Abstract
Mutation of the PRL1 gene, encoding a regulatory WD protein, results in glucose hypersensitivity and derepression of glucose-regulated genes in Arabidopsis. The yeast SNF1 protein kinase, a key regulator of glucose signaling, and Arabidopsis SNF1 homologs AKIN10 and AKIN11, which can complement the Δsnf1 mutation, were found to interact with an N-terminal domain of the PRL1 protein in the two-hybrid system and in vitro. AKIN10 and AKIN11 suppress the yeast Δsnf4 mutation and interact with the SNF4p-activating subunit of SNF1. PRL1 and SNF4 bind independently to adjacent C-terminal domains of AKIN10 and AKIN11, and these protein interactions are negatively regulated by glucose in yeast. AKIN10 and AKIN11, purified in fusion with glutathione S-transferase, undergo autophosphorylation and phosphorylate a peptide of sucrose phosphate synthase in vitro. The sucrose phosphate synthase-peptide kinase activity of AKIN complexes detected by immunoprecipitation is stimulated by sucrose in light-grown Arabidopsis plants. In comparison with wild type, the activation level of AKIN immunocomplexes is higher in the prl1 mutant, suggesting that PRL1 is a negative regulator of Arabidopsis SNF1 homologs. This conclusion is supported by the observation that PRL1 is an inhibitor of AKIN10 and AKIN11 in vitro.
Members of the conserved SNF1 serine/threonine protein kinase family play essential roles in eukaryotic glucose and stress signaling (1). In budding yeast, SNF1 controls the transcription of glucose-regulated genes implicated in carbon source utilization, gluconeogenesis, respiration, sporulation, thermotolerance, peroxisome biogenesis, and cell cycle (2). A mammalian SNF1 homolog, AMP-activated protein kinase (AMPK), regulates the activity of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) and acetyl-CoA carboxylase by protecting the cells from ATP depletion during oxidative and heat stress (3). Plant SNF1-like kinases also participate in the control of key metabolic enzymes, including HMGR, nitrate reductase, sucrose synthase, and sucrose phosphate synthase (SPS) (4). Plant SNF1 homologs, such as rye RKIN1 and tobacco NPK5, complement the yeast Δsnf1 deficiency, indicating a remarkable, functional conservation of SNF1-like kinases and associated signaling factors (5, 6). In fact, yeast SNF1 and mammalian AMPK share significant homology between their catalytic subunits AMPK-α and SNF1p, positive regulatory subunits AMPK-γ and SNF4p, and C-terminal domains of AMPK-β and SNF1-interacting proteins SIP1p, SIP2p, and GAL83p (1).
The activity of SNF1 kinase is regulated by glucose in yeast. Under glucose limitation, SNF4p binds to a C-terminal regulatory domain of SNF1p, facilitating further glucose-independent interaction of SNF1p and SNF4p with bridging and targeting proteins, including GAL83 and SIP1–4 (7, 8). In the presence of glucose, SNF1 is inactivated by binding of a protein phosphatase 1 and its regulatory subunit, REG1, which induces a conformational change leading to autoinhibition of the catalytic domain by the regulatory domain of SNF1p (7, 9). Genetic and biochemical data indicate that sugar-dependent regulation of SNF1 kinases involves several other sensory and signaling components in yeast, plants, and animals (1, 2, 10). Recently, we reported that mutation of pleiotropic regulatory locus 1 (PRL1) increases the sensitivity of plants to glucose and results in transcriptional derepression of glucose-regulated genes in Arabidopsis. PRL1 encodes a novel regulatory WD protein that interacts with an Arabidopsis α-importin in the yeast two-hybrid system, displays nuclear import in plant and animal cells, and functions in vitro as a heterologous receptor for a human protein kinase C-βII that is implicated in insulin signaling (11). Here we demonstrate that PRL1 is an SNF1-binding protein that shows glucose-dependent interaction with the C-terminal regulatory domain of yeast SNF1 and Arabidopsis SNF1 homologs AKIN10 and AKIN11. Comparison of the activation level of AKIN kinase immunocomplexes in wild-type and prl1 mutant plants as well as in vitro enzyme assays with purified AKIN10 and AKIN11 indicate that PRL1 is a negative regulator of Arabidopsis SNF1 homologs.
MATERIALS AND METHODS
Characterization of Arabidopsis SNF1 Homologs.
cDNAs encoding AKIN10 and AKIN11 were isolated from a λZAPII library (donated by I. Somssich, Max-Planck Institut, Cologne) with the NPK5 cDNA probe (6) and sequenced by using an automatic ABI377 sequencer. AKIN11 was assigned to bacterial artificial chromosome (BAC) clones F15b4, F12o1, and F14p16 and mapped with recombinant inbreds (12) to chromosome 3-47 in the vicinity of marker mi413 by using a KpnI restriction fragment length polymorphism detected with the cDNA probe between Arabidopsis ecotypes Col and Ler. AKIN10 was mapped close to mi74b in chromosome 3-0 as well as to the yeast artificial chromosome (YAC) clone CIC11F3 and BACs F16j5, F18i4, F12 m4, F6a1, F24c21, and F18a20 by PCR amplification with primers AP1: 5′-TGCGCTTATAGGCTACAGGA-3′ and AP2: 5′-TAGATCTACTAGTACACAACACAC-3′, which did not amplify AKIN12 located on YAC CIC12D8 at chromosome 5-83 (mul8.9, GenBank accession no. AB009054).
