activities in inflammation. NO is a free radical molecule with important regulatory roles in vasodilation, neurotransmission, inflammation, and cell signaling (4–6). Inducible nitric oxide synthase (NOS2, iNOS) is induced during infection and inflammation in vivo, and in cell cultures treated with bacterial lipopolysaccharide (LPS) or cytokines (7), resulting in high levels of NO production in the cells. It is known that nitric oxide, and NO donors are capable of inhibiting the catalytic activities of hepatic microsomal P450s (8–14). For example, Chlamydia infection resulted in the reduction of CYP1A- and 2B-related metabolism by 49% in mouse liver (15), which was blocked by NOS inhibitors. Three mechanisms are suggested for inhibition of activity 1) reversible ligation of NO to (primarily ferrous) P450 heme (8); 2) oxidation of P450 protein thiols (16); and 3) nitration of specific tyrosine residues on the enzyme Roberts (17). CYP8A1 (Prostacyclin synthase) and CYP2B1 have been shown to undergo tyrosine nitration by peroxynitrite in cell culture and in vitro, respectively (13,17).
Although it is clear that NO production is not globally responsible for the down-regulation of CYP gene expression that occurs in inflammation and infection (18–20), several reports suggested that inhibition of NOS can attenuate the down-regulation of some hepatic CYP mRNAs or proteins (21–23), or that NO donors can down-regulate some CYP mRNAs (24,25). This evidence is particularly strong for CYP2B proteins (22,23,26). CYP2B6 plays a significant role in human drug metabolism and is involved in the detoxification and/or activation of many toxins and carcinogens (27). CYP2B1 and 2B2, which differ by only three amino acids, are the analogous isoforms in rat and are induced by phenobarbital (28). CYP2B1 mRNA expression predominates over CYP2B2 by factors of about 5- and 150-fold in the livers of untreated and PB-treated rat livers, respectively (29). Our laboratory has shown that CYP2B proteins are down-regulated in primary cultures of rat hepatocytes by two independent mechanisms in response to LPS: NO-independent mRNA suppression at lower concentrations, and NO-dependent protein suppression at higher concentrations (26). The mechanism of this regulation of CYP2B protein by NO derived from NOS enzymes is poorly understood. It has been suggested that the modification of tyrosine and/or cysteine residues by reactive nitrogen species may affect protein stability, enzyme activity or cell signaling (4). S-nitrosylation of iron regulatory protein 2 resulted in ubiquitination of the protein followed by its proteasomal degradation (30). For these reasons, we hypothesized that modification of amino acid residues, of the CYP2B1/2 enzymes by reactive nitrogen species (RNS) is a signal for their ubiquitination and proteasomal degradation. Consistent with this idea, we show here that post-transcriptional down-regulation of native CYP2B1/2 in hepatocytes and of myc-tagged CYP2B1 in HeLa cells are proteasome-dependent, and that CYP2B1 undergoes NO-stimulated ubiquitination in HeLa cells and in an in vitro ubiquitination system.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Interleukin (IL)-1β and interleukin-6 (IL-6) were purchased from Sigma (St. Louis, MO), and GSNO, SNAP, and NOC-18 and ALLN and E64-d were purchased from Alexis Biochemicals (San Diego, CA). NMA, AG, Gliotoxin, chloroquine, Williams media, Krebs-Ringer buffer, collagenase, and other general chemicals were acquired from Sigma (St Louis, MO). Peroxynitrite was purchased from Cayman chemical (Ann Arbor, MI). HA-ubiquitin (HA-Ub), Ub-aldehyde (Ub-CHO), ATP-regenerating system, and proteasome inhibitors, MG132 and lactacystin were purchased from Boston Biochemicals (Cambridge, MA). Antibodies to CYP2B1 and NADPH-cytochrome P450 reductase (CPR) were kindly provided by Drs. James Halpert (University of Texas Medical Branch, Galveston, TX) and Bettie Sue Masters (University of Texas Health Science Center at San Antonio, TX), respectively. Anti-CYP2C11was generated in our laboratory as described previously (31). Anti-Myc and anti-hemagglutinin (HA) antibodies were acquired from Santa Cruz Biotech (Santa Cruz ,CA). The KXUal-eGFP vector was provided by Dr. T. J. Murphy (Emory School of Medicine, Atlanta, GA).
Rat Hepatocyte Isolation and Treatment
Rat hepatocytes were isolated by a two-step in situ collagenase perfusion procedure (19) with minor modification. Male Sprague-Dawley rats (230–280 g) were anesthetized with ketamine-xylazine solution, and then the liver was perfused with 400 to 500 ml of Krebs-Ringer Bicarbonate buffer for 8 to 10 min. This was followed by perfusion with 0.3 mg/ml collagenase type IV for 10 min. The liver was harvested and gently chopped to release cells, which were filtered through 90 μm mesh. Cells were plated on collagen plates with plating medium containing 5% Nu-Serum (Sigma) and then, three hours later, cells were overlaid with new media containing Matrigel (0.234 mg/ml, BD Biosciences, San Jose, CA). After 24 h, the medium was changed to CYP2B medium (Williams E containing 10mM Hepes pH 7.4, 10 nM insulin, 25 nM dexamethasone and 10mg/ml Pen/Strep). Cultures were maintained for 6 to 7 days at 37°C in 5% CO2 with regular media changes. Beginning on day 4, cells were treated with 1 mM PB to induce CYP2B expression, and the inducer was present for the rest of the experiment. Other treatments were begun 48 h after initiation of PB induction, as indicated in Results. At the conclusion of the experiment, media were removed and reserved for NO assay using the Griess reaction (32).
Microsome Preparation, Immunoblotting, and Measurement of Microsomal Activities
Livers of male Sprague-Dawley rats (230–280 g) or hepatocytes were homogenized or sonicated, respectively, in homogenization buffer (0.1 M potassium phosphate buffer, pH 7.4, containing 0.125 M potassium chloride, 1.0 mM EDTA and a protease inhibitor cocktail (Sigma, St Louis, MO), and then centrifuged at 10,000g for 10 min to obtain the post-mitochondrial fraction. This fraction was used for all immunoblot assays unless otherwise noted. For preparation of microsomes, the post-mitochondrial fraction was centrifuged at 160,000g for 45 min. The supernatant from ultracentrifugation was considered the cytosolic fraction and the pellet was washed with microsome storage buffer (0.25M sucrose containing protease inhibitor cocktail) and homogenized in the same buffer. SDS-PAGE and Western blotting were carried out as described by Ferrari et al. (26). Primary antibody (rat CYP2B1 antibody, diluted 1:20,000) was incubated overnight at 4 °C, washed and then horseradish peroxidase (HRP)-conjugated goat-anti-rabbit IgG was incubated for 1 h at room-temperature. Chemiluminescence was detected with ECL substrate (Amersham Pharmacia Biotech) on X-ray film. All assays were performed within a linear range and the intensity of stained bands was measured by Kodak imaging software. Pentoxyresorufin O-dealkylase activity of hepatic microsomes from PB-treated rats was measured from the rate of appearance of the fluorescent product resorufin as described previously (33).
