Abstract
Two aryl hydrocarbon receptors (AhRs), rtAhR2α and rtAhR2β, were cloned from rainbow trout (rt) cDNA libraries. The distribution of sequence differences, genomic Southern blot analysis, and the presence of both transcripts in all individual rainbow trout examined suggest that the two forms of rtAhR2 are derived from separate genes. The two rtAhR2s have significant sequence similarity with AhRs cloned from mammalian species, especially in the basic helix-loop-helix and PAS functional domains located in the amino-terminal 400 amino acids of the protein. In contrast, the Gln-rich transactivation domain found in the carboxyl-terminal half of mammalian AhRs is absent from both rtAhR2s. Both clones were expressed by in vitro transcription/translation and proteins of approximately 125 kDa were produced. These proteins bind 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and are able to bind dioxin response elements in gel shift assays. rtAhR2α and rtAhR2β are expressed in a tissue-specific manner with the highest expression of rtAhR2β in the heart. Expression of rtAhR2α and rtAhR2β mRNAs is positively regulated by TCDD. Both rtAhR2α and rtAhR2β produced TCDD-dependent activation of a reporter gene driven by dioxin response elements. Surprisingly, the two receptors showed distinct preferences for different enhancer sequences. These results suggest that the two receptor forms may regulate different sets of genes, and may play different roles in the toxic responses produced by AhR agonists such as TCDD.
Trout and other salmonids are especially sensitive to polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls (1-3). The toxicity of planar polychlorinated dibenzo-p-dioxins, dibenzofuran, and biphenyl congeners is mediated by the aryl hydrocarbon receptor (AhR).1 While this is well studied in mammals (4), the AhR pathway is less well characterized in fish (5, 6). Experiments with a photoaffinity ligand have identified a 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-binding protein in trout with a molecular mass of 145 kDa, somewhat similar to the 95–110-kDa AhR proteins found in mammals (7, 8). In addition, the AhR dimerization partner, ARNT, has been cloned from rainbow trout (9) and more recently, AhR homologs have been cloned in other fish species (10-12).
The AhR is a ligand-activated transcription factor in the PAS family of proteins. Transcriptional activity requires dimerization with another PAS protein, ARNT. The unliganded AhR resides in the cytosol as a multiprotein complex that includes hsp90 and AhR interacting protein (13). Ligand binding causes the AhR to move to the nucleus. During transport to the nucleus, AhR dissociates from hsp90 dimer and AhR interacting protein and subsequently forms a dimer with ARNT. This new complex binds to specific double-stranded DNA sequences, generally referred to as dioxin response elements (DREs), to alter the expression of genes (14, 15). The DRE consensus sequence has been defined as TnGCGTG for the mouse and human AhR (14). This sequence has been identified and shown to be transcriptionally active in the 5′-flanking region of genes such as CYP1A1 in mammals. This sequence has also been identified in the 5′-flanking region of the rainbow trout and Atlantic tomcod CYP1A genes, but has not been directly tested for functionality (16, 17).
The structure-activity relationship for both early life stage mortality and induction of CYP1A by polychlorinated dibenzo-p-dioxins and dibenzofuran congeners in rainbow trout is generally similar to that for mammals, but for planar polychlorinated biphenyls the structure activity relationship in fish does not correspond to that observed in mammals (3, 18, 19). The lower potency of the mono-ortho-polychlorinated biphenyls in fish may be attributable to differences between mammalian and fish AhRs, but proof of this hypothesis requires direct comparison of the receptor proteins from each species. As a step toward this analysis, we set out to clone AhR cDNAs from a rainbow trout gonad cell line (RTG-2). In this report, we describe the cloning of two distinct rainbow trout AhR cDNAs. The proteins encoded by the two clones are very similar in sequence. However, they show surprising differences in tissue distribution, regulation of expression, and transcriptional activity. These results suggest that the two types of receptors may mediate distinct responses in different tissues.
EXPERIMENTAL PROCEDURES
Rainbow Trout
Six female juvenile rainbow trout were obtained from the Aquaculture Program at the University of Wisconsin and held in flowing water at 8 °C for 3 days to acclimate. Trout were injected intraperitoneally with TCDD (10 μg/kg) or the same volume (2 ml/kg) of vehicle (5% acetone, 95% corn oil) as control. After 3 days tissues were harvested and immediately frozen in liquid nitrogen and stored at −70 °C until RNA was isolated.
Cell Culture
RTG-2 cells were obtained from ATCC (Manassas, VA) and cultured in modified Eagle's medium supplemented with 10% fetal bovine serum at 21 °C in an atmosphere of normal air. Cells were split and final media changes were completed at least 24 h prior to dosing. Cells were exposed to graded concentrations of TCDD (0.0005–1.0 nm) dissolved in Me2SO with a final Me2SO concentration in the culture media of 0.1%. TCDD exposure continued for 6 h before cells were harvested for RNA. COS-7 (monkey kidney epithelial cells) were obtained from ATCC and were cultured in Dulbecco's modified eagle's medium supplemented with 10% fetal bovine serum in an atmosphere of 5% CO2. Cells were split the day prior to transfection.
Library Construction and Screening
Two cDNA libraries were constructed using RNA isolated from RTG-2 cells. A random primed λ ZAP cDNA library (library 1) was constructed by Stratagene (La Jolla, CA). cDNAs were created using a random hexamer and a library which contained 5.8 × 106 primary clones was produced. A second library (library 2) was constructed in our laboratory using a λ ZAP express kit from Stratagene and a poly(dT) primer. Libraries were screened as described by Stratagene. Briefly, 50,000 plaque forming units were diluted in fresh XL1-Blue MRF bacteria, mixed with 0.7% agarose, and plated on 150-mm LB plates. After overnight incubation, nylon membranes were used to lift plaques. The membranes were hybridized with probe in a 50% formamide hybridization solution at 42 °C. Initial screens of library 1 were made using a portion of the Fundulus heteroclitus AhR2 (20) as a probe; 8.0 × 106 plaques were screened. Subsequent screens of library 2 used portions of the rainbow trout AhR as a probe; 6 × 106 plaques were screened. Probes were labeled with [32P]dCTP by random priming. Phagemids (pBluescript II SK and pBK-CMV) were excised from purified plaques with helper phage as described by the manufacturer. For use as a loading control, we cloned rainbow trout glyceraldehyde-3-phosphate dehydrogenase from library 2 using a probe generated by RT-PCR amplification of a conserved portion of glyceraldehyde-3-phosphate dehydrogenase (GenBank accession number AF027130).
Construction of Full-length AhR Clones
Two independent clones were isolated from library 1 which encompass the 5′ UTR and part of the coding sequence of each rtAhR2 and two more independent clones were isolated from library 2. After complete sequencing, overlapping sequences were found and a single full-length cDNA was constructed corresponding to each mRNA. The nucleotide sequence for the rainbow trout AhR2α cDNA has been submitted to GenBank with accession number AF065137. The nucleotide sequence for the rainbow trout AhR2β cDNA has been submitted to GenBank with accession number AF065138. For expression studies these fragments were ligated into pBK-CMV that had been digested with XbaI and NotI.
Sequencing
Sequencing of isolated clones was completed by manual sequencing using a Sequenase kit (U. S. Biochemical Corp., Cleveland, OH), and by automated sequencing using ABI ready reaction mixture and an ABI 377 automated sequencer (Perkin Elmer, Foster City, CA). Sequences were determined for each strand and each strand was sequenced at least 3 times.