The cDNAs were cloned into EcoRI-SalI sites of the yeast vector pNEV (13), yielding pNEVakin10 and pNEVakin11, and transformed into yeast strains MCY1846 and MCY1853 carrying, respectively, the snf1Δ10 and snf4Δ2 mutations (ref. 14; Fig. 1A). The transformants were grown on synthetic minimal SD medium containing either 2% glucose or 2% glycerol as sole carbon source. A Myc epitope followed by an NdeI site was placed upstream of the AKIN10 coding region in pMycAKIN10 by PCR amplification with primers P1: 5′-GGAGAATTCATGCCGGAGCAGAAGCTGATATCCGAGGAGGACCTGGCCCATATGGATGAGGGATCAGGCACAGG-3′ and P2: 5′-CCAAGCTTGTAAGGTAGAATCG-3′, and replacing the 5′ end of the cDNA by the PCR product using EcoRI-HindIII. The AKIN11 coding region in pMycAKIN11 was modified similarly by PCR using the pBluescript universal primer (Stratagene) and primer P3: 5′-GGAGAATTCATGCCGGAGCAGAAGCTGATATCCGAGGAGGACCTGGCCCATATGGATCATAG-3′ followed by cloning of an EcoRI-HindIII fragment of 120 bp from the PCR product in pAKIN11.
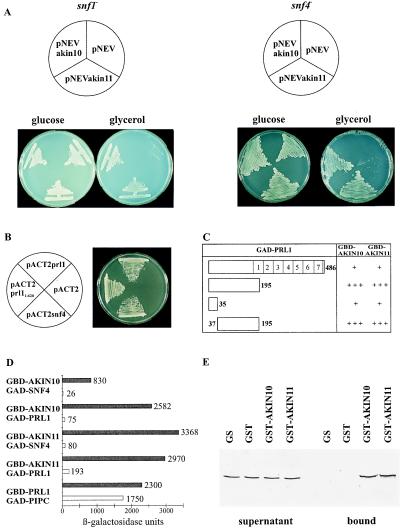
Figure 1.
AKIN10 and AKIN11 complement the snf1Δ10 deficiency, suppress the snf4Δ2 mutation, and interact with SNF4p and PRL1 in yeast. (A) Unlike controls with the empty vector pNEV, yeast strains MCY1846 snf1Δ10 (Left) and MCY1853 snf4Δ2 (Right), carrying pNEVakin10 and pNEVakin11, grew equally well on glucose and glycerol. (B) Interaction of GBD-SNF1p with GAD-PRL1 and GAD-SNF4p. Yeast carrying pAS1snf1 was transformed with pACT2prl1, pACT2prl11–620, pACT2snf4, and pACT2 as control and grew on SD medium in the presence of 50 mM 3-aminotriazole. (C) Mapping of PRL1 domain involved in binding of AKIN10 and AKIN11. GBD-AKIN10 and GBD-AKIN11 were combined with GAD-PRL1 and GAD fusions of PRL1 peptides located between positions 1 and 35, 1 and 195, and 37 and 195. LacZ-filter lift assays were performed with yeast colonies grown on SC medium with 2% glucose for 3 days at 30°C. In open bars symbolizing the GAD-PRL1 constructs (open bars), numbers indicate the boundaries of deletions and WD-40 repeats. 5-Bromo-4-chloro-3-indolyl β-d-galactoside staining after 4 h is marked by +++, whereas weaker staining observed after 16 h is labeled by +. (D) Glucose regulation of protein interactions of AKIN10 and AKIN11 with SNF4p and PRL1 in yeast. GBD-AKIN10 and GBD-AKIN11 were combined with either GAD-SNF4 or GAD-PRL1 in yeast Y190 grown either in SC medium with 2% glucose (open bars) or in SC-Gal/Gly/EtOH medium with 0.05% glucose (solid bars). β-Galactosidase activities were determined by using an o-nitrophenylgalactosidase assay (15). As control, interaction of GBD-PRL1 with the PRL1-binding protein GAD-PIPC was assayed similarly. The standard deviation of each assay was less than 10% of the maximum value measured. (E) Interaction of PRL1 with GST-AKIN10 and GST-AKIN11 in vitro. 35S-labeled PRL1 was incubated with immobilized GST, GST-AKIN10, GST-AKIN11, and control glutathione-Sepharose 4B matrix. The supernatant (Left) and matrix-bound (Right) fractions were separated by SDS/PAGE to detect the labeled PRL1 protein by autoradiography.
Detection of Protein Interactions in the Yeast Two-Hybrid System.