Biotin-Switch Detection of Nitrosylated Proteins
S-nitrosocysteine-containing proteins were specifically detected by the biotin-switch method with minor modification (34,35). Briefly, hepatocyte microsomes were incubated with HEPES-EDTA buffer containing 0.1 mM neocuproine, 0.4% CHAPS, and 20 mM methyl methanethiosulfonate at 50 °C in the dark for 20 min to block free unmodified thiols. After removing salts from the reaction, the S-nitrosothiols were biotinylated by reaction with 12.5 mM N-[6-(biotinamido)hexyl]-3-(2 pyridyldithio)propionamide and 0.75 mM ascorbate for 1 h at room temperature. The biotinylated protein was pulled down with immobilized Neutravidin (Pierce, Rockford, IL) and separated on SDS-PAGE for immunoblotting.
In Vitro Ubiquitination Assay
The in vitro ubiquitination assay was carried out by the method of Song and DeBose-Boyd (36) with modifications. For preparation of the cytosolic fraction, livers of male Sprague-Dawley rats (230–250 g) were excised, cut into small pieces, and homogenized in ice-cold 50 mM HEPES-KOH buffer, pH 7.2, containing 250 mM sorbitol, 70 mM potassium acetate, 5 mM potassium EGTA, 2.5 mM magnesium acetate, and a protease inhibitor cocktail (P8340, Sigma). Homogenates were centrifuged at 12,000 x g for 10 min, then the supernatants were subjected to centrifugation at 186,000 x g for 45 min. This was repeated, and the final cytosolic supernatant (80 mg/ml protein) was aliquotted and stored at −80 °C. For experiments, tubes were thawed in a 37 °C water bath and placed on ice until use.
For preparation of microsomes, male Sprague-Dawley rats (230–250 g) were injected i.p. with 100 mg/kg PB daily for three days, and microsomes were prepared 24 h after the last injection from the livers as described above. Microsomes (20 μg) were treated with the indicated chemicals and then incubated with 10 μl Cytosolic fraction, 6 μl of Ub stock (containing 1 mg/ml HA-Ub and 0.01 mg/ml Ub-CHO, protease inhibitor cocktail (Sigma), 100 μM MG132) and 2 μl of ATP regenerating system at 37 °C for 20 to 30 min. Reactions were terminated by adding 300 μl of 2X IP buffer (0.2 M Tris-Cl, pH 7.5, 1.8% NaCl, 0.2% Sodium deoxycholate, 2% NP-40, 0.2% SDS, and 0.02% NaN2).
Immunoprecipitation
Immunoprecipitations were carried out in 1X IP buffer containing protease inhibitor cocktail and polyclonal CYP2B antibody at 4 °C overnight with continuous mixing. After incubation, 30 μl of ImmunoPure® Immobilized Protein A Plus (Pierce) was added to the incubation mixture and then incubated an additional 2 h at room temperature. The incubation mixtures were washed 4 times with 1X IP buffer by centrifugation, precipitates were boiled at 95 °C for 5 min with SDS-loading buffer, and then analyzed by SDS-PAGE.
Regulation and Ubiquitination of CYP2B1 in HeLa Cells
HeLa cells from the American Type Culture Collection were cultured in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum in 5% CO2 incubator. Full-length rat CYP2B1 and CYP2C11 cDNAs were subcloned into the pCMV-myc vector (Clontech, Mountain View, CA) using the EcoRI and Sal I sites, and named pCMV-2B1 and pCMV-2C11, respectively. The clones were confirmed by DNA sequencing. pCMV-2B1 or pCMV2C11 were transfected, or cotransfected with pCDMA3.1-HA-Ub WT (37) into HeLa cells using Lipofectamine 2000 (Invitrogen) by the manufacturer’s instructions. Briefly, 2 μg of pCMV2B1 and 1 μg of pCDMA3.1-HA-Ub were mixed with 6 μl of Lipofectamine 2000 in Opti-MEM media (Invitrogen) and drop-wise applied onto HeLa cell cultures on 60 mm plates. After 24 h, cells were treated with NOC-18 and/or proteasome inhibitors for the indicated times. Cells were harvested and sonicated for 10 sec in cell lysis buffer (50 mM Tris-Cl, pH 7.5; 0.1% SDS; 0.5% NP-40; 1 mM EDTA). The cell sonicates were centrifuged at 10,000g for 10 min and the pellets discarded. CYP2B1protein in total cell lysates was measured using either an anti-Myc antibody (Santa Cruz) or anti-CYP2B antibody. To immunoprecipitate CYP2B protein, three microliters of monoclonal anti-Myc antibody was added to the supernatant and incubated overnight at 4 °C with continuous mixing. Thirty microliters of ImmunoPure® Immobilized Protein A Plus (Pierce) was added, followed by IP as described above. After SDS-PAGE and blotting, the membrane was incubated with monoclonal anti-HA antibody (1:1000 dilution, Santa Cruz Biotechnology) 4 °C overnight, and goat anti-mouse IgG labeled with HRP was incubated as a secondary antibody. The ubiquitination signal was detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Immunocytochemistry
HeLa cells were cultured on coverslips with 10% FBS/DMEM in a 5% CO2 incubator. Cells were transfected with pCMV-2B1 using Lipofectamine 2000. After 24 h, the cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, and then washed with PBS. The fixed coverslips were incubated with monoclonal anti-myc antibody and rabbit anti-CPR antibodies (1:500 dilution) in PBS containing 0.1% saponin (Sigma) and 0.1% BSA, overnight at 4 °C. After washing three times with PBS, the slides were incubated with Texas Red- and FITC-conjugated secondary antibodies (1:1000 dilution, Jackson Immunoresearch Laboratories, West Grove, PA) for 1 h at room temperature. Following a final wash, coverslips were mounted using Flouromount-G (Southern Biotechnology Associates, AL). Fluorescence images were acquired on a Zeiss LSM510 confocal fluorescence microscope.
RNA extraction and Real-Time PCR
Total RNA was extracted with TRIzol® Reagent (Invitrogen) according to the manufacturer’s instruction. Two micrograms of total RNA were used for cDNA synthesis with the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA), and real-time PCR was carried out with SYBR® GREEN PCR Master Mix (Applied Biosystem) using ABI 7300 RT-PCR equipment. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as the normalization control. The primers for CYP 2B1 were antisense, TCACACCGGCTACCAACCCT; and sense primers, CTGTGGGTCATGGAGAGCTG (38) and the primers for GAPDH were TGCCAAGTATGATGACATCAAGAAG and AGCCCAGGATGCCCTTTAGT. Analysis of real-time PCR were carried out by the ΔΔCt method as described by Livak and Schmittgen (39).
RESULTS
Post-transcriptional CYP2B suppression by IL-1 stimulation
We previously observed that LPS treatment of rat hepatocytes resulted in a rapid NO-dependent down-regulation of CYP2B proteins (26). One experiment with IL-1 as the inflammatory stimulus in that study suggested a similar phenomenon (26). To further probe the mechanism of this regulation, we decided to use IL-1 because in preliminary experiments the magnitude of the response to LPS proved more variable than that of IL-1.
First, we analyzed the time course of CYP2B protein and mRNA regulation by IL-1 in primary rat hepatocytes. Unlike their human homologue CYP2B6, rat CYPs 2B1 and 2B2 have very low basal expression. Therefore, all our experiments were performed in hepatocytes that had been induced with phenobarbital for 48 h prior to treatment, and the inducer was present throughout the experiments. The primary hepatocytes responded to IL-1 with a robust production of NO, as detected by NO2 + NO3 (NOx) in the culture medium (Fig. 1). NOx production was detected within 6 h and achieved near-maximal levels by 12 h of treatment (Fig. 1B). CYP2B protein was significantly down-regulated by 40% at 6 h and reached 10% of control within 24 h of treatment (Fig. 1A and B). In contrast CYP2B1 mRNA down-regulation was not significantly affected after 6h or 12 h of treatment, and was maximally suppressed to about 41% of control cultures at 24 h (Fig. 1B). Thus, the down-regulation of CYP2B protein was observed prior to CYP2B1 mRNA down-regulation, demonstrating that the rapid IL-1 - stimulated CYP2B down-regulation is post-transcriptional.
