Abstract
Kinesin is a two-headed motor protein that transports cargo inside cells by moving stepwise on microtubules. Its exact trajectory along the microtubule is unknown: alternative pathway models predict either uniform 8-nm steps or alternating 7- and 9-nm steps. By analyzing single-molecule stepping traces from “limping” kinesin molecules, we were able to distinguish alternate fast- and slow-phase steps and thereby to calculate the step sizes associated with the motions of each of the two heads. We also compiled step distances from nonlimping kinesin molecules and compared these distributions against models predicting uniform or alternating step sizes. In both cases, we find that kinesin takes uniform 8-nm steps, a result that strongly constrains the allowed models.
Conventional kinesin is a homodimeric motor protein with two microtubule-binding head domains linked to a common, coiled-coil stalk. It moves processively, taking up to hundreds of steps along microtubules before dissociating (1). Kinesin steps are produced by an asymmetric, hand-over-hand walk carried out by its heads (2–4) as it follows a path parallel to the microtubule protofilaments (5). However, the trajectories followed by the heads during stepping have long been a source of controversy (6–9), and remain an outstanding issue in the field (10). Using a high-resolution optical trapping assay, we measured the positions of microscopic beads attached to the stalks of single kinesin molecules, and from these data inferred the motions of the heads. It is not yet well established whether kinesin spends time during stepping in a predominantly one-head-bound state (8,10) or in a two-heads-bound state (4,11), so both possibilities were considered. For the case where kinesin molecules dwell mainly in a one-head-bound state, there are two plausible stepping scenarios (Fig. 1): the “tightrope” pathway, where successive microtubule binding sites and stalk positions both lie along a common line coincident with a single protofilament of the microtubule surface lattice, and the “straddle” pathway, where successive microtubule binding sites alternate between adjacent protofilaments and the stalk position follows a zigzag pathway among these positions. In the tightrope pathway, the stalk advances by uniform, 8-nm steps as the heads move from one tubulin dimer to the next. In the straddle pathway, however, due to the ∼1-nm longitudinal offset between adjacent protofilaments (12), the stalk advances alternately in ∼7- and ∼9-nm steps, measured as projections along the microtubule axis. For the case where kinesin molecules dwell predominantly in a two-heads-bound state, the tightrope pathway generates uniform, 8-nm steps. In contrast, the straddle pathway can lead, in principle, either to uniform 8-nm steps (the “normal straddle”, corresponding to the situation where the stalk position reports the average location of the two bound heads) or to a zigzag motion with alternating step sizes, just as above (the “asymmetric straddle”, corresponding to the situation where the kinesin stalk is pulled closer to one head than to the other).
Figure 1.
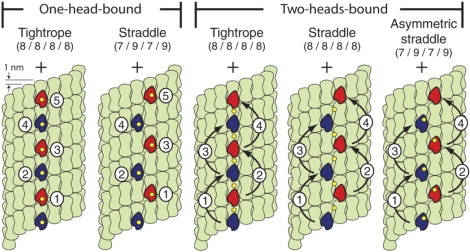
Stepping pathways. Candidate trajectories for kinesin stepping predict consecutive 8-nm steps or alternating 7- and 9-nm steps, measured along the microtubule axis from a point on the stalk. The surface lattice for a 13-protofilament microtubule and successive positions (numbered) occupied by the walking heads (red and blue) of a dimeric kinesin molecule are shown. Stalk position is indicated (yellow), along with head motions (black arrows). Tubulin α-β heterodimers (green dumbbells) form longitudinal protofilaments that are offset by 0.94 nm. In one-head-bound models, the position of the free head is not displayed; the stalk reports a position near the bound head. For the tightrope and straddle models, the stalk position is assumed to be located at the midpoint between the bound heads; in the asymmetric straddle model, the stalk is associated with a single head.
We used an optical force-clamp apparatus with high spatiotemporal resolution to measure the stepping motions of single kinesin molecules attached to 0.5-μm diameter beads, which were trapped in solution, then placed near coverslip-immobilized microtubules (2). Once a kinesin molecule bound the microtubule and began moving processively, a feedback loop was automatically engaged to maintain a fixed separation between the bead and the trap center, thereby applying constant load to the kinesin molecule. Records of the positions of beads obtained under such conditions displayed a clear series of molecular steps, with abrupt transitions lasting <2 ms. Operationally, the step distance was calculated from the difference in the mean positions of dwell intervals located on either side of a given transition (Supplementary Material). In principle, a careful comparison of the distances subtended by the even- and odd-numbered steps within a single, long record of kinesin motion could be used to discriminate among the competing pathway models. In practice, however, positional noise within individual records (SD ∼2 nm; N = 1,063) and the reduced processivity of kinesin molecules under load (which limits the number of steps before dissociation) preclude such an approach. Instead, statistical accuracy was improved by combining data from many different runs and molecules. The challenge, then, is to find a way to keep track of the phases of alternating steps between records. This challenge was met by collecting data from force-clamped recombinant kinesin molecules (load = −4 pN; [ATP] = 2 mM) that display an intrinsic asymmetry in their stepping behavior, and therefore provide a means of distinguishing their alternating steps, i.e., by using molecules that limp (2).