To construct AKIN fusions with the GAL4 DNA-binding domain (GBD), the cDNAs from pMycAKIN10 and pMycAKIN11 were cloned by NdeI-XhoI in the NdeI-SalI sites of pAS2 (15). An NdeI-BglII fragment of 1,145 bp from pAS2-AKIN10 was inserted into the NdeI-BamHI sites of pAS2 to construct GBD-AKIN101–381, carrying 381 N-terminal amino acids of AKIN10. GAD-AKIN10349–512, containing AKIN10 sequences between position 349 and the C terminus, was obtained by cloning an NcoI-XhoI fragment of AKIN10 cDNA in pACT2 (15). In this construct, a deletion was introduced in the SNF4p-binding domain situated between positions 334 and 404 of AKIN10 (Fig. 2). The PRL1 cDNA was cloned by EcoRI in pACTprl1 to express a PRL1 fusion with the GAL4-activation domain (GAD). From pACTprl1, the cDNA was cloned by NdeI-XhoI in pAS2prl1 to construct a GBD-PRL1 fusion. An NcoI cDNA fragment inserted in pACT2 yielded a GAD fusion carrying 35 N-terminal amino acids of PRL1 in pACTprl11–123. A BalI-PvuII cDNA fragment cloned in pACTprl1127–620 by SmaI provided a GAD fusion with a PRL1 peptide between amino acid positions 37 and 195. pACTprl1 was cleaved by XhoI-PvuII and religated after filling the ends to construct pACTprl11–620 encoding a GAD fusion with PRL1 sequences between the N terminus and amino acid position 195. The pAS2-AKIN baits and pAS1snf1, expressing a GBD-SNF1p fusion, were transformed into yeast Y190 that expressed either GAD-SNF4 from pACT2snf4 or GAD-PRL1 fusions encoded by pACT2prl1 and pACT2prl11–620 (Fig. 1B). pACT2 and pACT2-PIPC, encoding a GAD fusion of PRL1-interacting factor PIPC (GenBank accession no. AJ006021), were used as controls. Protein interactions were assayed as described (16, 17) by using LacZ-filter lift assays and monitoring growth on SD medium containing 25, 50, or 100 mM 3-aminotriazole. Glucose regulation of protein interactions (Fig. 1D) was assayed by measuring β-galactosidase levels in at least three independent transformants grown either in synthetic complete SC medium containing 2% glucose or in SC-Gal/Gly/EtOH medium containing 2% galactose, glycerol, and ethanol and 0.05% glucose (7, 8).
Figure 2.
Amino acid sequence comparison of AKIN10, AKIN11, AKIN12, and NPK5. Arrows mark boundaries of the kinase catalytic domain, and numbers indicate the conserved subdomains (6). An arrow marks sequences that show homology with the SNF4p-binding domain of SNF1p. A conserved threonine residue required for SNF1 kinase activation is framed.
Protein Binding and AKIN Kinase Assays in Vitro.
EcoRI-XhoI cDNA fragments from pMycAKIN10 and pMycAKIN11 were cloned in pGEX-5X1 (Pharmacia), transformed into Escherichia coli B834, and used for purification of glutathione S-transferase (GST)-AKIN fusion proteins (18). A PCR-amplified PRL1 cDNA linked 5′ upstream to a T7 polymerase promoter served as template to synthesize [35S]methionine-labeled PRL1 protein in a coupled transcription–translation system (Promega). Equal amounts of 35S-labeled PRL1 protein were incubated with glutathione-Sepharose coupled to GST-AKIN10, GST-AKIN11, and control GST proteins (5 μg of each) and with the empty matrix in 150 μl of binding buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/2 mM EDTA/0.1% Nonidet P-40) for 1 h at 4°C. After removal of the supernatant, the beads were washed extensively with binding buffer, the matrix-bound proteins were eluted with 6× SDS-loading buffer, and, together with the supernatant fractions, separated on a 10% SDS-polyacrylamide gel to detect the 35S-labeled PRL1 protein by autoradiography (19).
The coding sequence of SNF1-substrate peptide GRMRRISSVEMMDNWANTFK from sucrose phosphate synthase (GenBank accession no. S54379; ref. 20) was synthesized by T4-DNA polymerase-mediated strand extension after annealing of oligonucleotides P4: 5′-TCCGAATTCGGGTCGTATGCGTCGTATCTCCTCCGTTGAAATGATGGACAA-3′ and P5: 5′-CCGCTCGAGTTTGAAGGTGTTAGCCCAGTTGTCCATCATTTCAACGGAGGAGAT-3′ and cloned in pET201 (provided by J. Lidholm, Pharmacia, Sweden) by EcoRI-XhoI. The resulting plasmid, pET201-KD, was transformed into E. coli B843-T7 to purify the thioredoxin (TRX)-KD substrate peptide in fusion with N-terminal TRX and C-terminal His6 tags by using Qiagen Ni/NTA-agarose (19). To construct TRX-PRL1, a coding sequence for His6 tag was created by cloning of annealed T1: 5′-CCAGCTAGCCTTCGTCGTGCATCTGTTCATCATCATCATCATCATGAATTCCACC-3′ and T2 (complementary to T1) oligonucleotides into the SmaI site of pBluescript, yielding p6HIS-Blue. A C-terminal PRL1-His6 tag fusion was obtained by inserting the PRL1 cDNA with EcoRI-XhoI in p6HIS-Blue. Then, the cDNA was moved by NheI-XhoI into pET201 to purify an N-terminal thioredoxin PRL1 fusion carrying a C-terminal His6 tag, as described for TRX-KD.