FIGURE 1. Rapid down-regulation of CYP2B proteins in response to IL-1.
After 48 h treatment with 1 mM PB, hepatocytes were treated with fresh medium containing PB without (Con) or with IL-1 (5 ng/ml). Cells were harvested at the indicated time after IL-1 treatment. A. Western blot of CYP2B protein from post-mitochondrial fractions. B. Relative levels of CYP2B proteins and CYP2B1 mRNA in the cells as well as NOx levels in the media. NOx production was measured by Griess reaction. For NOx, a relative level of 100 corresponds to 42.5 μM in the media from the IL-1 24 h sample. Values represent the means ± SEM, of three individual cell culture samples per experimental group. a, significantly different from control, p<0.05 (t-test).
NO dependence of IL-1-stimulated down-regulation of CYP2B proteins
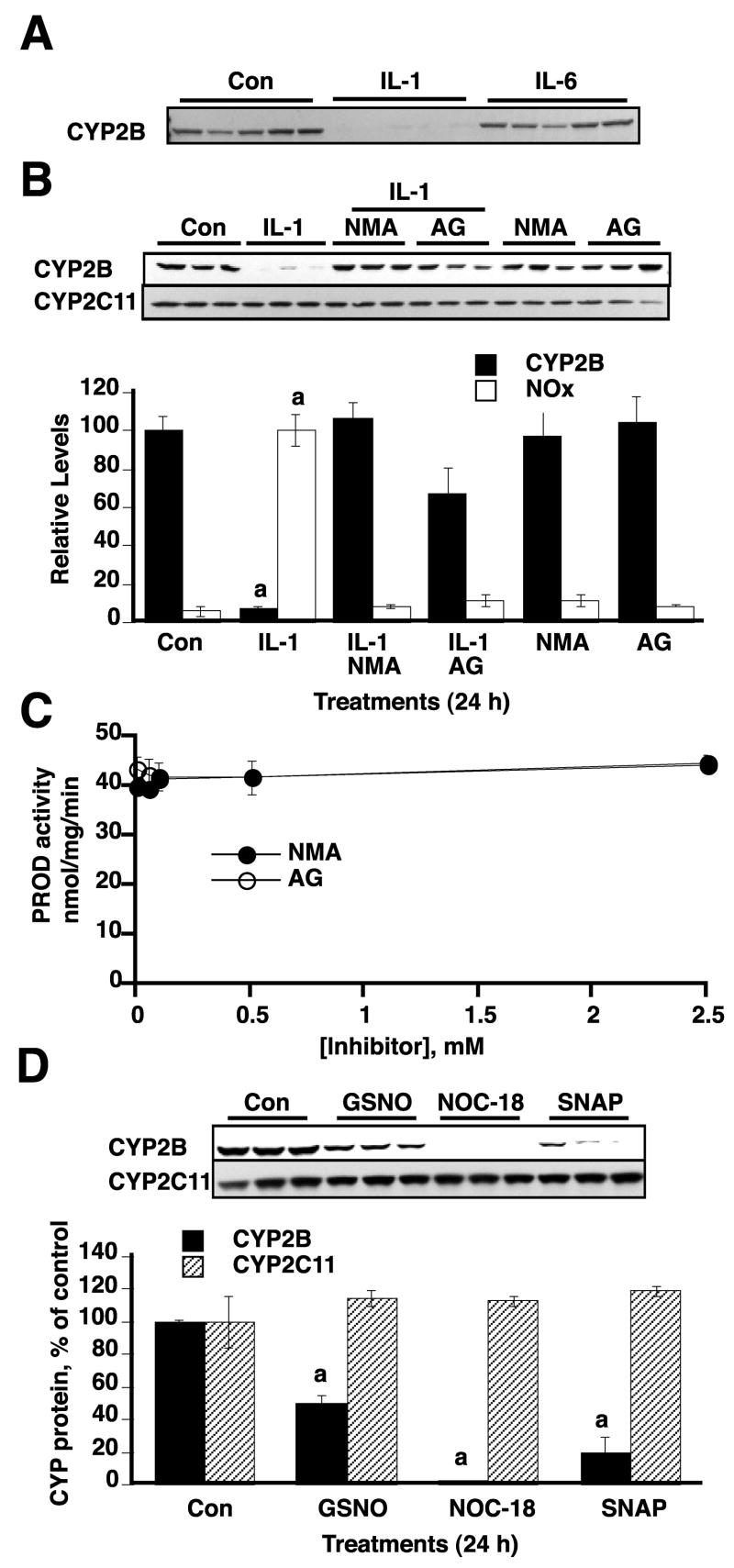
We next analyzed whether CYP2B protein down-regulation by IL-1 is NO-dependent. As a control, we also measured CYP2C11, a protein that we have previously shown is down-regulated by a slower, pretranslational and NO-independent process (19). Cells were treated with the inflammatory cytokines, IL-1 and IL-6. In our experimental system, IL-1 stimulated NO production (see fig. 1B), while IL-6 did not cause NO production within 24 h in primary hepatocyte culture (not shown). The CYP2B protein was down-regulated only in IL-1 treated samples, but not by IL-6 treatment (Fig. 2A). Treatment of cells with the NOS inhibitors NMA and AG resulted in complete blockade of NO production in IL-1 -stimulated samples (Fig. 2B). These treatments also resulted in complete inhibition of IL-1- stimulated CYP2B protein down-regulation, suggesting that IL-1-stimulated CYP2B down-regulation is NO-dependent. In contrast, none of the treatments affected CYP2C11 protein (Fig. 2B). Since some CYP enzymes can be stabilized by ligand binding, we tested the abilities of AG and NMA to inhibit the pentoxyresorufin O-dealkyase activity (a selective CYP2B substrate (40)) of microsomes from PB-treated rat liver. Neither NMA nor AG inhibited this activity at concentrations up to 2.5 mM (Fig. 2C). Thus, NMA and AG did not inhibit CYP2B1 degradation by binding to and stabilizing the protein.
FIGURE 2. NO-dependent down-regulation of CYP2B proteins.
After 48 h treatment with 1 mM PB, hepatocytes were treated with fresh medium containing PB and the indicated inflammatory cytokines, NO donors and/or NOS inhibitors. Cells were harvested 24 h after treatment. A. CYP2B Western blots of samples from cells treated with vehicle, IL-1 (5 ng/ml), or IL-6 (10 ng/ml). B. Effect of NOS inhibitors NMA (300 μM) and AG (300 μM) on down-regulation of CYP2B proteins in response to NOS. Western blots of CYP2B and CYP2C11 are shown. The lower panel shows the quantitative analysis of the data together with NOx levels measured in the media. C. Lack of inhibition of CYP2B1 by the NOS inhibitors NMA and AG. CYP2B1-specific pentoxyresorufin O-dealkylase (PROD) activity of hepatic microsomes from PB-treated rats was measured by the rate of appearance of the fluorescent product resorufin. NMA or AG were included in the incubations at the indicated concentrations. Each point is the mean of triplicate determinations. D. The effects of the NO donors GSNO, NOC-18, and SNAP (500 μM each) on CYP2B and 2C11 proteins were assessed by Western blotting. Values in B and D represent the means ± SEM, of three individual cell culture samples per experimental group a, significantly differ from control, p<0.05 (one-way ANOVA and Dunnett’s test).