In Vitro Transcription/Translation
In vitro transcription/translation reactions were completed using a TNT kit from Promega essentially as directed by the manufacturer. For each reaction 1 μg of plasmid DNA was used in a 50-μl total reaction volume. Amino acid mixtures lacking methionine were added to each reaction and reactions were supplemented with translational grade [35S]methionine (NEN Life Science Products Inc., Boston, MA). 10 μl of each reaction was diluted with an equal volume of 2 × loading dye and separated on 10% SDS-polyacrylamide gel electrophoresis. Gels were fixed, dried onto filter paper, and exposed to PhosphorImager screens overnight.
Velocity Sedimentation Analysis
2,3,7,8-Tetrachloro[1,6-3H]dibenzo-p-dioxin (35 Ci/mmol) was obtained from Chemsyn Science Laboratories (Lenexa, KS) and purified to ≈95% by high performance liquid chromatography according to the method of Gasiewicz and Neal (21). 2,3,7,8-TCDF was obtained from Ultra Scientific (Hope, RI). Methylated-[methyl-14C]ovalbumin was from NEN Life Science Products Inc. (Boston, MA). Methylated-[methyl-14C]catalase was synthesized as described previously (22). AhR proteins were expressed by in vitro transcription and translation (TnT) and analyzed by velocity sedimentation on sucrose gradients in a vertical tube rotor by the method of Tsui and Okey (23). For each AhR, two identical TnT reactions (100 μl total) were combined, diluted 1:1 with MEEDMG buffer (8) (25 mm MOPS, pH 7.5, 20 °C, containing 1 mm dithiothreitol, 1 mm EDTA, 5 mm EGTA, 0.02% NaN3, 20 mm Na2MoO4, 10% (v:v) glycerol), split into two 100-μl aliquots, and incubated with [3H]TCDD (2 nm) ± TCDF (400 nm) for 1–2 h at 4 °C. [3H]TCDD concentration was verified by sampling each tube for total counts. No charcoal-dextran treatment was used to remove unbound [3H]TCDD, as trout AhR has been shown to be sensitive to small amounts of charcoal (24). After incubation, 90 μl of each incubation was applied to 10–30% sucrose gradients prepared using the method of Coombs and Watts (25); tubes were then spun for 140 min at 60,000 rpm at 4 °C in a VTi 65.2 rotor. Gradients were fractionated (150 μl per fraction) and counted using a Beckman LS5000TD scintillation counter. Specific binding is defined as the difference between total binding (incubations containing [3H]TCDD) and nonspecific binding (incubations containing [3H]TCDD plus a 200-fold excess of TCDF). [14C]Catalase (11.3 S) and [14C]ovalbumin (3.6 S) were added as internal sedimentation markers; they eluted in fractions ∼15–16 and ∼4, respectively, as indicated. Sedimentation coefficients were determined by the method of Martin and Ames (26).
In Vitro Electrophoretic Mobility Shift Assay
All sense strand oligonucleotides are listed 5′ to 3′, complementary strands are not shown. The consensus DRE core is underlined and mutated bases are in bold. Oligonucleotide wt rtDRE1 (ACCTTTGCACGCTATCGAAAT) was 5′-end labeled with 32P using T4 polynucleotide kinase and annealed to a 3-fold molar excess of the complementary oligonucleotide followed by probe purification. Unlabeled competitor DNAs were similarly produced by annealing unlabeled wt rtDRE1 with its complementary oligonucleotide, mut rtDRE1 (ACCTTTGCGCGCTATCGAAAT and its complementary oligonucleotide. For in vitro DNA binding assays, approximately equal amounts of in vitro produced rtAhR2α or rtAhR2β and rtARNTb proteins were incubated in the presence of 10 nm TCDD in 0.2% Me2SO or Me2SO alone for 90 min at 22 °C. Following incubation, 1.5 μg of poly(dI-dC) and binding buffer (20 mm Hepes, pH 7.9, 100 mm NaCl, 1 mm dithiothreitol, 6% (v:v) glycerol) were added and the incubation continued for an additional 20 min at 22 °C before the addition of 100,000 cpm of the wt rtDRE probe and 10-fold molar excess of unlabeled wild type rtDRE, or mutated rtDRE competitor DNAs. After 20 min incubation at 22 °C, complexes were resolved on a 0.5 × TBE (90 mm Tris, 64.6 mm boric acid, and 2.5 mm EDTA, pH 8.3) 4.5% acrylamide gel at 4 °C. The dried gels were exposed to a phosphor screen overnight before analysis.
Genomic Southern Blots
Genomic DNA was isolated from rainbow trout livers and Southern blots were prepared using standard techniques (27). Membranes were hybridized overnight at 42 °C in 50% formamide hybridization solution with random primed 32P-labeled DNA probes. Membranes were washed with high stringency (0.1 × SSC, 0.1% SDS). Probe 1 was made by digesting rtAhR2α with SpeI and HindIII to release a 520-bp fragment of the 3′-UTR which specifically bound to rtAhR2α sequences. Probe 2 was made by digesting rtAhR2β with PvuII and gel isolating a 598-bp fragment from the 5′ end of the coding sequence. Probe 2 cross-hybridizes with both rtAhR2α and rtAhR2β sequences.
Total RNA and Poly(A+) RNA Isolation
Total RNA was isolated using two methods. Plates of cells exposed to TCDD in Me2SO or Me2SO alone were rinsed with culture media to remove dosing solution, lysed, and scraped into Qiashredder homogenizers (Qiagen, Chatsworth, CA). RNA was isolated from the lysates using Qiagen RNeasy kits. RNA was isolated from cells and organ tissues using TRI reagent (Molecular Research Laboratories, Cincinnati, OH) as directed by the manufacturer. Poly(A+) RNA was isolated from total RNA using PolyATtract™ mRNA isolation kits (Promega, Madison, WI).
Northern Blots
RNA was separated in 1.2% denaturing formaldehyde agarose gels, blotted to nylon with 20 × SSC, and hybridized overnight at 42 °C with random primed 32P-labeled DNA probes in 50% formamide hybridization solution (19). Relative message levels were determined using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Reporter Vectors
prt1Aluc was constructed using PCR amplification of rainbow trout genomic DNA (forward primer 5′-AGGTTGGTTGAGTGAGATG-3′; reverse primer 5′-TGCAGGGAGATCGAAGAAG-3′) to amplify a 1540-bp portion of the 5′-flanking region of the rainbow trout CYP1A gene promoter from base pair 139–1678 (GenBank accession number S69277). This includes 2 DREs and the transcriptional start site (position 1594). The PCR product was TA cloned into pGemT-Easy. The resulting plasmid was digested in its multiple cloning region with SacI and NcoI and ligated into pGL3-Basic. This plasmid was named prt1Aluc and provided a TCDD responsive firefly luciferase reporter vector under control of the rainbow trout CYP1A gene promoter.
The pGudluc 1.1 reporter vector (28) was obtained from Dr. Michael Denison (University of California, Davis, CA). This reporter vector is based on pGL2-Basic and has the firefly luciferase gene under control of a 484-bp fragment of the mouse CYP1A1 enhancer, that contains 4 DREs and the murine mammary tumor virus promoter. The pRL-TK vector (Promega) was used in all experiments as a control for transfection efficiency. This vector contains the Renilla reniformis luciferase gene under the control of a herpes simplex virus thymidine kinase promoter.