The timing of the successive steps taken by a recombinant, homodimeric kinesin molecule (DmK401) has previously been shown to alternate between fast and slow rates (2). Assuming that the slow and fast dwell times correspond to the alternating motions of its two heads in a hand-over-hand walk (2–4), we can sort kinesin steps on this basis and thereby combine data from multiple records. We separately computed the average duration of all even- and odd-numbered dwell times within every record, assigning those times with shorter average duration to the “fast” phase and times with longer average duration to the “slow” phase. As found previously, the distributions of the fast and slow phases assigned in this way were fit by exponentials with different time constants (2), implying that the two classes of step arise from distinct stochastic processes. The severity of limping for each record was assessed by its limp factor, L, defined as the ratio of the average slow step duration to the average fast step duration for records containing ≥6 dwell intervals. Stepping traces were analyzed as described, and records with L ≥ 5 were retained for analysis (Supplementary Material; N = 107 records; 10 beads). Average step sizes associated with either the fast or slow phases were statistically indistinguishable (two-tailed t-test; t = 0.27; α = 0.05; P < 0.001) and well-fit by Gaussians with means of 8.20 nm (Fig. 2 a). The average step size here is identical to previous measurements for kinesin based on the motions produced by both heads (13).
Figure 2.
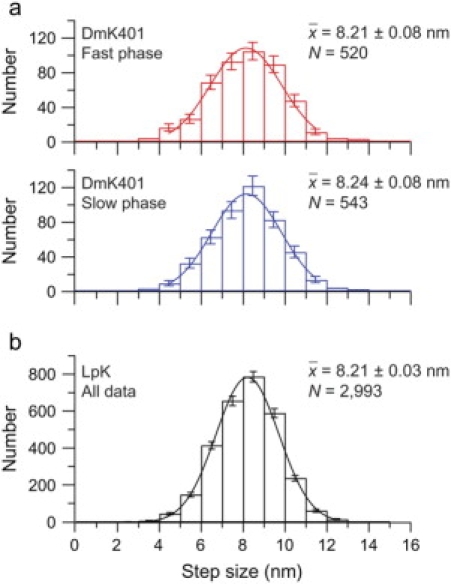
Step size distributions and Gaussian fits. (a) DmK401 histograms of sizes for the fast (red bars) and slow (blue bars) phases with superposed Gaussian fits (solid lines); bin width 1 nm; statistical errors as indicated. Best fit values (μ ± σμ) are 8.19 ± 0.09 nm (red) and 8.22 ± 0.08 nm (blue). (b) LpK distribution of all step sizes (black bars with statistical errors). Data are fit to a Gaussian (solid line) with 8.22 ± 0.03 nm (μ ± σμ). All fits were restricted to bins with ≥10 counts. Legends display sample averages mean ± SE.
We also compiled step data from nonlimping, wild-type kinesin molecules (LpK) purified from squid optic lobe under force-clamped conditions (load = –5 pN; [ATP] = 2 mM). Because phase assignments cannot be made in the absence of limping, the histogram of all step distances was tested against fits to alternative models (Fig. 2 b): 1), a single Gaussian distribution or 2), a sum of two Gaussian distributions with fixed means (7.28 nm; 9.16 nm), equal to the experimental best-fit kinesin step size (8.22 nm) increased and decreased by the longitudinal offset between adjacent protofilaments (the stagger distance). For microtubules with a B-type helical lattice, the offset is given by (3/13) times the axial monomer spacing for a 13-protofilament, 3-start helical microtubule (14). Based on x-ray and electron diffraction, estimates of the monomer spacing range from 4.05 to 4.09 nm (12,15,16), corresponding to a stagger distance of ∼0.94 nm. Fitting returned ( = 1.68; ν = 7; P ∼ 0.15) for a single Gaussian and ( = 5.02; ν = 6; P < 0.001) for two offset Gaussians. Comparing these results by an F-test (17), we find that the experimental data from native kinesin are more likely to represent a single step size than two (alternating) step sizes (F = 3.0; P = 0.09).
Control experiments confirmed that both analytical methods report alternating step sizes of ∼7 and ∼9 nm when actually present. Kinesin-coated beads were immobilized on microtubules using a nonhydrolyzable ATP analog. To simulate stepping, the microscope piezo stage was advanced in alternating 7- and 9-nm increments at random times chosen from exponential distributions. Alternating step sizes were faithfully recovered (Supplementary Material). These simulated data were also fit to either one or two Gaussians, as described in the foregoing: in this instance, the fit to two Gaussians was superior (F-test: F = 5.2; P = 0.04).
Previous work that tracked at nanometer-level accuracy the position of a single fluorophore attached to one of the two kinesin heads revealed steps of ∼16 nm during processive movement (i.e., alternating steps of 0 and 16 nm for a given labeled head) (4). This result may be taken as evidence that both kinesin heads are likely to be localized to different sites on the microtubule throughout most of the kinetic cycle, as opposed to one being freely tethered or closely associated with its partner. Additional support for this interpretation came from FRET-based experiments conducted with dyes placed on the kinesin stalk and one head, which were most consistent with a two-heads-bound intermediate state during stepping (18). Recent evidence that the rear head of kinesin may be able to synthesize ATP under certain conditions also suggests that both its heads remain predominantly bound to the microtubule during the stepping cycle (11). These observations argue against one-head-bound pathway models (Fig. 1).