We also determined the effect of NO donors on CYP2B expression. The NO-donors GSNO, NOC-18, and SNAP each caused CYP2B down-regulation within 24 h of treatment, with NOC-18 being the most efficacious compound, and GSNO the least effective (Fig. 2D). Thus, NO donors mimic the effects of IL-1 and, taken together, our data show that IL-1 - stimulated down-regulation of CYP2Bs is NO-dependent.
Elucidation of the proteases involved in CYP2B down-regulation
Modifications of proteins by reactive nitrogen species can alter protein functions in various ways by changing their enzymatic activities, stabilities, or conformations (4). Our data suggest that high cellular NO levels stimulate the degradation of CYP2B proteins. Therefore, we analyzed the degradation fate of CYP2B by co-treating cells with IL-1 and various protease inhibitors. Induction of iNOS by IL-1 via NFκB is dependent on IκB degradation by the proteasome (41). Therefore, treatment with MG132 at the same time as IL-1 blocked IL-1 - stimulated NO production and CYP2B down-regulation (Fig. 3A). To analyze the direct role of the proteasome in NO-dependent CYP2B degradation evoked by IL-1, some cells were treated with MG132 beginning 3 h after IL-1 treatment to allow initiation of iNOS transcription (and thus NO production). Delayed MG132 treatment resulted in levels of IL-1-stimulated NO production over a 24 h period that were not different from those in the absence of MG132 (Fig. 3A). However, the same treatment blocked the degradation of CYP2B protein in response to IL-1 at 24 h after treatment, (Fig. 3A), or at 6 h after treatment (supplemental Fig. 1) suggesting that IL-1-stimulated CYP2B degradation is proteasome-dependent. This conclusion was tested using more specific proteasomal inhibitors. Both proteasomal inhibitors, MG132 and lactacystin, were added to cells 3 h after IL-1 treatments and showed NO production and blockade of CYP2B degradation stimulated by IL-1 (Fig. 3B). In another experiment (not shown), we found that another specific proteasome inhibitor, gliotoxin, had the same effect as lactacystin. CYP2C11 was not affected by IL-1 stimulation or proteasomal inhibitors (Fig. 3B). Lastly, we treated hepatocytes with the NO donor, NOC-18 in the presence or absence of MG132. Again, NOC-18 treatment stimulated CYP2B1 degradation, and co-treatments with MG132 and NOC-18 prevented this effect (Fig. 3C).
FIGURE 3. Proteasome-dependent down-regulation of CYP2B proteins.
After 48 h treatment with 1 mM PB, hepatocytes were treated with fresh medium containing PB and IL-1 (5 ng/ml), proteasome inhibitors, and/or NO donors as indicated. Cells were harvested 24 h after treatment. A. Effect of simultaneous or delayed addition of MG132 (25 μM) to the cultures. The upper panel is a Western blot of CYP2B proteins. The lower panel shows the quantitative analysis of the data together with NOx levels measured in the media. All co-treatments were simultaneous additions to the media, except for those samples marked MG132, 3 h, in which MG132 was added 3 h after the addition of IL-1. B. MG132 or lactacystin (Lac) were added to the cultures 3 h after treatment with IL-1. C. Effect of proteasome inhibition on down-regulation by an NO donor. Cells were treated with the NO donor NOC-18 (500 μM) in the presence or absence of MG132 (10 μM) and cells were harvested 24 h after treatment. In A and B, values represent the means ± SEM, of three individual cell culture samples per experimental group. a, significantly different from control; b, significantly different from IL-1-treated group, p<0.05 (one-way ANOVA and Tukey’s test).
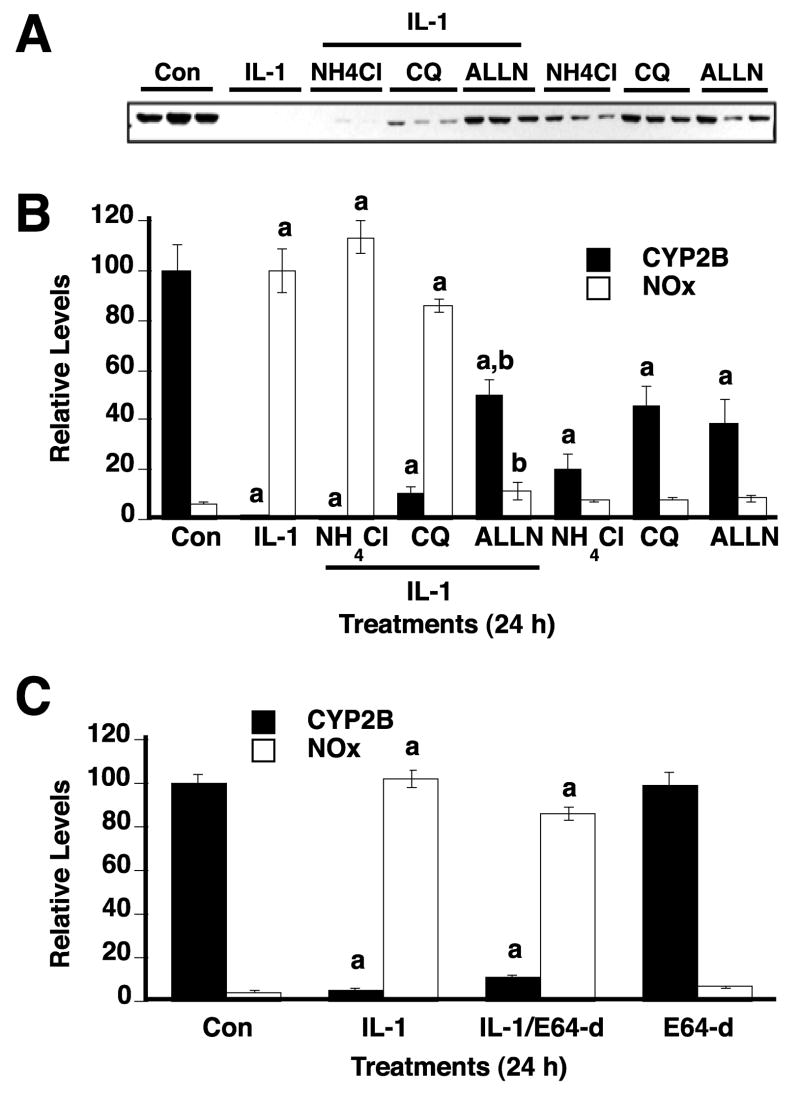
We also evaluated the contribution of calpains and lysosomal proteases to CYP2B degradation by IL-1 stimulation. Cells were treated with IL-1 in the presence or absence of the Ca++ - dependent calpain inhibitor E64-d (42) or lysosomotrophic agents NH4Cl and chloroquine (43). Neither NH4Cl, chloroquine, nor E64-d inhibited NO production in media by IL-1 stimulation (Fig 4B, C), although NH4Cl, and chloroquine caused some cell toxicity as judged by altered cell morphology and release of some cells from the plate. Neither NH4Cl, nor E64-d suppressed the down-regulation of CYP2B (Fig. 4A, B, C). By comparison ALLN, a calpain and proteasome inhibitor (44) did block the effect (Fig 4A, B). Chloroquine partially reversed CYP2B down-regulation by 10%, although this effect was slight compared to those of ALLN (Fig. 4A, B) and the other proteasome inhibitors (Fig. 3).