In protein kinase assays, the GST-AKIN10, GST-AKIN11, and GST proteins (1 μg of each) were immobilized on glutathione-Sepharose and incubated in kinase buffer (20 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/1 mM DTT) with 5 μCi [γ-32P]ATP for 30 min at 30°C to measure autophosphorylation. After washing with kinase buffer, the matrix-bound proteins were eluted with SDS-sample buffer, separated on a 15% SDS-polyacrylamide gel, and subjected to autoradiography, and the protein bands were excised to measure 32P incorporation. To assay PRL1-mediated inhibition of AKIN kinases, either TRX-PRL1 or a control thioredoxin-His6 fusion protein (1, 2, or 5 μg of each) was added to the assays containing immobilized GST-AKIN10 or GST-AKIN11, as well as TRX-KD substrate (1 μg of each), and incubated on ice for 30 min. The kinase reaction at 30°C was started by the addition of [γ-32P]ATP, and then equal aliquots were withdrawn after 5, 10, 20, 40, and 60 min of incubation, mixed with SDS-sample buffer, and separated by SDS/PAGE to measure 32P incorporation in TRX-KD, as well as autophosphorylation of GST-AKIN10 and GST-AKIN11.
SPS-Peptide Kinase Assays with AKIN Immunocomplexes.
IgG fraction of rabbit anti-GST-NPK5 antiserum prepared by 45% ammonium sulfate precipitation was depleted of anti-GST IgG on GST-Sepharose and purified on protein A-Sepharose by using the Gentle-Elute system (Pierce). Plant extracts were prepared by extracting 0.5 g plant tissue with solid CO2 by using 2 ml buffer (50 mM Tris⋅HCl, pH 7.8/1 mM EGTA/1 mM EDTA/2 mM DTT/0.05% Nonidet P-40/10% glycerol/0.5 mM PMSF/1 mM benzamide/2 μg/ml pepstatin/0.5 μg/ml each of aprotinin, leupeptin, and antipain), mixed with 0.1 g polyvinylpyrrolidone (PVP-40, Sigma), incubated for 10 min, and then cleared by centrifugation (100,000 × g for 30 min) at 4°C. The protein samples were precleared in 100 μl binding buffer (extraction buffer containing 0.1% Nonidet P-40/0.1 mg/ml BSA/150 mM NaCl) with 20 μl protein A-Sepharose coated with preimmune serum for 1 h at 4°C and then incubated with 10 μg anti-NPK5 antibody for 2 h. AKIN immunocomplexes were bound to 15 μl protein A-Sepharose, washed with TBS (25 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.1% Tween 20) and kinase buffer (50 mM Tris⋅HCl, pH 7.8/15 mM MgCl2/5 mM EGTA/1 mM DTT), and suspended in 15 μl kinase buffer containing 4.5 μg TRX-KD substrate. The kinase reactions were initiated by the addition of 5 μCi [γ-32P]ATP and, after incubation for 25 min at 30°C, terminated by the addition of SDS-sample buffer. Phosphorylation of the TRX-KD substrate was measured as described above.
RESULTS
Interaction of PRL1 with Yeast SNF1 in the Two-Hybrid System.
The prl1 mutation was observed to cause glucose hypersensitivity and derepression of several glucose regulated genes in Arabidopsis (11). Orthologs of PRL1 and SNF1, controlling the derepression of glucose-responsive genes in yeast, were found to be conserved in eukaryotes (1, 11). This prompted us to test whether PRL1 can interact with SNF1p and SNF4p, representing conserved subunits of eukaryotic SNF1 kinases, in the yeast two-hybrid system. A fusion of SNF1p with the GBD in pAS1snf1 was combined with pACT2prl1, encoding a PRL1 fusion with the GAD, in yeast Y190 (15, 16). A combination of the GBD-SNF1p bait with pACT2snf4, encoding GAD-SNF4p, was used as control (Fig. 1B). The yeast strain expressing GBD-SNF1 and GAD-PRL1 grew in the presence of 50 mM 3-aminotriazole HIS3 inhibitor, as well as the control, and displayed LacZ+ phenotype in filter lift assays (16, 17), indicating that an interaction between SNF1p and PRL1 led to the activation of the GAL4-controlled HIS3 and LacZ reporter genes. A similar assay with pACT2prl11–620 and pAS1snf1 showed that PRL1 sequences located between the N terminus and amino acid position 195 were involved in the binding of SNF1p (Fig. 1B). In contrast, the combination of pACT2snf4 and pAS2prl1 revealed no interaction between GAD-SNF4p and GBD-PRL1, suggesting that PRL1 binding to SNF1p did not require SNF4p as mediator in yeast (data not shown).
Identification of Functional SNF1 Homologs in Arabidopsis.
An Arabidopsis cDNA library was screened with a probe encoding the tobacco kinase NPK5 that was demonstrated to complement the Δsnf1 mutation and interact with SNF4p, SIP1p, and SIP2p in yeast (6–8). Two types of cDNAs were obtained. The first class represented a gene for a known Arabidopsis protein kinase AKIN10 (GenBank accession no. M93023; ref. 21) in chromosome 3-0, whereas the second identified a locus in chromosome 3-47 encoding a novel SNF1 homolog, AKIN11 (GenBank accession no. X99279). As compared with NPK5, the deduced amino acid sequences of AKIN10 and AKIN11, both of 512 aa, contained only a few amino acid exchanges in the conserved N-terminal kinase catalytic domain, but showed some sequence divergence within the C-terminal regulatory domains (Fig. 2). The cDNA screen failed to identify a less related Arabidopsis SNF1 homolog, AKIN12, which was found recently by genome sequencing in chromosome 5-83 (mul 8.9, GenBank accession no. AB009054). This hypothetical AKIN12 sequence differed from AKIN10 and AKIN11 by numerous N-terminal amino acid exchanges and deletions in the C-terminal regulatory domain (Fig. 2).