Reporter Gene Assays
Assays were performed 24 h after exposure to TCDD. A Dual Luciferase Assay (Promega) was used to determine firefly (AhR agonist-dependent) and Renilla (transfection control) luciferase activity for each well. Media was removed by vacuum aspiration, each well was washed with 1 × PBS and 100 μl of passive lysis buffer was added. Plates were incubated 20 min at room temperature on an orbital shaker. Cell lysis was confirmed microscopically and a 10-μl aliquot was transferred to a 96-well luminometer plate. Luminescence assays were completed using a Dynatech Laboratories ML-2250 luminometer (Chantilly, VA). Assays for luciferase activity were conducted as follows: 50 μl of luciferase assay buffer II was injected into each well, incubated 2 s, and the resulting luminescence integrated over the next 10 s. After reading each plate the assay buffer was changed to Stop & Glo and identical assay conditions were used to measure Renilla activity in the same wells. Because the Renilla luciferase control vector is susceptible to induction by trans effects when a second reporter construct with a strong promoter is activated, the amount of transfected control plasmid was reduced to 3 ng of pRL-TK/μg DNA in each well in order to avoid this problem.
Statistical Analysis
TCDD dose-response experiments in RTG-2 cells were completed in triplicate. AhR or CYP1A mRNA PhosphorImager signals were normalized by dividing by the corresponding β-actin mRNA signals. This normalized value obtained for each TCDD dose was divided by the mean value for the Me2SO control to determine the magnitude of induction by TCDD. For transactivation experiments in transiently transfected COS-7 cells, graded concentrations of TCDD were used to produce dose-response curves with each AhR/reporter gene pair and each experiment was repeated 3–5 times. A normalized luciferase activity number was determined for each assay well by dividing the firefly luciferase activity by Renilla luciferase activity. EC50 values were determined using a nonlinear estimation process for determination of half-maximal response in the Statistica software package (Stat-Soft, Tulsa, OK). Level of significance for all analyses was p < 0.05.
RESULTS
Cloning of Two Rainbow Trout AhR cDNAs
A random primed rainbow trout cDNA library was screened using a previously cloned F. heteroclitus AhR cDNA fragment as a probe (20). A total of 8.0 × 106 plaques were screened, and 16 clones were isolated. These clones fell into 2 groups, termed α and β, by restriction digest and sequence. Sequencing demonstrated that both classes of clones were homologous to mammalian AhRs; however, these sequences were incomplete: each clone contained a section of 5′-untranslated region (UTR), an initiation ATG codon and a single open reading frame which continued to the end of the insert without a stop codon. A second cDNA library was constructed using a poly(dT) primer so that cDNAs encompassing the 3′ end of mRNAs would be favored. Screening this library yielded clones corresponding to the α and β clones. These had regions overlapping the previously isolated clones at their 5′ ends, and as expected, these clones contained complete 3′ ends. From this it was deduced that two distinct AhR mRNAs are expressed in RTG-2 cells.
Comparison of Rainbow Trout AhRs to AhRs and PAS Proteins from Other Species
The full-length sequences for rtAhR2α and rtAhR2β were obtained by sequencing each strand at least 3 times. These sequences were each found to have a single open reading frame, encoding proteins of 1059 and 1058 amino acids, respectively. This is approximately 200 amino acids longer than most other AhR sequences previously reported. The predicted molecular mass of unmodified rtAhR2α and rtAhR2β is 115 kDa, somewhat smaller than the 145-kDa mass estimated for AhR in RTG-2 cells and trout liver by photoaffinity labeling (7, 8). The two predicted amino acid sequences show 95% identity with each other. Most of the differences between the two amino acid sequences occur in the first 345 amino acids, however, occasional differences appear throughout the length of the protein sequences.
Fig. 1 presents an alignment of the rtAhR2α and rtAhR2β predicted amino acid sequences with the human AhR sequence (29). The greatest similarity between the sequences is found within the conserved basic helix-loop-helix and the PAS A and B functional domains. The basic helix-loop-helix domain has been shown to mediate DNA binding and play a secondary role in AhR/ARNT dimerization in mammalian AhRs. The PAS region is important in ligand binding and AhR/ARNT dimerization (30, 31). Notably, the rainbow trout AhRs do not contain a Gln-rich region that is thought to play an important role in mediating transactivation by the AhR. The zebrafish AhR and the Atlantic tomcod AhR sequences also lack a Gln-rich domain (10, 11). BLASTP comparison of the rainbow trout AhRs to the GenBank data base showed that the rtAhR2 sequences are most similar to existing AhR sequences, with significant but lower similarity to other PAS family proteins. The amino-terminal halves of the protein sequences (including the basic helix-loop-helix and PAS domains) have the greatest similarity. Both sequences resemble the fish AhR2 sequences more closely than the AhR1 sequences reported by Hahn et al. (11) and are therefore designated AhR2s.
Fig. 1. Comparison of rtAhR2α, rtAhR2β, and human AhR deduced amino acid sequences.
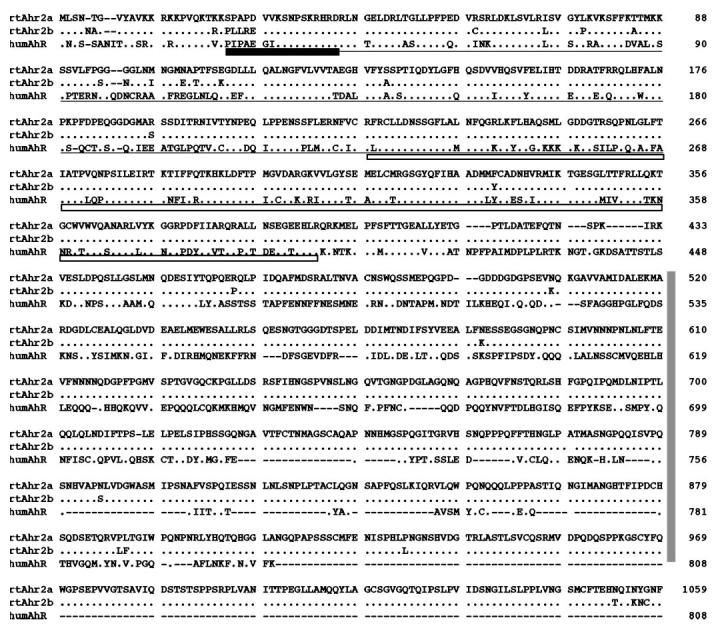
Periods indicate identities and dashes indicate gaps. Functional domains defined for the human AhR (29) are indicated (DNA-binding domains, dark box; ARNT dimerization, underlined; ligand binding, open box; transactivation, vertical box). Sequence alignment was completed using ClustalW (51). GenBank accession numbers for the sequences are as follows: rtAhR2α (AF065137), rtAhR2β (AF065138), and human AhR (S41124).
In Vitro Translation and Functional Characterization of rtAhR2α and rtAhR2β
To demonstrate the ability of the rtAhR2α and rtAhR2β cDNAs to encode proteins, we used a coupled in vitro transcription/translation system to make [35S]methionine-labeled translation products. Fig. 2 shows the phosphorimage of the SDS-PAGE separated in vitro translation products. A single band of approximately 125 kDa was produced with each cDNA. This is intermediate between the 115-kDa size predicted from the coding sequence and the estimated 145-kDa size observed in the photoaffinity ligand binding experiments.
Fig. 2. In vitro translation of rtAhR2α and rtAhR2β.
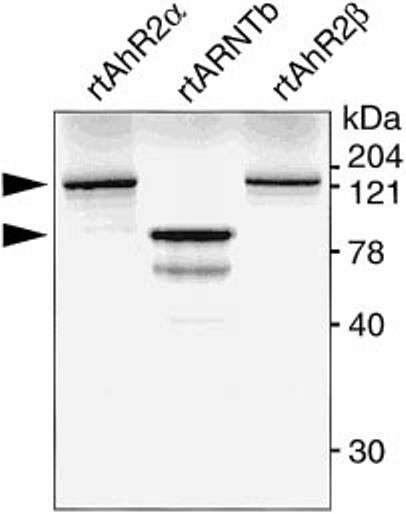
[35S]Methionine-labeled in vitro translated rtAhR2α, rtAhR2β, and rtARNTb proteins were resolved on an 8% SDS-polyacrylamide gel. A phosphorimage of the dried gel is shown. Arrows indicate position of the full-length proteins.