FIGURE 4. The effects of calpain inhibition or lysosomotrophic agents on CYP2B down-regulation.
After 48 h treatment with 1 mM PB, hepatocytes were treated with fresh medium containing PB and IL-1 (5 ng/ml) and/or the indicated chemicals. Cells were harvested 24 h after treatment. A. Western blot showing the effects of treatment with the lysosomotrophic agents, NH4Cl (20 mM) and chloroquine (CQ, 100 μM), as well as the mixed calpain and proteasome inhibitor ALLN (20 μM) on IL-1-stimulated CYP2B down-regulation. B. Quantitative analysis of the data from panel A together with NOx levels measured in the media. C. Effect of the Ca++ - dependent calpain inhibitor E64-d (20 μM). In B and C, values represent the means ± SEM, of three individual cell culture samples per experimental group.. a, significantly different from control; b, significantly different from IL-1-treated group, p<0.05 (one-way ANOVA and Tukey’s test).
Effect of cGMP-dependent protein kinase inhibition
Many of the biological effects of NO are mediated via stimulation of guanylyl cyclase, synthesis of cGMP, and the resultant stimulation of cGMP protein kinases (45). To test whether NO-dependent CYP2B protein down-regulation is dependent on this mechanism, we used the cGMP-dependent protein kinase inhibitor Rp-cGMPS (46). Rp-cGMPS had no effect on the down-regulation of CYP2B protein, or on NO production, in response to IL-1 (supplemental Fig. 2).
Regulation of Myc-CYP2B1 protein in HeLa cells
We next wanted to determine whether or not the proteasomal degradation of CYP2B in response to NO is preceded by ubiquitination of the protein. Because of the difficulty of detecting the ubiquitination signal in vivo, and limitations of the CYP2B antibodies, we carried out experiments in HeLa cells with heterologously expressed CYP2B1. First, we examined the cellular localization of Myc-CYP2B1 expressed in HeLa cells. Full length WT-CYP2B1 was cloned in a c-Myc-tag expression vector and then transfected into HeLa cells. Immunostaining and confocal microscopy demonstrated a perinuclear staining of the cells with the c-myc antibody, consistent with localization in the endoplasmic reticulum (Fig. 5A). This was confirmed by the co-localization with native CPR in the cells (Fig. 5A). Next, we determined that HeLa cells showed a similar pattern of CYP2B1 down-regulation in response to nitric oxide. NOC-18 was used as the NO donor since HeLa cells did not produce NO in response to IL-1 (data not shown). The expression of CYP2B1 protein and mRNA were measured at various time points after NOC-18 treatments (Fig. 5B). CYP2B1 protein was significantly reduced within 8 h of treatment, and the level of CYP2B1 protein was less than 10% compared to untreated cells at 12 h (Fig. 5B). In contrast, the mRNA levels of CYP2B1 in HeLa cells were not significantly different from control at any time points examined (Fig. 5B). Myc-CYP2B1was down-regulated by NOC-18 treatment in a dose-dependent manner, with an EC50 of ~50 μM at 24 h. The lowest concentration producing a maximal effect was 150 μM and again the level of CYP2B1 was less than 10% compared to control at 24 h (Fig. 5C). The down-regulation of CYP2B1 protein by 18 h of NOC-18 treatment was prevented by MG132 (Fig. 5D). In this experiment, the HeLa cells were co-transfected with Myc-CYP2B1 and KXUal-eGFP vector to account for potential differences in transfection efficiencies among the samples. The levels of eGFP were not statistically different among the treatment groups, suggesting that the effect of NOC-18 was specific for CYP2B1. We also showed that the level of GAPDH and CPR were not different among the treatments (data not shown).
FIGURE 5. CYP2B1 expression and regulation by NO in HeLa cells.
Cells were transfected with myc-CYP2B1 as described in the text. A. Confocal images of transfected myc-CYP2B1 and native CPR proteins in HeLa cells. B. Time course of CYP2B1 down-regulation by NOC-18. 24 h after transfection, HeLa cells were treated with NOC-18 (NOC, 500 μM) for the indicated time and then CYP2B1 mRNA and protein levels were determined by real-time PCR and immunoblotting, respectively. The upper panel shows a Western blot of the samples. The lower panel shows the quantified results, with the data expressed as a percentage of the control group at each time point. a, significantly different from control, p<0.05 (t-test). C. Concentration dependence of NOC-18 effect. 24 h after transfection, HeLa cells were treated with various NOC-18 concentrations. Cells were harvested after 24 h of treatment, and CYP2B1 levels were determined by immunoblotting. D. Effect of MG132 on down-regulation by NOC-18. HeLa cells were co-transfected with myc-CYP2B1 and the KXUal-eGFP vector. 24 h later, the cells were treated with NOC-18 (500 μM) or/and MG132 (10 μM) for 18 h. CYP2B1 and eGFP levels in the cell lysates were analyzed by immunoblotting. Values represent the means ± SEM after normalization to the level of eGFP, of three individual cell culture samples per experimental group. a, significantly different from control; b, significantly different from NOC-18 treated group, p<0.05 (one-way ANOVA and Tukey’s test).
Polyubiquitination of CYP2B1 in HeLa cells
To study the ubiquitination of CYP2B protein in response to NO, pCMV-Myc-CYP2B1 and pCMV-HA-Ub plasmids were co-transfected into HeLa cells, which were then treated for 6 h with NOC-18 and/or the proteasomal inhibitor MG132. After immunoprecipitation of CYP2B1 with an anti-Myc antibody, ubiquitination of CYP2B1 was detected by immunoblotting with anti-HA antibody (Fig. 6). Treatment with NOC-18 alone showed only a slight increase in CYP2B1 ubiquitination. This phenomenon may be a result of rapid degradation of ubiquitinated CYP2B1 via the proteasome. Treatment of cells with MG132 caused an increase in detectable CYP2B1 ubiquitination compared to mock treatment (DMSO treatment), and this was further increased by simultaneous treatment with NOC-18. As a negative control, myc-CYP2C11 did not show enhancement of ubiquitination by NOC-18 and MG132 treatment (data not shown). Down-regulation of total CYP2B1 by NOC-18 was detected in this experiment, reflecting the fact that the onset of down-regulation occurs between 4 and 8 h as seen in Fig. 5.
FIGURE 6. NOC-18-induced polyubiquitination of CYP2B1 protein in HeLa cells.

HeLa cells were co-transfected with myc-CYP2B1 and HA-Ub as described in the text. 24 h after transfection, the cells were treated with NOC-18 (500 μM) and/or MG132 (500μM) for 6 h. CYP2B1 proteins were immunoprecipitated with anti-Myc antibody. A. Ubiquitination was detected by immunoblotting with anti-HA antibody. B. CYP2B1 levels in the immunoprecipitate were detected by immunoblotting with anti-CYP2B1. The experiment was repeated three times with similar results.