The cDNAs were cloned in pNEV (13) to express AKIN10 and AKIN11 under the control of a plasma-membrane H+-ATPase promoter in yeast mutants that carried snf1Δ10 or snf4Δ2 deficiency, preventing growth on any other carbon source than glucose (14). Both yeast mutants, harboring either pNEVakin10 or pNEVakin11, grew equally well on media containing either glucose or glycerol at 30°C (Fig. 2A), indicating that AKIN10 and AKIN11 complemented the snf1Δ10 deficiency and acted as multicopy suppressors of snf4Δ2 deletion. Subsequently, AKIN10 and AKIN11 were fused to a GBD in pAS2 and transformed into the yeast two-hybrid host Y190 that carried the construct pACT2snf4, encoding a fusion of GAD with SNF4p. HIS3+ and LacZ+ phenotypes of these strains indicated that both AKIN10 and AKIN11 interacted with SNF4p in yeast (Table 1). β-Galactosidase assays monitoring the interaction of GBD-AKIN10 and GBD-AKIN11 with GAD-SNF4p revealed a 32- to 42-fold-higher activation of the lacZ reporter in cells growing under glucose limitation in comparison with cultures growing on glucose (Fig. 1D). This indicated that, as NPK5 (6, 7), the Arabidopsis SNF1p homologs were capable of responding to components of the glucose-signaling pathway controlling the SNF1–SNF4 interaction in yeast.
Table 1.
Interactions of AKIN10 and AKIN11 with SNF4p and PRL1 in the yeast two-hybrid system
| GBD fusions | GAD fusions | LacZ assay | His phenotype |
|---|---|---|---|
| GBD-AKIN10 | GAD-SFN4 | + | + |
| GBD-AKIN11 | GAD-SNF4 | + | + |
| GBD-AKIN101–381 | GAD-SNF4 | − | − |
| GBD-AKIN101–381 | GAD-PRL11–195 | − | − |
| GBD-SNF4 | GAD-AKIN10349–512 | − | − |
| GBD-PRL11–195 | GAD-AKIN10349–512 | + | + |
Duplicates of LacZ filters were stained after 4 and 12 h, and the growth of strains was assayed in the presence of 50 mM 3-aminiotriazole HIS3 inhibitor (15).
Mapping of Protein Domains Involved in Glucose-Regulated Interaction of PRL1 with Arabidopsis SNF1 Homologs.
To assay PRL1 binding of AKIN10 and AKIN11, GAD-PRL1 fusions carrying either full-length PRL1 or N-terminal PRL1 segments between amino acid positions 1 and 35, 1 and 195, and 37 and 195 were combined with the GBD-AKIN10 and GBD-AKIN11 baits. The results of these two-hybrid tests (Fig. 1C) indicated that both AKIN10 and AKIN11 interacted with an N-terminal segment of the PRL1 protein as observed for SNF1p. The N-terminal 35 aa of PRL1 alone conferred weak binding, but for a firm interaction with AKIN10 and AKIN11, PRL1 sequences between positions 37 and 195 also were required. The presence of WD-40 repeats in PRL1 appeared to reduce the strength of interactions with AKIN10 and AKIN11. This negative effect of WD-40 repeats correlated with the presence of glucose in the medium. The interaction of GBD-AKIN10 and GBD-AKIN11 baits with GAD-PRL1 resulted in a 15- to 32-fold increase of β-galactosidase levels when the glucose concentration in the medium was decreased from 2 to 0.05% (Fig. 1D). Because similar assays revealed no striking glucose dependence of PRL1 binding to control PRL1-interacting protein PIPC (GenBank accession no. AJ006021), the data suggested that AKIN10 and AKIN11 were responsible for glucose regulation of PRL1-AKIN protein interactions in the yeast two-hybrid system.
The PRL1-binding domain was mapped by using GBD-AKIN101–381 and GAD-AKIN10349–512. These constructs defined a deletion between positions 349 and 381 of AKIN10, which interrupted the conserved SNF4-binding domain located between amino acid positions 334 and 404 of both AKIN10 and AKIN11 (Fig. 2). The GBD-fusion protein carrying the AKIN10 catalytic domain between positions 1 and 381 did not interact either with GAD-PRL11–195, carrying 195 N-terminal amino acids from PRL1, or GAD-SNF4p. In contrast, GAD-AKIN10349–512, containing AKIN10 sequences extending from position 349 to the C terminus, failed to bind to GBD-SNF4p, but interacted firmly with GBD-PRL11–195 (Table 1). This data showed that neither the kinase catalytic domain nor the SNF4p-binding site was implicated in PRL1 binding, which required only a C-terminal segment of AKIN10 sharing limited sequence homology with the SIP1, SIP2, and Gal83-binding domain of yeast SNF1p (7, 8).