The in vitro translated proteins were tested for ability to bind TCDD (Fig. 3). Lysates containing unlabeled in vitro translated proteins were incubated with [3H]TCDD and analyzed by velocity sedimentation on sucrose density gradients by the method of Tsui and Okey (23). Both trout AhR2s exhibited a peak of [3H]TCDD specific binding (i.e. binding that was abolished by a 200-fold excess of 2,3,7,8-TCDF) with a sedimentation coefficient of 10.6 S. This peak was not seen when unprogrammed lysate was used in place of lysate containing the AhR translation products. This experiment shows that rtAhR2α and rtAhR2β both encode proteins capable of specific, high-affinity binding of TCDD.
Fig. 3. Specific binding of [3H]TCDD to rtAhR2α and rtAhR2β as analyzed by velocity sedimentation on sucrose gradients.
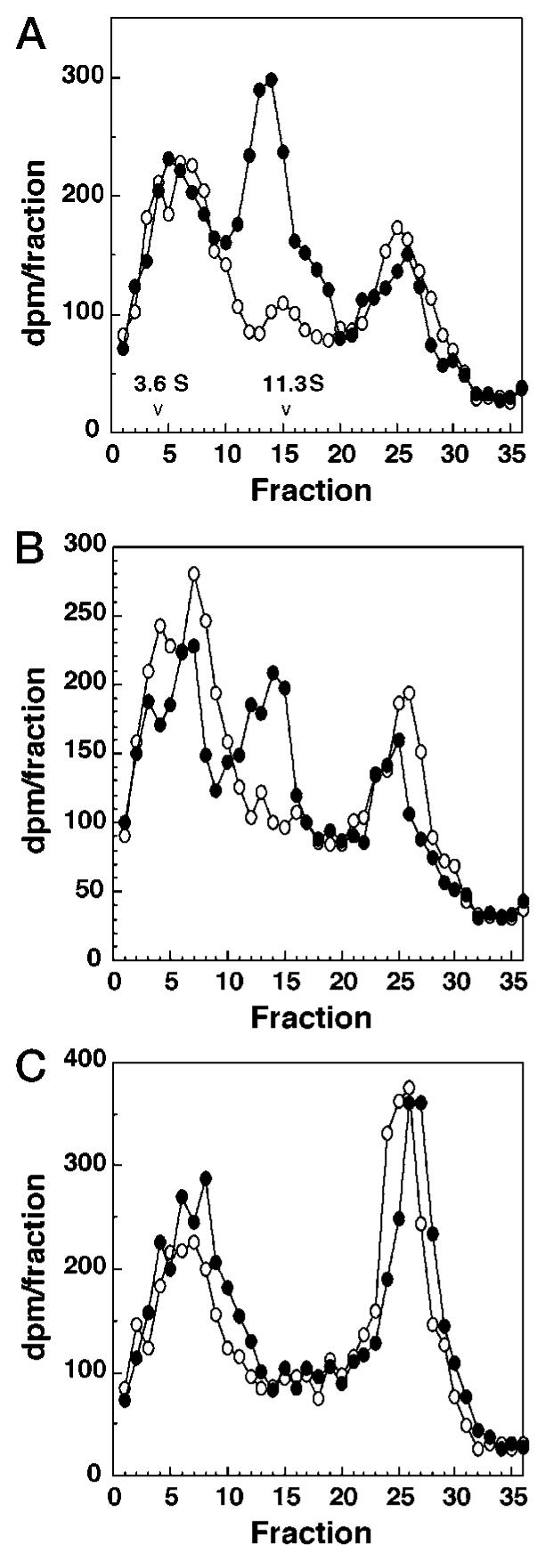
AhR proteins were expressed by in vitro transcription and translation (TnT), diluted 1:1 with MEEDMG buffer containing molybdate (see “Experimental Procedures”), and incubated with [3H]TCDD (2 nm) ± TCDF (400 nm) for 1–2 h at 4 °C. After each incubation, 90 μl was applied to 10–30% sucrose gradients and spun for 140 min at 60,000 rpm in a VTi 65.2 rotor. Gradients were fractionated (150 μl per fraction) and counted. Specific binding is the difference between total binding (−TCDF, solid circles) and nonspecific binding (+TCDF, open circles). [3H]TCDD concentration was verified by sampling each tube for total counts. [14C]Catalase (11.3 S) and [14C]ovalbumin (3.6 S) were added as sedimentation markers; their elution positions are indicated. A, rtAhR2α; B, rtAhR2β; and C, unprogrammed lysate.
To further confirm that the clones encode functional AhR proteins, gel shift experiments were used to demonstrate specific binding to double-stranded DNA fragments containing a DRE from the rainbow trout CYP1A promoter. In vitro translated proteins were preincubated with in vitro translated rainbow trout ARNT along with 10 nm TCDD in order to form an active, DNA binding complex. After activation, a 32P-labeled double-stranded oligonucleotide containing the DRE (core TAGCGTG) was added, and the bound probe was separated from free oligonucleotide probe by native gel electrophoresis (Fig. 4). This experiment shows that both the α and β forms of the receptor produced a shifted band (solid arrow) that could be specifically competed by a 10-fold excess of unlabeled oligonucleotide. This competition was substantially reduced when the unlabeled oligonucleotide was mutated at a single position (core TAGCGCG; mutated base underlined). Addition of either ARNT alone or the AhRs without ARNT, failed to produce specific bands, although nonspecific bands (open arrows) were observed in these lanes. Although both forms of the receptor produced a readily visible shifted band, the α form of the receptor consistently produced a stronger band with this probe than the β form. These results also demonstrate that in vitro rtAhR2/rtARNTb DRE binding is independent of added ligand. Ligand-independent DNA binding has also been reported for the mammalian AhR (32).
Fig. 4. Gel-shift analysis of rtAhR2s and rtARNTb interactions in vitro.
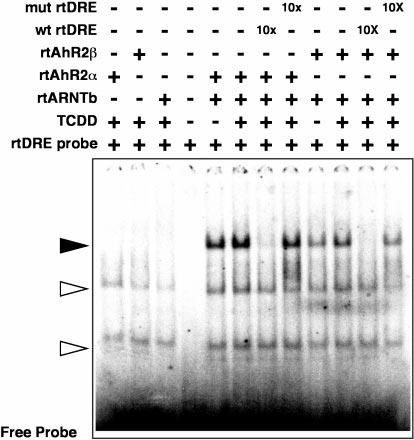
Equal amounts of in vitro translated rtAhR2α or rtAhR2β proteins were incubated with equal molar amounts of rtARNTb with, or without, 10 nm TCDD. The samples were then incubated with a 32P-labeled oligonucleotide probe derived from a DRE in the rainbow trout CYP1A enhancer. In some lanes a 10-fold molar excess of unlabeled competitor oligonucleotides was also added as indicated. The bound and free oligonucleotides were separated on a native acrylamide gel, and a phosphorimage of the dried gel is shown. The solid arrow indicates the rtAhR2·rtARNTb·DRE complexes. Open arrows indicate positions of nonspecific complexes.