By contrast, Cross and co-workers (19) recently concluded that dimeric kinesin molecules can bind to individual α-β tubulin dimers not formed into protofilaments. Results of their biochemical kinetic experiments, conducted with such tubulin-bound motors, suggested that one kinesin head may be able to regulate the biochemical cycle of its partner even under conditions where both heads are not simultaneously bound to a common substrate. These findings were therefore interpreted as lending support to a one-head-bound trajectory, where a tethered head spends significant time “docked” against its bound partner during the stepping cycle. However, it is not clear whether the conclusions reached by Cross and colleagues relate to normal processive stepping, or perhaps may represent a new form of head “gating” peculiar to tubulin dimers, as pointed out in an accompanying commentary (20).
We conclude that kinesin molecules step invariably by 8 nm during processive motion, as measured from a point on the common stalk: this point does not alternate between 7- and 9-nm advancements. Our finding therefore excludes pathway models that require such alternation, i.e., the one-head-bound straddle model and the two-heads-bound asymmetric straddle model (Fig. 1). Taking the available experimental evidence into consideration, we tend to favor a two-heads-bound pathway, and therefore propose that kinesin steps either by a two-heads-bound tightrope or by a two-heads-bound straddle mechanism. Future experiments may be able to discern additional features of kinesin motion that would distinguish between these alternatives.
Supplementary material
To view the supplemental file associated with this article, visit www.biophysj.org.
Appendix A.
Supplement
Acknowledgments
We acknowledge support from a National Science Foundation Graduate Research Fellowship (A.N.F.) and National Institutes of Health grant R01-GM51453 (S.M.B.).
Footnotes
Editor: Yale E. Goldman.
References and footnotes
- 1.Howard J., Hudspeth A.J., Vale R.D. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 2.Asbury C.L., Fehr A.N., Block S.M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaseda K., Higuchi H., Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz A., Tomishige M., Vale R.D., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 5.Ray S., Meyhofer E., Milligan R.A., Howard J. Kinesin follows the microtubule's protofilament axis. J. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Block S.M., Svoboda K. Analysis of high resolution recordings of motor movement. Biophys. J. 1995;68 2305S–2395S (discussion 2395S–2415S) [PMC free article] [PubMed] [Google Scholar]
- 7.Lockhart A., Crevel I.M.T.C., Cross R.A. Kinesin and NCD bind through a single head to microtubules and compete for a shared MT binding site. J. Mol. Biol. 1995;249:763–771. doi: 10.1006/jmbi.1995.0335. [DOI] [PubMed] [Google Scholar]
- 8.Cross R.A. On the hand-over-hand footsteps of kinesin heads. J. Muscle Res. Cell Motil. 1995;16:91–94. doi: 10.1007/BF00122526. [DOI] [PubMed] [Google Scholar]
- 9.Block S. Kinesin: what gives? Cell. 1998;93:5–8. doi: 10.1016/s0092-8674(00)81138-1. [DOI] [PubMed] [Google Scholar]
- 10.Cross R.A. Molecular motors: kinesin's interesting limp. Curr. Biol. 2004;14:R158–R159. [PubMed] [Google Scholar]
- 11.Hackney D.D. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc. Natl. Acad. Sci. USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelkow E., Thomas J., Cohen C. Microtubule structure at low resolution by x-ray diffraction. Proc. Natl. Acad. Sci. USA. 1977;74:3370–3374. doi: 10.1073/pnas.74.8.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svoboda K., Schmidt C.F., Schnapp B.J., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 14.Chretien D., Wade R.H. New data on the microtubule surface lattice. Biol. Cell. 1991;71:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 15.Li H., DeRosier D.J., Nicholson W.V., Nogales E., Downing K.H. Microtubule structure at 8Å resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 16.Wais-Steider C., White N.S., Gilbert D.S., Eagles P.A. X-ray diffraction patterns from microtubules and neurofilaments in axoplasm. J. Mol. Biol. 1987;197:205–218. doi: 10.1016/0022-2836(87)90119-7. [DOI] [PubMed] [Google Scholar]
- 17.Bevington P.R., Robinson D.K. WCB/McGraw-Hill; New York: 1992. Data Reduction and Error Analysis for the Physical Sciences. [Google Scholar]
- 18.Tomishige M., Stuurman N., Vale R.D. Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat. Struct. Mol. Biol. 2006;13:887–894. doi: 10.1038/nsmb1151. [DOI] [PubMed] [Google Scholar]
- 19.Alonso M.C., Drummond D.R., Kain S., Hoeng J., Amos L., Cross R.A. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackney D.D. Biochemistry. Processive motor movement. Science. 2007;316:58–59. doi: 10.1126/science.1141549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement