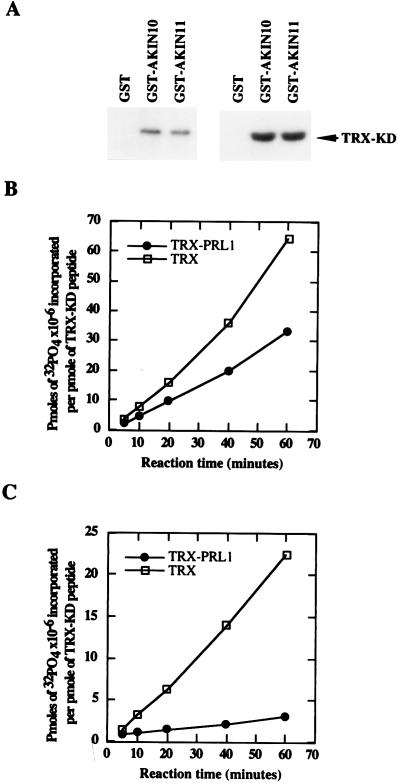
To confirm that no yeast proteins were involved as mediators in the PRL1-AKIN interactions observed in the two-hybrid system, N-terminal GST fusions with AKIN10 and AKIN11 were constructed, purified, and immobilized on glutathione-Sepharose. [35S]Methione-labeled PRL1 protein was synthesized by coupled transcription–translation by using the cDNA as template and incubated with GST-AKIN10 and GST-AKIN11, as well as with control GST, and glutathione-Sepharose matrices. After stringent washes, the supernatant fractions and matrix-bound proteins were resolved by SDS/PAGE and detected by autoradiography, demonstrating specific binding of PRL1 to both GST-AKIN10 and GST-AKIN11 in vitro (Fig. 1E). In protein kinase assays, GST-AKIN10 and GST-AKIN11 showed autophosphorylation and phosphorylated a thioredoxin-fusion protein, TRX-KD, that carried a peptide from SPS (Fig. 3 A and B).
Figure 3.
Autophosphorylation, substrate specificity, and inhibition of AKIN10 and AKIN11 by PRL1 in vitro. (A Left) GST-AKIN10 and GST-AKIN11 on glutathione-Sepharose were incubated with [γ-32P]ATP. After elution and SDS/PAGE separation, the autophosphorylation was detected by autoradiography. (Right) Phosphorylation of the TRX-KD substrate by immobilized GST-AKIN10 and GST-AKIN11 was detected by SDS/PAGE followed by autoradiography. (B) Immobilized GST-AKIN10 (1 μg) was preincubated for 30 min with either 1 μg TRX-PRL1 (●) or 1 μg His6-thioredoxin (□) protein before the addition of the TRX-KD substrate and [γ-32P]ATP. Equal amounts of samples withdrawn at different time points were separated by SDS/PAGE, and the phosphorylated TRX-KD bands were excised to measure the incorporated radioactivity. (C) PRL1-inhibition assay with GST-AKIN11 (as described in B). Phosphate incorporation per picomole of TRX-KD substrate is plotted against the reaction time.
The prl1 Mutation and Sucrose Stimulate the Activation of AKIN Kinase Complexes in Arabidopsis.
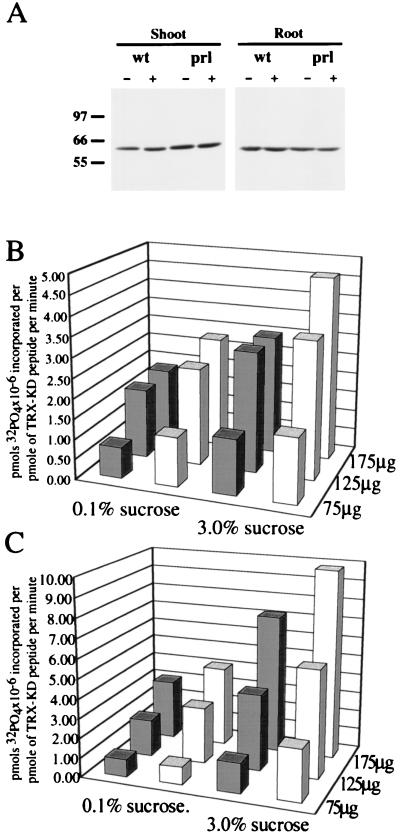
Purified GST fusions of AKIN10 and AKIN11 were recognized in immunoblotting experiments by an antibody raised against the homologous tobacco NPK5 kinase to the same extent (data not shown). To measure the activity of AKIN complexes immunoprecipitated from Arabidopsis, protein extracts were prepared from shoots and roots of wild-type and prl1 mutant seedlings grown in the presence of either 0.1 or 3% sucrose in the light. The concentration of AKIN kinases was adjusted to equal in the protein extracts based on Western blotting with the anti-NPK5 antibody (Fig. 4A). Increasing amounts of protein extracts were mixed with 10 μg anti-NPK5 IgG, bound to protein A-Sepharose, and used in kinase assays to measure the phosphorylation of the SPS-peptide substrate TRX-KD (Fig. 4 B and C). This titration showed that the linearity of kinase assay was distorted by nonspecific saturation of the IgG when the amount of protein extract applied to the IgG matrix exceeded 150 μg from shoots and 200 μg from roots. Below these limits, the assays showed a proportional increase of enzyme activity with the amount of immunoprecipitated proteins (Fig. 4 B and C). The data summarized in Table 2 showed that AKIN activities were increased to 1.7-fold in shoots and 2.2- to 2.4-fold in roots of wild-type and prl1 mutant plants by increasing the concentration of sucrose from 0.1 to 3% in the growth medium. In contrast to glucose inhibition of yeast SNF1 (14), the activity of Arabidopsis AKIN kinases in light-grown plants thus was stimulated by sucrose, and this regulation was unaffected by the prl1 mutation. The AKIN kinase activities measured in the prl1 mutant were 1.5-fold higher in shoots and about 1.4-fold higher in roots as compared with the wild type in the presence of both 0.1 and 3.0% sucrose. This enhancement of AKIN kinase activation in the prl1 mutant suggested that PRL1 may negatively regulate AKIN10 or AKIN11, or both.