Evidence for Distinct Genes
Several pieces of evidence suggest that the α and β clones are encoded by distinct genes. First, the differences between the two sequences are widely scattered throughout the sequence, making differential splicing an unlikely mechanism for producing the two messages. Second, probing a genomic Southern blot with a cDNA expected to hybridize with both sequences consistently produced at least two bands (Fig. 5A). In contrast, a probe expected to hybridize with only rtAhR2α sequences consistently produced a more simple pattern (Fig. 5B). Finally, RT-PCR analysis of mRNA from RTG-2 cells, 6 individual juvenile rainbow trout, and 24 individual rainbow trout fry demonstrated that the two different transcripts were present in all samples (data not shown). If these two transcripts were allelic variants, it would be predicted that some of the 30 individuals tested would be homozygous for one of the alleles. Taken together, our results suggest that these transcripts are likely to be the products of different genes.
Fig. 5. Southern blot with rainbow trout genomic DNA.
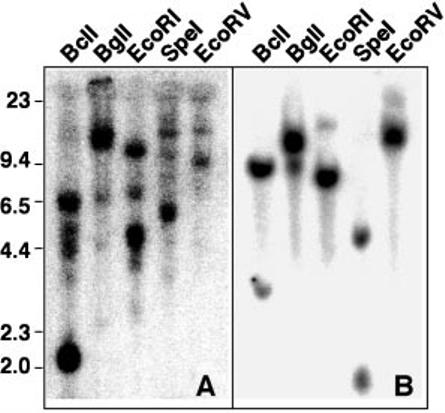
Genomic DNA, 25 μg/lane, was digested with the indicated enzyme, separated on a 0.8% agarose gel, and blotted. A, the blot was probed with probe 2 which cross-reacts with both rtAhR2α and rtAhR2β sequences. B, the blot was stripped and probed with probe 1, which hybridizes specifically with rtAhR2α.
TCDD Induction of rtAhR2 mRNA Abundance
RTG-2 cells were treated with graded concentrations of TCDD in Me2SO, and total RNA was isolated 6 h later for Northern blotting (Fig. 6). Using a probe that hybridizes with both the α and β sequences (probe 2), we observed two bands on the RTG-2 total RNA blot. These bands correspond to rtAhR2β and rtAhR2α mRNA in mobility. The blot was also probed with a fragment complimentary to 3′-untranslated portions of the rtAhR2α message (probe 1), sequences that are not found in the rtAhR2β mRNA. With this probe, we observed only a single band, consistent with the expected position of rtAhR2α. We therefore conclude that the upper band represents rtAhR2α mRNA, and that TCDD produces a dose-related increase in both rtAhR2β and rtAhR2α mRNA abundance. The blot was also probed with a fragment complimentary to the rainbow trout CYP1A mRNA (19), demonstrating that the TCDD dose-responsive induction of rtAhR2β, rtAhR2α, and CYP1A mRNAs occurs over a similar range of TCDD concentrations.
Fig. 6. Induction of rtAhR2β and rtAhR2α mRNA by TCDD in a rainbow trout gonadal cell line (RTG-2).
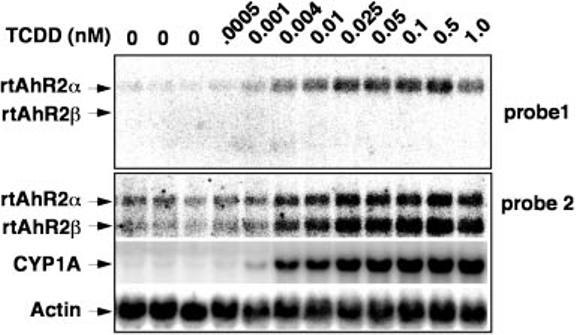
RTG-2 cells were treated with graded concentrations of TCDD as indicated, and RNA was isolated after 6 h. Phosphorimage of a representative Northern blot, with 20 μg of total RNA, hybridized with probe 1, probe 2, β-actin, and CYP1A probes is indicated.
TCDD treatment also induced both rtAhR2α and rtAhR2β mRNAs in kidney of juvenile rainbow trout. Trout were injected intraperitoneally with TCDD, (10 μg/kg) or vehicle as a control, and tissues were collected for Northern blotting 3 days after injection. As shown in Fig. 7, TCDD induced both rtAhR2α and rtAhR2β mRNAs in kidney, but failed to change these message levels in spleen and liver. This result confirms that the TCDD induction of rtAhR2α and rtAhR2β seen in RTG-2 cells also occurs in a least some cells in the whole organism.
Fig. 7. TCDD induction of rtAhR2α and rtAhR2β mRNA in kidneys of juvenile rainbow trout.
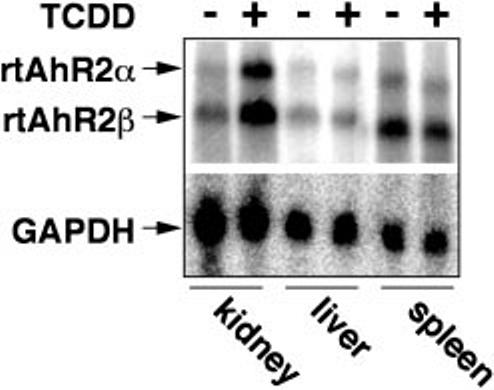
Rainbow trout were treated with vehicle or 10 μg/kg TCDD and kidneys, livers, and spleens were isolated for Northern blotting as described under “Experimental Procedures.” The blot was probed with probe 2, which hybridizes with both rtAhR2α and rtAhR2β as indicated by the arrows. The blot was stripped and a probe for glyceraldehyde-3-phosphate dehydrogenase was used to control for variations in loading and transfer.
Differences in rtAhR2α and rtAhR2β mRNA Abundance in Rainbow Trout Tissues
A Northern blot using RNA samples isolated from different rainbow trout organs showed tissue-specific differences in the expression of rtAhR2α and rtAhR2β. A Northern blot using poly(A+) RNA samples from the indicated rainbow trout organs was probed with a fragment that hybridizes with both of the AhR transcripts (Fig. 8A). The blot shows that rtAhR2α mRNA is consistently expressed at lower levels than the rtAhR2β message. However, the ratio of AhR2β to AhR2α message varied between tissues (Fig. 8B). The rank order for organs with the highest AhR2β message abundance relative to rtAhR2α was heart > liver= brain > kidney = blood = intestine = spleen. Expression of both messages was low and similar in the ovary. This tissue-specific expression suggests that the two forms of AhR may play distinct roles in different tissues.
Fig. 8. Organ-specific expression of rtAhR2α and rtAhR2β in juvenile rainbow trout.
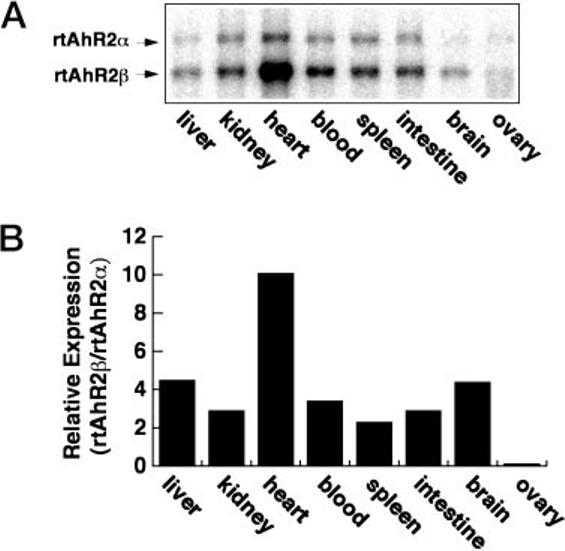
Poly(A+) RNA (3 μg/lane) from the indicated organs of juvenile rainbow trout was separated in 1.2% denaturing agarose, blotted to nylon, and hybridized with a probe that recognizes both rtAhR2α and rtAhR2β transcripts.
Comparison of Transcriptional Activity of rtAhR2α and rtAhR2β with Different Enhancers
To further demonstrate functional activity of rtAhR2α and rtAhR2β, we used a transfection assay to measure transactivation. This assay system uses transfected COS-7 monkey kidney cells to express the different AhR/ARNT proteins, and a luciferase reporter vector driven by promoter fragments from either the rainbow trout CYP1A or the mouse CYP1A1 genes to measure transactivation activity. This allowed us to compare the activity of different AhR/ARNT proteins from 3 different species, rainbow trout, zebrafish, and human, in a setting where the cellular background remained constant, and only the receptor molecules and the enhancer elements were varied.
In these experiments, we compared the activity of rtAhR2α and rtAhR2β from rainbow trout to that of human AhR (huAhR) and a zebrafish AhR (zfAhR2). The AhR cDNAs were transfected together with ARNT cDNAs from the same species. In the case of rainbow trout, the same form of ARNT (rtARNTb) was paired with both the α and β forms of the AhR. Transcription of the AhR and ARNT constructs was verified by Northern blot (data not shown). COS cells do not express endogenous AhR, but do express a small amount of endogenous ARNT protein (33). The COS cells were transfected with the constructs, exposed to graded concentrations of TCDD, and as sayed for luciferase activity as a measure of transactivation. The transfection efficiency in each assay well was monitored by inclusion of a control construct, pRL-TK, expressing Renilla luciferase activity in all transfections.
When the reporter construct contained the rainbow trout CYP1A enhancer, along with huAhR, zfAhR2, or rtAhR2α, addition of TCDD gave a dose-dependent increase in luciferase expression (Fig. 9, top). In contrast, cells transfected with rtAhR2β showed little if any transcriptional response to TCDD. However, rtAhR2β is not transcriptionally inactive in all settings. When cells containing the reporter driven by the mouse enhancer elements were transfected with rtAhR2β, there was a TCDD dose-related increase in luciferase expression (Fig. 9, bottom). On the other hand, zfAhR2 was less active with the mouse enhancer fragment. Thus, in addition to tissue differences in message expression pattern, the two forms of rainbow trout AhR differ in their preference for enhancers. This suggests that the two receptors may regulate distinct sets of genes.
Fig. 9. Transactivation activity of rtAhR2α, rtAhR2β, zfAhR2, and huAhR with two different AhR-responsive luciferase reporter vectors.
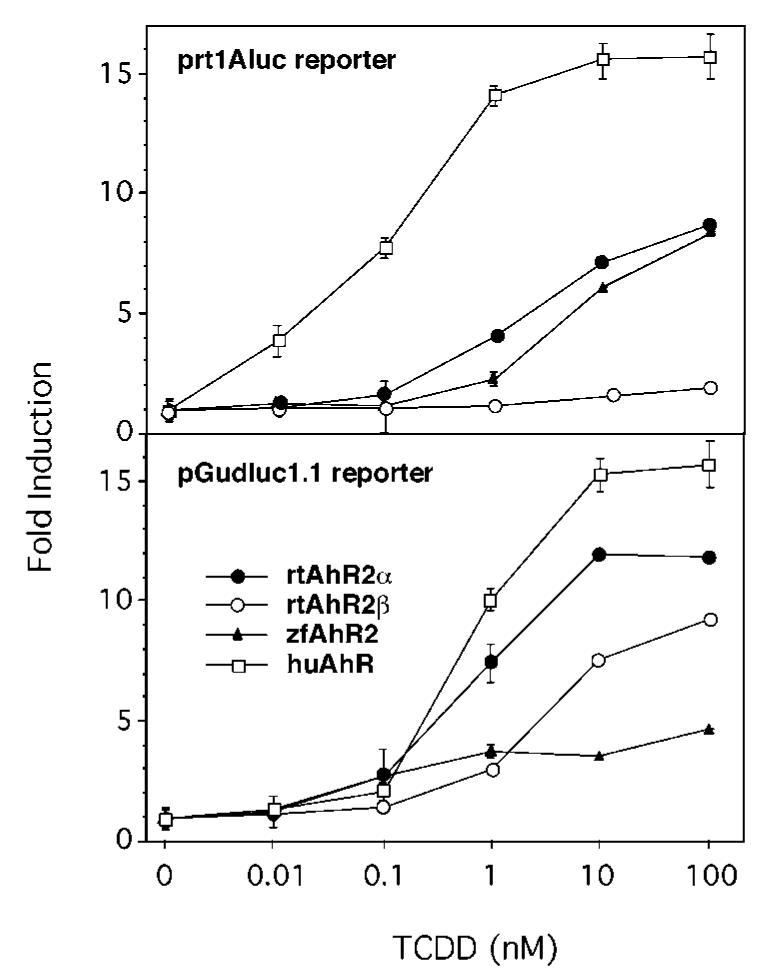
COS-7 cells were transfected with the indicated AhR and the species-specific ARNT expression construct, along with pRL-TK and one of the reporter vectors, either prt1Aluc or pGudluc 1.1. Points represent fold induction of luciferase activity relative to Me2SO control (mean ± S.E., n = 3).
DISCUSSION
Rainbow Trout Express Two Distinct AhR Transcripts
We have isolated two different AhR2 clones from rainbow trout cDNA libraries. These two cDNAs encode proteins of 1058 and 1059 amino acids that are 95% identical at the amino acid level. The rainbow trout AhRs are substantially longer than mammalian AhR proteins, which range from 805 amino acids for mouse, to 853 amino acids for the rat AhR. For comparison, the Atlantic tomcod (Microgadus tomcod) receptor contains 823 amino acids (10), and the zebrafish AhR is 1027 amino acids long (11). The rtAhR2s show strong similarities to mammalian receptors over the first 400 amino acids, but the carboxyl termini are much less conserved. Notably, the rainbow trout AhRs do not contain the Gln-rich domain made up of repeated glutamines that are thought to play an important role in mediating transactivation (34). The zebrafish and Atlantic tomcod sequences also lack Gln-rich domains (10, 11). While the stretches of polyglutamine are absent in the fish receptors, there appear to be corresponding segments of polyasparagine in approximately the same locations. In addition to the repeated glutamines, several other regions in the carboxyl-terminal half of the mammalian AhR are also important in transactivation (35). Three motifs have been found to generally mediate transactivation: acidic amino acid-rich, glutamine-rich, or areas rich in a mixture of proline/serine/threonine (36). Examining the distal end of the rainbow trout AhRs we find several regions that match these specifications. The most striking of these in rtAhR2α runs from amino acid 985 to 994 as … STSTSPPSRP … with 9 of 10 residues falling into this category. Serine/threonine-specific phosphatases have been found to alter the rate of AhR transcriptional enhancement at a step subsequent to AhR/ARNT DNA binding (37). Confirmation of transactivation activity in these sequences will require further study.
For both the α and β clones, open reading frames were found out of frame and just upstream of the coding sequence. Multiple resequencing experiments confirm that these are truly out of frame with the AhR ORFs, do not encode part of the receptor, and are highly similar to each other. The strong conservation between these two upstream sequences suggests that they serve an important function. One possibility is that they encode a functional product. If so, this would be a novel protein, in that the predicted amino acid sequences encoded by these regions failed to match any known protein sequences in the data bases. Perhaps a more likely possibility is that these sequences are involved in some form of regulation of AhR gene expression, either controlling transcription or translation.