Induction of in vitro ubiquitination of CYP2B by NO donors
To further investigate the mechanism of NO-dependent CYP2B ubiquitination, we carried out in vitro ubiquitination assays. Hepatic microsomes from PB-treated rats were treated with GSNO (an S-nitrosylation agent) or peroxynitrite (a tyrosine nitrating agent). We found that GSNO treatment in vitro caused the S-nitrosylation of CYP2B proteins using the biotin-switch method (Fig. 7A). After GSNO and/or peroxynitrite treatment, microsomes were incubated with a rat liver cytosolic fraction containing HA-Ub with or without an ATP regenerating system. As a positive reaction control, the total reaction mixture was immunoblotted with anti-HA antibody (Fig. 7B, left panel). Strong smearing signals of high molecular mass indicative of ubiquitination were detected in the presence of the ATP regenerating system. A detectable but much lower signal was also found in the absence of the ATP-regenerating system, suggesting that endogenous ATP was sufficient to support a low level of ubiquitination. These results confirmed that our in vitro ubiquitination assay system is functional. Next, CYP2B proteins were immunoprecipitated from the reaction mixture with anti-CYP2B antibodies, and the HA-Ub conjugated to CYP2B was detected by immunoblotting with anti-HA antibody. GSNO treated microsomes showed a significant ATP-dependent enhancement of high molecular mass, HA-tagged ubiquitinated CYP2B proteins compared to control microsomes (Fig. 7B, right panel). The peroxynitrite-treated sample also showed a slight increase of HA-Ub-conjugated high molecular mass signals, but co-treatments of GSNO and peroxynitrite did not show significant induction of the HA-Ub signal compared to microsomes treated with GSNO alone.
FIGURE 7. In vitro ubiquitination of CYP2B protein induced by NO donors.
A. In vitro S-nitrosylation of CYP2B by GSNO treatment. Microsomes from PB-induced rat liver were treated 50 μM of GSH or GSNO for 1 h in the dark and then S-nitrosylation was assessed by the Biotin-Switch method as described in Experimental Procedures. After pull-down of the biotinylated protein with Neutravidin, CYP2B proteins were detected by immunoblotting. B. Microsomes from PB-induced rat liver were treated with GSNO (50 μM) and/or peroxynitrite (300 μM) for 1 h in the dark, and then mixed with rat liver cytosolic fraction containing Ub-CHO and HA-Ub with or without ATP for 30 min as described in Experimental Procedures. After the reaction, aliquots of the reaction mixture were either applied directly to the gel, or used for immunoprecipitation of CYP2B proteins with anti-CYP2B antibody. Ubiquitinated proteins were detected by Western blotting with an anti-HA antibody.
DISCUSSION
The work presented here shows that the IL-1-stimulated and NO-dependent rapid degradation of CYP2B1 in rat hepatocytes proceeds via the proteasomal pathway, and that exposure of cells or microsomal CYP2B to NO donors results in increased ubiquitination of CYP2B. These conclusions are supported by the following findings: 1) CYP2B down-regulation in hepatocytes evoked by IL-1 or NO donors is blocked by several different protease inhibitors, but not by inhibitors of calpains or lysosomal proteolysis; 2) NO-stimulated down-regulation of myc-tagged CYP2B1 in HeLa cells is also blocked by proteasome inhibition. 3) The NO donor NOC-18 increases the extent of polyubiquitination of myc-tagged CYP2B1 in HeLa cells; and 4) treatment of microsomes with the nitrosyl donor GSNO or the nitration agent peroxynitrite increases the polyubiquitination of CYP2B proteins in vitro.
Our previous work had conclusively established the role of NO in down-regulation of CYP2B1 in hepatocytes in response to LPS (26). However, the evidence that down-regulation by IL-1 proceeded by the same mechanism was less extensive. Here, we demonstrate that the proteasomal degradation of CYP2B elicited by IL-1 is NO-dependent, and this is supported by strong evidence including: 1) CYP2B protein was down-regulated within 6 hours of treatment, prior to mRNA down-regulation; 2) The down-regulation was blocked by NOS inhibitors AG or NMA; 3) CYP2B protein was unaffected by IL-6 that did not stimulate NO production; 4) NO-donors also down-regulated CYP2B proteins.
Although it was not the goal of this study to determine the origin of the IL-1-induced NO production in hepatocytes, several lines of evidence point to NOS2 as the source of NO. First, NOS2 is the only NOS isoform that is cytokine-inducible in hepatocytes. Second, the selective NOS2 inhibitor AG (47) blocked the production of NO. Third, we reported previously that LY83583, which inhibits iNOS induction rather than its enzymatic activity, blocked both NO production and 2B down-regulation in response to IL-1 (26).
The reported constitutive half-life of CYP2B1 varies by 19 to 25 hours (48,49), and the degradation pathway of CYP2B1 seemed to be in lysosomes based on the presence of CYP2B1 in lysosomal compartments both in the presence or absence of leupeptin treatment of the rats(50,51). This was supported by genetic evidence in yeast, in which transfected rat CYP2B1 was found to be degraded in the vacuolar/lysosomal compartment (52). In contrast, CYP2B1 stably expressed in HeLa cells under control of the tet operon had a constitutive half-life of only 8.7 h and was blocked by proteasomal inhibitors (43). However, this proteasomal degradation was apparently ubiquitin-independent, and was proposed to proceed via the 20S proteasome (43). Thus, it appears that native CYP2B1 protein may be degraded differently in yeast and HeLa cells. Ubiquitination and proteasomal degradation of cumene hydroperoxide-inactivated CYP2B1 has been demonstrated in an in vitro system (53), but has yet to be shown in cell culture. However, our studies show that IL-1, via NO production, stimulates CYP2B1 down-regulation by the proteasome in hepatocytes, and that this is accompanied by ubiquitination in HeLa cells and in an in vitro system from rat liver. This is the first demonstration of the regulated proteasomal degradation of a CYP enzyme by a pathophysiological signal. We previously found that the concentration of IL-1 used in this study, 5 ng/ml, was the lowest concentration that caused optimal induction of the hepatic acute phase genes β-fibrinogen and α1-acid glycoprotein and maximal down-regulation of CYP2C11 in cultured rat hepatocytes (54) It does not change cell viability over a 24 h time course (19). Much higher concentrations (25–30 ng IL-1/g liver) have been detected in the livers of rats undergoing an acute phase response to cecal ligation and puncture (55). These findings are likely to be relevant to the down-regulation of CYP2B1 in the LPS model of sepsis because a) the maximal NOx concentration detected in the culture medium after IL-1 treatment in the present studies was 70 μM, which is considerably lower than the 500 μM concentrations that can be detected in rat plasma after LPS injection (20); b) we showed that CYP2B1 protein can be rapidly down-regulated within 8 h of LPS injection in rats (56).