Figure 4.
Enhancement of AKIN kinase activation by the prl1 mutation and sucrose. (A) Protein extracts were prepared from shoots and roots of wild-type (wt) and prl1 mutant (prl) seedlings grown in the presence of either 0.1% (−) or 3% sucrose (+) in the light. The amount of kinase catalytic subunits was adjusted to equal in the samples by Western blot titration with the anti-NPK5 IgG followed by enhanced chemiluminescence detection. (B and C) Titration of kinase assay with AKIN immunocomplexes. Protein extract (75, 125, and 175 μg) from shoots (in B) and roots (in C) of wild-type (solid bars) and prl1 mutant (open bars) seedlings treated with 0.1 or 3% sucrose were immunoprecipitated with 10 μg anti-NPK5 IgG, bound to protein A-Sepharose, and used in kinase assays to measure AKIN activities. Units show pmol phosphate incorporation × 10−6 per pmol TRX-KD substrate per min.
Table 2.
Activation of AKIN kinase complexes in shoots and roots of light-grown wild-type and prl1 mutant seedlings by sucrose
| Shoots
|
Roots
|
|||||
|---|---|---|---|---|---|---|
| 0.1% sucrose | 3.0% sucrose | Activation | 0.1% sucrose | 3.0% sucrose | Activation | |
| Wild type | 10.5 ± 0.7 | 17.5 ± 0.7 | 1.66 | 16.0 ± 1.4 | 36.2 ± 6.6 | 2.26 |
| prl1 mutant | 15.95 ± 0.7 | 26.7 ± 0.4 | 1.67 | 23.5 ± 0.7 | 50.2 ± 8.2 | 2.45 |
| Activation | 1.52 | 1.52 | 1.47 | 1.39 | ||
AKIN enzyme activities are given in milliunits defined as pmol of PO4 × 10−9 incorporated in pmol of TRX-KD peptide per min per μg of protein.
PRL1 Is an Inhibitor of AKIN10 and AKIN11 in Vitro.
To test how PRL1 binding affects the kinase activity of AKIN10 and AKIN11 in vitro, a His6-tagged thioredoxin-PRL1 fusion protein, TRX-PRL1, was constructed and purified to homogeneity. TRX-PRL1 was incubated at different concentrations with GST-AKIN10 and GST-AKIN11, assaying the phosphorylation of TRX-KD substrate. In comparison with control assays performed with a His6-tagged thioredoxin protein, the TRX-PRL1 fusion protein resulted in an approximately 2-fold inhibition of GST-AKIN10 (Fig. 3B) and an 8-fold inhibition of GST-AKIN11-mediated phosphorylation of TRX-KD (Fig. 3C). TRX-PRL1 itself was not phosphorylated in these assays, indicating that PRL1 was not a substrate of AKIN10 and AKIN11 (data not shown).
DISCUSSION
The data described above suggest that PRL1 is a potential subunit of Arabidopsis SNF1 kinase homologs that binds to conserved C-terminal sequences of these proteins in yeast and in vitro. PRL1 does not interact with the SNF4p-activating subunit of SNF1 in the two-hybrid system, suggesting that PRL1 differs from the yeast GAL83 and SIP factors that are thought to stabilize the SNF1p-SNF4p complex and target the kinase to different substrates (2). Arabidopsis contains at least two functional SNF1 homologs, which, similar to rye RKIN1 and tobacco NPK5 (5, 6), complement the Δsnf1 mutation and interact with SNF4p in yeast. In addition, as SNF1p does in yeast, AKIN10 and AKIN11 function as multicopy suppressors of the Δsnf4 deficiency. Although it is not yet proven that AKIN10 and AKIN11 are implicated in glucose signaling, based on these results, we favor a model suggesting that plant SNF1 homologs perform a function similar to SNF1 in yeast. A recent observation showing that antisense inhibition of an SNF1-like gene prevents the activation of a sucrose-inducible sucrose synthase gene in potato supports this model (22). Furthermore, these data suggest that plant SNF1 homologs also may be required for activation of gene expression in response to sucrose and glucose, whereas SNF1 in yeast is thought to be essential only for derepression of glucose-repressible genes under glucose starvation (2).
We have found that the activity of AKIN kinase immunocomplexes, standardized for equal amounts of kinase catalytic subunit, is stimulated by sucrose in light-grown Arabidopsis plants. This result may be particularly important, if a current model of yeast glucose signaling is being applied to guide studies of plant-glucose repression (10). For example, hexokinases are known to be conserved from yeast to man and proposed to function as potential primary sensors in glucose signaling. Thus, overexpression of a yeast hexokinase augments the glucose-stimulated insulin production in pancreatic beta cells (23), whereas antisense inhibition of a hexokinase prevents glucose repression of light-regulated genes in Arabidopsis (24). Hexokinase inhibition causes glucose insensitivity, whereas hexokinase overexpression confers glucose hypersensitivity in Arabidopsis. In yeast, activation of glucose signaling via hexokinase leads to glucose repression and inhibition of the downstream SNF1 kinase (2). When applying the yeast model to plants, one therefore may predict that mutations causing glucose hypersensitivity should reduce the activity of plant SNF1 kinases. Our data show that this is probably not the case in light-grown plants. Similar to hexokinase overexpression, the prl1 mutation results in glucose hypersensitivity. Instead of stimulating glucose repression, the prl1 mutation leads to derepression of glucose-repressible and glucose-inducible genes. Therefore, we proposed that PRL1 may be a negative regulator of a function that counteracts the activity of signaling factors mediating glucose repression in Arabidopsis (11). This hypothesis is supported by our results of yeast two-hybrid and in vitro protein interaction assays, showing that PRL1, indeed, is an interacting partner of the SNF1 kinase required for relieving glucose repression in yeast.