There are several possible explanations for our finding two distinct AhR clones: they could represent products from two different genes, they might be splice variants from a single gene, or they might represent allelic variants of a single gene. The wide distribution of substitutions between the two clones is difficult to account for by splicing, requiring either a very large number of splices, or almost total gene duplication to produce the two different messages. In addition, when a genomic Southern blot was probed with a labeled fragment that was expected to cross-hybridize with both sequences, a consistent pattern of at least two bands was observed. Because the probe was relatively short (598 bp) it is unlikely that in all cases multiple bands were produced by cleavage of a single gene within the region corresponding to the probe. Indeed, when the blot was reprobed with a similar sized fragment corresponding to the 3′-UTR of rtAhR2α, with no similarities to the rtAhR2β clone, a much less complex pattern was consistently observed. In addition to the genomic Southern blot, an experiment using reverse transcriptase/PCR amplification of sequences from the mRNA of 30 individual fish demonstrated the presence of both mRNAs in all of the samples. If the two sequences are allelic variants, this would indicate that 30 out of 30 fish are heterozygotes, with 0 out of 30 fish homozygous for either allele. Since with two alleles, an individual fish has a 50% chance of being homozygous for one allele or the other, the odds of obtaining no homozygotes in 30 fish are very small. Our results are most consistent with at least two distinct AhR genes in rainbow trout.
Multiple AhRs in Fish
Other fish species also appear to have more than a single AhR gene. The cloning of multiple AhR types from several fish species (rainbow trout, zebrafish, killifish, and smooth dogfish) raises questions regarding the function of these multiple genes (12). It is hypothesized that vertebrates underwent an ancient genome duplication event and that some fish, including salmonids and catastomids, underwent a second such duplication more recently (38-40). Comparison of rtAhR2α and rtAhR2β to the partial sequences of AhR1 and AhR2 from killifish and smooth dogfish show that both rtAhR2s are more similar to the AhR2 forms than either is to AhR1. Therefore, an AhR1 gene may exist in rainbow trout. A portion of a third AhR-like transcript in rainbow trout has been isolated but not fully characterized (41). It has been suggested that complete redundancy of function after gene duplication is unstable, and over time one of the duplicated genes will become inactive, or will diverge in function. We have demonstrated that the two rtAhR2s are differentially expressed between organs, suggesting divergence in function.
Differences between rtAhR2α and rtAhR2β
The two forms of AhR from rainbow trout show remarkable similarity in size and sequence. They both bind TCDD and interact with DRE containing sequences. Despite these similarities, the two receptor forms show surprising differences in expression pattern and transactivation properties. The two rtAhR2s are expressed in different tissues and at different levels. rtAhR2β mRNA was identified in all organs examined, albeit at very low levels in the ovary, and at strikingly high levels in the heart. The high level of rtAhR2β mRNA in the heart is of interest toxicologically because the heart has been identified as a key target organ for TCDD developmental toxicity in early life stages of rainbow trout (42-44).
We have found that TCDD induces rtAhR2α and rtAhR2β mRNA levels in RTG-2 cells as well as in kidney cells in juvenile trout. The elevation in rtAhR2β and rtAhR2α mRNA in RTG-2 cells mirrors the induction of CYP1A mRNA, which is believed to be the result of AhR activation. The TCDD EC50 values for induction of the three transcripts in RTG-2 cells are about 0.025 nm TCDD and are maximally elevated at 6 h. These findings suggest that rtAhR2α and rtAhR2β expression is directly induced by ligand activation of the rainbow trout AhR. This appears to be distinct from the regulation of AhR expression observed in mammals. In mammalian cell lines and organs, AhR protein is rapidly depleted after acute exposure to TCDD (45-48). Exposure to TCDD during mouse embryonic development caused a decrease in the amount of AhR mRNA and AhR protein in the palate of embryonic mice (49). The difference between our results with rtAhR2α, rtAhR2β, and the previous results might be ascribed to a fundamental difference between fish and mammalian systems, as the zebrafish AhR2 mRNA is also induced by TCDD (11). However, the Atlantic tomcod AhR mRNA in liver does not appear to be regulated by aryl hydrocarbons (10).
Our finding that AhR isoforms from the same species have different specificities for enhancer sequences indicates that the rtAhR2α and rtAhR2β isoforms may regulate distinct sets of genes. This is surprising given the high degree of sequence conservation between the two receptor forms. The zebrafish receptor also showed a distinct preference for one reporter construct over another. In contrast to rtAhR2β, the zfAhR2 recognizes the trout enhancer, but is only weakly active with the mouse reporter. We have previously shown that the zebrafish AhR2/ARNT dimer only weakly binds to a DRE based on the mouse CYP1A1 gene enhancer, although it does bind a similar oligonucleotide based on the rainbow trout CYP1A gene enhancer (11). This is despite the fact that both enhancers contain nearly identical core consensus DRE sequences. Other researchers have reported reduced activity of the rainbow trout CYP1A promoter in a mammalian cell line when compared with mammalian CYP1A1 promoter sequences (50). The possibility that the two trout AhRs induce distinct sets of genes, taken with the observed variations in tissue distribution, suggests that the two isoforms may mediate different biological responses. Whether one form or the other contributes more to mediating TCDD toxicity in rainbow trout remains to be seen. These results also suggest that there may be more than one form of the AhR in other vertebrate classes and should serve as an impetus for their discovery and functional characterization.
Acknowledgments
We thank Dr. Sibel Karchner for assistance in the cloning of the partial F. heteroclitus AhR, Eric Andreasen for help with RTG-2 experiments, and Zhengjin Cao and Dr. Judd Aiken for assistance with in vivo rtAhR2 expression experiments.
Footnotes
This work was supported in part by the University of Wisconsin Sea Grant Institute under the National Sea Grant College Program, National Oceanic and Atmospheric Administration, United States Department of Commerce, and the State of Wisconsin, Federal Grant NA46RG0481 and Sea Grant project number R/MW-58 (to W. H. and R. E. P.), National Institutes of Health NIEHS Grant R29 ES06272 and Sea Grant College Program Office Grant NA46RG0470, Sea Grant project number R/B-124, and the Woods Hole Oceanographic Institute (to M. E. H.). This is contribution 9742 from the Woods Hole Oceanographic Institution and contribution 324 of the University of Wisconsin, Environmental Toxicology Center.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AF027130, AF065137, and AF065138.
The abbreviations used are: AhR, aryl hydrocarbon receptor; rt, rainbow trout; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; ARNT, aryl hydrocarbon receptor nuclear translocator; P450lA, cytochrome P4501A protein; cyp1A cytochrome P4501A; DRE, dioxin response element; RTG-2, rainbow trout gonadal cells; UTR, untranslated region; GCG, genetics computer group; EC50, half-maximal response; Me2SO, dimethyl sulfoxide; RT-PCR, reverse transcriptase-polymerase chain reaction; bp, base pair(s); MOPS, 4-morpholinepropanesulfonic acid; TCDF, 2,3,7,8-tetrachlorodibenzofuran.