As demonstrated using the cGMP-dependent protein kinase inhibitor Rp-cGMPS, the down-regulation of CYP2B1 is cGMP-independent. Therefore, the most parsimonious hypothesis arising from our data is that modification of Cys or Tyr residues on CYP2B1 by reactive nitrogen species causes a structural modification of the protein that renders it a good substrate for ubiquitination. There are two other examples of proteins that undergo ubiquitination and proteasomal degradation in response to modification by NO-derived species. NO causes S-nitrosylation of iron regulatory protein 2, which leads to IRP2 ubiquitination, followed by its proteasomal degradation (30). Also, NO stimulated proteasomal activity and the degradation of tyrosine nitrated transferrin receptor via ubiquitination (57). Prior work by Roberts et al showed that the apoprotein (not heme) of purified CYP2B1 was modified by in vitro treatment with 300 μM of peroxynitrite (17) and that stoichiometric nitration on tyrosines 190 and 203 was associated with a corresponding reduction in catalytic activity. Peroxynitrite is formed in vivo by the reaction of NO with superoxide and is a powerful oxidizing agent that can initiate lipid peroxidation, oxidize sulfhydryls, and nitrate the aromatic residues of proteins (4,5,58). Studies with mutated proteins later demonstrated that nitration of Tyr 190 was the cause of the reduced catalytic activity of CYP2B1(59). However, it is not known if nitration is responsible for proteolytic degradation of the enzyme in the cell. It must also be considered whether or not peroxynitrite concentrations sufficient to nitrate the enzyme can be achieved physiologically or pathophysiologically. In our experiments, we have so far been unable to confirm that tyrosine nitration of CYP2B proteins occurs in microsomes treated with peroxynitrite. This may be due to the quality of the antibodies: we tried four different published antibodies but were only able to observe significant induction of any immunoreactive bands in peroxynitrite treated microsomal samples by using antibody that was highly immunopurified by affinity chromatography on a column of 3-nitro-L-tyrosine. However, even with this purified antibody we could not identify CYP2B1 as a nitrated protein.
We were able to detect S-nitrosylation of CYP2B1 by in vitro treatment with GSNO, which is a nitrosonium ion donor mainly causing protein nitrosylation (60) using the biotin-switch method. Our in vitro ubiquitination assay showed that treatment with GSNO is more effective than the nitrating agent peroxynitrite in stimulating the ubiquitination of CYB2B1. This suggests that, if modification of CYP2B1 triggers the ubiquitination and degradation, then Cys modification may be more important than Tyr nitration. So far, we have been unable to reproducibly detect IL-1-stimulated nitrosylation or nitration of CYP2B1 in vivo or in rat hepatocytes. The fast turnover rate of the modified enzyme and further modification of S-nitrosocysteine residues could contribute to these problems. For example, S-nitrosylated residues can be further oxidized to sulfinic and sulfenic acids. The lack of quality antibodies (either S-nitrosylation or nitrotyrosine), and difficulty of immunoprecipitation of CYP proteins (61) is another contributing factor. An alternative approach will be to mutate the putative target amino acid residues and compare the degradation rate to that of the WT protein. Another possibility that we are currently considering is that the NO dependency of the degradation arises not from the modification of the CYP2B proteins, but from NO regulation of the endoplasmic reticulum associated degradation/ubiquitination-proteasomal machinery.
In summary, this work is the first to demonstrate the pathophysiologically regulated, ubiquitin-dependent proteasomal degradation of a CYP enzyme. Our findings add to our limited knowledge of proteins whose ubiquitination and degradation is regulated by NO. Until now, the major focus on regulation of CYP enzyme and drug metabolism during inflammation has been on transcriptional mechanisms. However, this work suggests that attention should be paid to possible post-transcriptional regulation of other CYPs under conditions of inflammation.
Supplementary Material
Footnotes
We thank Kimberly Pierce, Malik Raynor, and Dr. Alison Aitken for expert technical assistance; Dr. T.J. Murphy for the KXUa1-eGFP vector, Dr. James Halpert for antibodies to CYP2B1, Dr. Bettie Sue Masters for antibodies to CPR, and Drs. Keith Wilkinson, Emory University and Dennis Koop, Oregon Health and Science University, for helpful consultations. This work was supported by a grant from the National Institutes of Health (GM066971, E.T.M). C.-M.L was supported by a Training Grant from the National Institutes of Health (T32ES01287).
The abbreviations used are: AG, aminoguanidine; ALLN, Acetyl-L-Leucyl-L-Leucyl-L-Norleucinal; CPR, NADPH-cytochrome P450 reductase; CYP, cytochrome P450; GSNO, S-nitrosoglutathione; HA, hemagglutinin; HRP, horseradish peroxidase; IL-1, interleukin-1β; IL-6, interleukin-6; IP, immunoprecipitation; NO, nitric oxide; NOC-18, (Z)-1-[2-(2-Aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate; NMA, NG-methyl-L-arginine; NOx, nitrate plus nitrite; Rp-cGMPS, Rp isomer of 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphorothioate; SNAP, S-nitroso N-acetylpenicillamine ; Ub, ubiquitin; Ub-CHO, Ubiquitin aldehyde
References
- 1.Guengerich FP. Drug Metab Rev. 2004;36:159–197. doi: 10.1081/dmr-120033996. [DOI] [PubMed] [Google Scholar]
- 2.Morgan ET. Drug Metab Rev. 1997;29:1129–1188. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- 3.Aitken AE, Richardson TA, Morgan ET. Annu Rev Pharmacol Toxicol. 2006;46:123–149. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- 4.Mannick JB, Schonhoff CM. Free Radic Res. 2004;38:1–7. doi: 10.1080/10715760310001629065. [DOI] [PubMed] [Google Scholar]
- 5.Radi R. Proc Natl Acad Sci USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedon PC, Tannenbaum SR. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Geller DA, Freeswick PD, Nguyen D, Nussler AK, Di Silvio M, Shapiro RA, Wang SC, Simmons RL, Billiar TR. Arch Surg. 1994;129:165–171. doi: 10.1001/archsurg.1994.01420260061008. [DOI] [PubMed] [Google Scholar]
- 8.Wink DA, Osawa Y, Darbyshire JF, Jones CR, Eshenaur SC, Nims RW. Arch Biochem Biophys. 1993;300:115–123. doi: 10.1006/abbi.1993.1016. [DOI] [PubMed] [Google Scholar]
- 9.Kim YM, Bergonia HA, Muller C, Pitt BR, Watkins WD, Lancaster JR., Jr J Biol Chem. 1995;270:5710–5713. doi: 10.1074/jbc.270.11.5710. [DOI] [PubMed] [Google Scholar]
- 10.Minamiyama Y, Takemura S, Imaoka S, Funae Y, Tanimoto Y, Inoue M. J Pharmacol Exp Ther. 1997;283:1479–1485. [PubMed] [Google Scholar]