Binding of PRL1 to SNF1p, AKIN10, and AKIN11 is inhibited by glucose in yeast. Assuming that a similar regulation exists in Arabidopsis, inactivation of PRL1, which functions in vitro as an inhibitor of AKIN10 and AKIN11, is expected to lead to an increase of AKIN kinase activities. In fact, the activity of AKIN complexes is higher in the prl1 mutant, but the mutation does not seem to affect the regulation of kinase activities by sucrose. This suggests that either PRL1 is not involved directly in glucose signaling or that additional factors modulate the activity of AKIN kinase complexes and their interaction with PRL1 in vivo. In fact, light is known to play an important role in the regulation of plant SNF1 kinases that also are controlled metabolically because plant cells themselves are capable of producing glucose by photosynthetic CO2 fixation (25). Plant glucose responses thus probably are modulated by a cross-talk with light and hormone signaling. It therefore is not surprising that, in addition to changes in glucose responses, the prl1 mutation results in pleiotropic defects, increasing the sensitivity of Arabidopsis to plant hormones, including cytokinin, ethylene, auxin, and abscisic acid (11).
Biochemical analysis of the role played by PRL1 in glucose signaling is faced with the problem that Arabidopsis, as other plants, contains more than one closely related SNF1 homolog that may perform either complementary or redundant functions in different organs during various phases of development (4). Substrate specificity and subunit composition of different SNF1 kinase complexes represent additional unresolved issues requiring the generation of specific antibodies and expression of epitope-tagged proteins in transgenic plants. As a multiprotein-interacting factor implicated in the binding of α-importin, AKIN10, and AKIN11, PRL1 therefore may provide a useful tool to explore the function of plant SNF1 kinases and their associated factors in the regulation of glucose, hormone, and stress signaling.
Acknowledgments
We thank Drs. Francesco Salamini, Mark Stitt, and Peter Geingenberger for their helpful comments on the manuscript. This work was supported as part of a joint research project between the Max-Planck Institute (Cologne) and the Biological Research Center (Szeged), supported by the Deutsche Forschungsgemeinschaft and the Hungarian Academy of Sciences, as well as by grants from the Deutsches Zentrum für Luft- und Raumfahrt (DLR UNG-027-97), Deutsche Forschungsgemeinschaft Arabidopsis Schwerpunkt (II B1-1438/1-3), and Országos Tudományos Kutatási Alap (OTKA) T3182.
ABBREVIATIONS
- PRL1
Arabidopsis pleiotropic regulatory locus
- AKIN
Arabidopsis SNF1 kinase homolog
- GST
glutathione S-transferase, GBD, GAL4 DNA-binding domain
- GAD
GAL4 activation domain, TRX, thioredoxin
- SPS
sucrose phosphate synthase
Footnotes
References
- 1.Hardie G D, Carling D, Carlson M. Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Carlson M. Curr Opin Genet Dev. 1998;8:560–564. doi: 10.1016/s0959-437x(98)80011-7. [DOI] [PubMed] [Google Scholar]
- 3.Hardie G D, Carling D. Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 4.Halford N G, Hardie G D. Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- 5.Alderson A, Sabelli P A, Dickinson R J, Cole D, Richardson M, Kreis M, Shewry P R, Halford N G. Proc Natl Acad Sci USA. 1991;88:8602–8605. doi: 10.1073/pnas.88.19.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muranaka T, Banno H, Machida Y. Mol Cell Biol. 1994;14:2958–2965. doi: 10.1128/mcb.14.5.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang R, Carlson M. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 8.Jiang R, Carlson M. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludin K, Jiang R, Carlson M. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smeekens S, Rook F. Plant Physiol. 1997;115:7–13. doi: 10.1104/pp.115.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S, et al. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 13.Sauer N, Stolz J. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- 14.Celenza J L, Carlson M. Mol Cell Biol. 1989;9:5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legrain C, Dokhelar M C, Trancy C. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Hubbard E J A, Carlson M. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- 17.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 19.Harper J W, Adam G R, Wie N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 20.Dale S, Wilson W A, Edelman A M, Hardie D G. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 21.Le Guen L, Thomas M, Bianchi M, Halford N G, Kreis M. Gene. 1992;120:249–254. doi: 10.1016/0378-1119(92)90100-4. [DOI] [PubMed] [Google Scholar]
- 22.Purcell P C, Smith A M, Halford N G. Plant J. 1998;14:195–202. [Google Scholar]
- 23.Epstein P N, Boschero A C, Atwater I, Cai X, Overbeek P A. Proc Natl Acad Sci USA. 1992;89:12038–12042. doi: 10.1073/pnas.89.24.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang J-C, León P, Sheen J. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber S C, Huber J L, Michael R W. Int Rev Cytol. 1994;149:47–98. [Google Scholar]