REFERENCES
- 1.Walker MK, Spitsbergen JM, Olson JR, Peterson RE. Can. J. Fish Aquat. Sci. 1991;48:875–883. [Google Scholar]
- 2.Elonen GE, Sphear RL, Holcombe GW, Johnson RD. Environ. Toxicol. Chem. 1998;17:472–483. [Google Scholar]
- 3.Cook PM, Zabel EW, Peterson RE. In: Chemically-induced Alterations in the Functional Development and Reproduction of Fishes. Rolland RM, Gilbertson M, Peterson RE, editors. SETAC Technical Publications Series; 1997. pp. 9–27. [Google Scholar]
- 4.Rowlands JC, Gustafsson JA. Crit. Rev. Toxicol. 1997;27:109–134. doi: 10.3109/10408449709021615. [DOI] [PubMed] [Google Scholar]
- 5.Walker MK, Peterson RE. In: Dioxins and Health. Schecter A, editor. Plenum Press; New York: 1994. pp. 347–387. [Google Scholar]
- 6.Hahn ME. Comp. Biochem. Physiol. C. 1998;121:23–53. doi: 10.1016/s0742-8413(98)10028-2. [DOI] [PubMed] [Google Scholar]
- 7.Swanson HI, Perdew GH. Toxicol. Lett. 1991;58:85–95. doi: 10.1016/0378-4274(91)90194-b. [DOI] [PubMed] [Google Scholar]
- 8.Hahn ME, Poland A, Glover E, Stegeman JJ. Arch. Biochem. Biophys. 1994;310:218–228. doi: 10.1006/abbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 9.Pollenz RS, Sullivan HR, Holmes J, Necela B, Peterson RE. J. Biol. Chem. 1996;271:30886–30896. doi: 10.1074/jbc.271.48.30886. [DOI] [PubMed] [Google Scholar]
- 10.Roy NK, Wirgin I. Arch. Biochem. Biophys. 1997;344:373–386. doi: 10.1006/abbi.1997.0238. [DOI] [PubMed] [Google Scholar]
- 11.Tanguay RL, Abnet CC, Heideman W, Peterson RE. Biochim. Biophys. Acta. 1999;1444:35–48. doi: 10.1016/s0167-4781(98)00252-8. [DOI] [PubMed] [Google Scholar]
- 12.Hahn ME, Karchner SI, Shapiro MA, Perera SA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Q, Whitlock JP., Jr. J. Biol. Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- 14.Denison MS, Fisher JM, Whitlock JP., Jr. J. Biol. Chem. 1988;263:17221–17224. [PubMed] [Google Scholar]
- 15.Krishnan V, Porter W, Santostefano M, Wang X, Safe S. Mol. Cell. Biol. 1995;15:6710–6719. doi: 10.1128/mcb.15.12.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berndtson AK, Chen TT. Arch. Biochem. Biophys. 1994;310:187–195. doi: 10.1006/abbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 17.Roy NK, Kreamer GL, Konkle B, Grunwald C, Wirgin I. Arch. Biochem. Biophys. 1995;322:204–213. doi: 10.1006/abbi.1995.1453. [DOI] [PubMed] [Google Scholar]
- 18.Zabel EW, Walker MK, Hornung MW, Clayton MK, Peterson RE. Toxicol. Appl. Pharmacol. 1995;134:204–213. doi: 10.1006/taap.1995.1185. [DOI] [PubMed] [Google Scholar]
- 19.Zabel EW, Pollenz R, Peterson RE. Environ. Toxicol. Chem. 1996;15:2310–2318. [Google Scholar]
- 20.Hahn ME, Karchner SI. Biochem. J. 1995;310:383–387. doi: 10.1042/bj3100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasiewicz TA, Neal RA. Toxicol. Appl. Pharmacol. 1979;51:329–339. doi: 10.1016/0041-008x(79)90475-7. [DOI] [PubMed] [Google Scholar]
- 22.Dottavio-Martin D, Ravel JM. Anal. Biochem. 1978;87:562–565. doi: 10.1016/0003-2697(78)90706-6. [DOI] [PubMed] [Google Scholar]
- 23.Tsui HW, Okey AB. Can. J. Physiol. Pharmacol. 1981;59:927–931. doi: 10.1139/y81-143. [DOI] [PubMed] [Google Scholar]
- 24.Lorenzen A, Okey AB. Toxicol. Appl. Pharmacol. 1990;106:53–62. doi: 10.1016/0041-008x(90)90105-4. [DOI] [PubMed] [Google Scholar]
- 25.Coombs DH, Watts NR. Anal. Biochem. 1985;148:254–259. doi: 10.1016/0003-2697(85)90654-2. [DOI] [PubMed] [Google Scholar]
- 26.Martin RG, Ames BN. J. Biol. Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 27.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. In: Current Protocols in Molecular Biology. Chanda VB, editor. Vol. 1. John Wiley & Sons, Inc.; New York: 1997. [Google Scholar]
- 28.Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. Fundam. Appl. Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- 29.Dolwick KM, Schmidt JV, Carver LA, Swanson HI, Bradfield CA. Mol. Pharmacol. 1993;44:911–917. [PubMed] [Google Scholar]
- 30.Whitelaw ML, Gottlicher M, Gustafsson JA, Poellinger L. EMBO J. 1993;12:4169–4179. doi: 10.1002/j.1460-2075.1993.tb06101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolwick KM, Swanson HI, Bradfield CA. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. J. Biol. Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 33.Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S, Fujii-Kuriyama Y. J. Biol. Chem. 1994;269:27337–27343. [PubMed] [Google Scholar]
- 34.Jain S, Dolwick KM, Schmidt JV, Bradfield CA. J. Biol. Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- 35.Ma Q, Dong L, Whitlock JP., Jr. J. Biol. Chem. 1995;270:12697–12703. doi: 10.1074/jbc.270.21.12697. [DOI] [PubMed] [Google Scholar]
- 36.Ptashne M. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 37.Li SY, Dougherty JJ. Arch. Biochem. Biophys. 1997;340:73–82. doi: 10.1006/abbi.1997.9905. [DOI] [PubMed] [Google Scholar]
- 38.Bailey GS, Poulter RT, Stockwell PA. Proc. Natl. Acad. Sci. U. S. A. 1978;75:5575–5579. doi: 10.1073/pnas.75.11.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allendorf FW, Thorgaard GH. In: Evolutionary Genetics of Fishes. Turner BJ, editor. Plenum Publishing Corp.; New York: 1984. pp. 1–53. [Google Scholar]
- 40.Ferguson MM, Allendorf FW. In: Biochemistry and Molecular Biology of Fishes, Phylogentic and Biochemical Perspectives. Mommsen TP, editor. Vol. 1. Elsevier; New York: 1991. pp. 25–42. [Google Scholar]
- 41.Abnet CC, Peterson RE. Toxicol. Sci. 1998;42:385. [Google Scholar]
- 42.Hornung MW, Spitsbergen JM, Peterson RE. Toxicol. Sci. 1999;47:40–51. doi: 10.1093/toxsci/47.1.40. [DOI] [PubMed] [Google Scholar]
- 43.Cheung MO, Gilbert EF, Peterson RE. Toxicol. Appl. Pharmacol. 1981;61:197–204. doi: 10.1016/0041-008x(81)90409-9. [DOI] [PubMed] [Google Scholar]
- 44.Walker MK, Pollenz RS, Smith SM. Toxicol. Appl. Pharmacol. 1997;143:407–419. doi: 10.1006/taap.1996.8068. [DOI] [PubMed] [Google Scholar]
- 45.Prokipcak RD, Okey AB. Can. J. Physiol. Pharmacol. 1991;69:1204–1210. doi: 10.1139/y91-176. [DOI] [PubMed] [Google Scholar]
- 46.Pollenz RS. Mol. Pharmacol. 1996;49:391–398. [PubMed] [Google Scholar]
- 47.Roman BL, Pollenz RS, Peterson RE. Toxicol. Appl. Pharmacol. 1998;150:228–239. doi: 10.1006/taap.1998.8388. [DOI] [PubMed] [Google Scholar]
- 48.Giannone JV, Li W, Probst M, Okey AB. Biochem. Pharmacol. 1998;55:489–497. doi: 10.1016/s0006-2952(97)00493-0. [DOI] [PubMed] [Google Scholar]
- 49.Abbott BD, Perdew GH, Birnbaum LS. Toxicol. Appl. Pharmacol. 1994;126:16–25. doi: 10.1006/taap.1994.1085. [DOI] [PubMed] [Google Scholar]
- 50.Carvan MJ, III, Solis WA, Nebert DW. Toxicol. Sci. 1998;42:359. [Google Scholar]
- 51.Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]