- 11.Eum HA, Yeom DH, Lee SM. Nitric Oxide. 2006;15:423–431. doi: 10.1016/j.niox.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Tunctan B, Yaghini FA, Estes A, Malik KU. Nitric Oxide. 2006;14:51–57. doi: 10.1016/j.niox.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Zou MH, Daiber A, Peterson JA, Shoun H, Ullrich V. Arch Biochem Biophys. 2000;376:149–155. doi: 10.1006/abbi.2000.1699. [DOI] [PubMed] [Google Scholar]
- 14.Vuppugalla R, Mehvar R. J Pharmacol Exp Ther. 2004;310:718–727. doi: 10.1124/jpet.104.065557. [DOI] [PubMed] [Google Scholar]
- 15.Khatsenko OG, Barteneva NS, de la Maza LM, Kikkawa Y. Biochem Pharmacol. 1998;55:1835–1842. doi: 10.1016/s0006-2952(98)00066-5. [DOI] [PubMed] [Google Scholar]
- 16.Takemura S, Minamiyama Y, Imaoka S, Funae Y, Hirohashi K, Inoue M, Kinoshita H. J Hepatol. 1999;30:1035–1044. doi: 10.1016/s0168-8278(99)80257-8. [DOI] [PubMed] [Google Scholar]
- 17.Roberts ES, Lin H, Crowley JR, Vuletich JL, Osawa Y, Hollenberg PF. Chem Res Toxicol. 1998;11:1067–1074. doi: 10.1021/tx980099b. [DOI] [PubMed] [Google Scholar]
- 18.Sewer MB, Barclay TB, Morgan ET. Mol Pharmacol. 1998;54:273–279. doi: 10.1124/mol.54.2.273. [DOI] [PubMed] [Google Scholar]
- 19.Sewer MB, Morgan ET. Biochem Pharmacol. 1997;54:729–737. doi: 10.1016/s0006-2952(97)00226-8. [DOI] [PubMed] [Google Scholar]
- 20.Sewer MB, Morgan ET. J Pharmacol Exp Ther. 1998;287:352–358. [PubMed] [Google Scholar]
- 21.Stadler J, Trockfeld J, Schmalix WA, Brill T, Siewert JR, Greim H, Doehmer J. Proc Natl Acad Sci U S A. 1994;91:3559–3563. doi: 10.1073/pnas.91.9.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatsenko OG, Boobis AR, Gross SS. Toxicol Lett. 1997;90:207–216. doi: 10.1016/s0378-4274(96)03857-x. [DOI] [PubMed] [Google Scholar]
- 23.Carlson TJ, Billings RE. Mol Pharmacol. 1996;49:796–801. [PubMed] [Google Scholar]
- 24.Hara H, Mitani N, Adachi T. Free Radic Res. 2000;33:279–285. doi: 10.1080/10715760000301441. [DOI] [PubMed] [Google Scholar]
- 25.Hara H, Adachi T. Mol Pharmacol. 2002;61:194–200. doi: 10.1124/mol.61.1.194. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari L, Peng N, Halpert JR, Morgan ET. Mol Pharmacol. 2001;60:209–216. doi: 10.1124/mol.60.1.209. [DOI] [PubMed] [Google Scholar]
- 27.Ingelman-Sundberg M. Trends Pharmacol Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal AK, Shapiro BH. Mol Pharmacol. 1996;49:523–531. [PubMed] [Google Scholar]
- 29.Baldwin SJ, Bramhall JL, Ashby CA, Yue L, Murdock PR, Hood SR, Ayrton AD, Clarke SE. Drug Metab Dispos. 2006;34:1063–1069. doi: 10.1124/dmd.105.008185. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Wing SS, Ponka P. Mol Cell Biol. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan ET, MacGeoch C, Gustafsson JA. Mol Pharmacol. 1985;27:471–479. [PubMed] [Google Scholar]
- 32.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 33.Li-Masters T, Morgan ET. Drug Metab Dispos. 2001;29:252–257. [PubMed] [Google Scholar]
- 34.Jaffrey SR, Snyder SH. Sci STKE 2001. 2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YH, Keszler A, Broniowska KA, Hogg N. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Song BL, DeBose-Boyd RA. J Biol Chem. 2004;279:28798–28806. doi: 10.1074/jbc.M402442200. [DOI] [PubMed] [Google Scholar]
- 37.Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 38.Li HC, Kupfer D. J Biochem Mol Toxicol. 1998;12:315–323. doi: 10.1002/(sici)1099-0461(1998)12:6<315::aid-jbt1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Honkakoski P, Kojo A, Lang MA. Biochem J. 1992;285:979–983. doi: 10.1042/bj2850979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 42.McGowan EB, Becker E, Detwiler TC. Biochem Biophys Res Commun. 1989;158:432–435. doi: 10.1016/s0006-291x(89)80065-8. [DOI] [PubMed] [Google Scholar]
- 43.Huan JY, Streicher JM, Bleyle LA, Koop DR. Toxicol Appl Pharmacol. 2004;199:332–343. doi: 10.1016/j.taap.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Milligan SA, Owens MW, Grisham MB. Arch Biochem Biophys. 1996;335:388–395. doi: 10.1006/abbi.1996.9998. [DOI] [PubMed] [Google Scholar]
- 45.Bredt DS, Snyder SH. Annu Rev Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 46.Willmott N, Sethi JK, Walseth TF, Lee HC, White AM, Galione A. J Biol Chem. 1996;271:3699–3705. doi: 10.1074/jbc.271.7.3699. [DOI] [PubMed] [Google Scholar]
- 47.Misko TP, Moore WM, Kasten TP, Nickols GA, Corbett JA, Tilton RG, McDaniel ML, Williamson JR, Currie MG. Eur J Pharmacol. 1993;233:119–125. doi: 10.1016/0014-2999(93)90357-n. [DOI] [PubMed] [Google Scholar]
- 48.Shiraki H, Guengerich FP. Arch Biochem Biophys. 1984;235:86–96. doi: 10.1016/0003-9861(84)90257-1. [DOI] [PubMed] [Google Scholar]
- 49.Roberts BJ. J Biol Chem. 1997;272:9771–9778. doi: 10.1074/jbc.272.15.9771. [DOI] [PubMed] [Google Scholar]
- 50.Masaki R, Yamamoto A, Tashiro Y. J Cell Biol. 1987;104:1207–1215. doi: 10.1083/jcb.104.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronis MJ, Johansson I, Hultenby K, Lagercrantz J, Glaumann H, Ingelman-Sundberg M. Eur J Biochem. 1991;198:383–389. doi: 10.1111/j.1432-1033.1991.tb16026.x. [DOI] [PubMed] [Google Scholar]
- 52.Liao M, Zgoda VG, Murray BP, Correia MA. Mol Pharmacol. 2005;67:1460–1469. doi: 10.1124/mol.104.009654. [DOI] [PubMed] [Google Scholar]
- 53.Korsmeyer KK, Davoll S, Figueiredo-Pereira ME, Correia MA. Arch Biochem Biophys. 1999;365:31–44. doi: 10.1006/abbi.1999.1138. [DOI] [PubMed] [Google Scholar]
- 54.Chen JQ, Strom A, Gustafsson JA, Morgan ET. Mol Pharmacol. 1995;47:940–947. [PubMed] [Google Scholar]
- 55.Villa P, Sartor G, Angelini M, Sironi M, Conni M, Gnocchi P, Isetta AM, Grau G, Buurman W, van Tits LJ, et al. Clin Diag Lab Immunol. 1995;2:549–553. doi: 10.1128/cdli.2.5.549-553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li-Masters T, Morgan ET. Biochem Pharmacol. 2002;64:1703–1711. doi: 10.1016/s0006-2952(02)01423-5. [DOI] [PubMed] [Google Scholar]
- 57.Kotamraju S, Tampo Y, Keszler A, Chitambar CR, Joseph J, Haas AL, Kalyanaraman B. Proc Natl Acad Sci U S A. 2003;100:10653–10658. doi: 10.1073/pnas.1933581100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beckman JS, Koppenol WH. Am J Physiol. 1996;271:C1424–1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 59.Lin HL, Kent UM, Zhang H, Waskell L, Hollenberg PF. Chem Res Toxicol. 2003;16:129–136. doi: 10.1021/tx020040b. [DOI] [PubMed] [Google Scholar]
- 60.Spencer NY, Zeng H, Patel RP, Hogg N. J Biol Chem. 2000;275:36562–36567. doi: 10.1074/jbc.M005347200. [DOI] [PubMed] [Google Scholar]
- 61.Correia MA, Sadeghi S, Mundo-Paredes E. Annu Rev Pharmacol Toxicol. 2005;45:439–464. doi: 10.1146/annurev.pharmtox.45.120403.100127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.