Abstract
The cytoplasmic Syk kinase plays key roles in immune responses and comprises two N-terminal regulatory Src homology 2 (SH2) domains followed by a catalytic region. Atomic structures of these domains have only been solved in isolation. To gain insights into the three-dimensional structure of full-length Syk, we have used single-particle electron microscopy. Syk acquires a closed conformation resembling the inhibited structure of Zap-70, another member of the Syk family. Such configuration suggests an inhibition of the N-terminal domains on its catalytic activity. The phosphotyrosine binding pockets of both SH2 domains are not occluded and they could interact with other phosphoproteins.
Keywords: single-particle electron microscopy, EM, Syk, ZAP-70, kinases
1. Introduction
Spleen tyrosine kinase (Syk) family proteins are non-receptor protein tyrosine kinases (PTKs) that associate with activated antigen receptors at the inner side of the plasma membrane [1, 2]. Over 10 distinct families of non-receptor PTKs exist including the Src, Tec, JAK, Csk, Abl and Syk families [1, 3]. A common structural feature of the Syk family is the presence of two N-terminal SH2 domains located in tandem and a C-terminal catalytic region (Fig. 1A). These three domains are separated physically by intervening amino acid sequences that are usually referred to as interdomain A (located between the two SH2 regions) and B (located between the second SH2 and the kinase domain) (Fig. 1A) [4]. In vertebrates, the Syk family is composed of two members, Syk and Zap70.
Fig. 1.
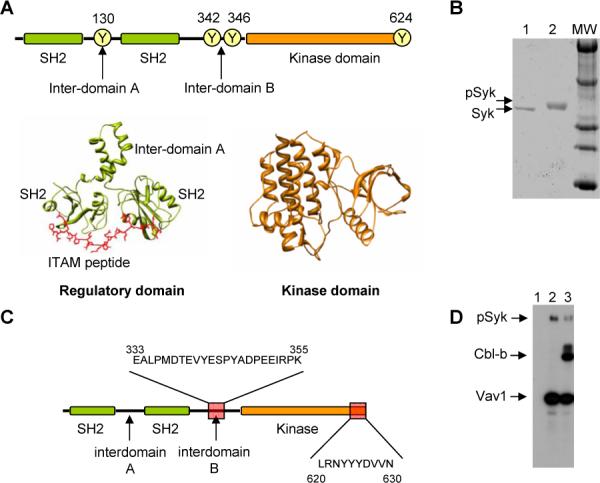
Structure and purification of GST-Syk. (A) Schematic representation of the domain organization of Syk highlighting some of the tyrosines that can become phosphorylated. At the bottom, atomic structure of the regulatory domains of Syk bound to an ITAM peptide (PDF file 1A81) [4] and the kinase domain (PDB file 1XBA) [16]. (B) SDS-PAGE gel of non-phosphorylated (0.5 μg, lane 1) and phosphorylated (1 μg, lane 2) Syk-GST. Phosphorylation assays were performed by incubation 1 mg/ml of Syk-GST with 1 mM ATP and 2.5 mM MgCl2 for 60 min at 25 °C. (C) Scheme of the two phosphorylated peptides detected using mass spectrometry. (D) Syk can autophosphorylate and also phosphorylate their targets as shown by autoradiography using radioactive ATP. Lane 1, Syk + Vav1 CH-Ac before the reaction; Lane 2, Syk + Vav1 CH-Ac after the kinase reaction; Lane 3, Syk + Vav1 CH-Ac + N-Cbl-b after the kinase reaction.
Previous reports have shown that the activation of Syk proteins during the stimulation of antigen receptors requires a complex cascade of signalling events. Those involve the previous stimulation of upstream kinases of the Src family by the antigen-bound receptors, the Src family-dependent phosphorylation of two tyrosine residues located in the immunoreceptor tyrosine-based activating motifs (ITAMs) of ancillary molecules to the antigen receptors, the translocation of Syk family members to the plasma membrane via the interaction of their SH2 domains with those phosphorylated ITAM motifs, and the eventual activation of Syk proteins by either trans-phosphorylation by Src family kinases (in the case of Zap70) or by auto-phosphorylation (in the case of Syk) [5]. Syk undergoes conformational changes upon auto-phosphorylation and/or binding to phosphorylated ITAMs that likely contribute to activate the protein kinase [6-8]. Upon activation, Syk family proteins promote the stimulation of downstream signal transduction cascades that are essential for the developmental and effector responses of several haematopoietic cell lineages and, when deregulated, for the progression of a several human pathologies [9, 10]. Despite the important roles of Syk in cell signalling and pathophysiological events, their mechanism of activation at the structural level remains still obscure. Experimental evidence indicates specific and non-overlapping Syk phosphorylation sites are important for either the stimulation of the intrinsic kinase activity of Syk or for serving as docking sites for the recognition of Syk downstream targets. Thus, tyrosine residues located in the interdomain B have been implicated in the regulation of Syk kinase activity [11]. Furthermore, phosphorylated Tyr342 is recognized by the SH2 domain of Vav1, a phosphorylation-dependent exchange factor that plays crucial roles in lymphocyte signalling [12]. The phosphorylation of Tyr342 and Tyr346 is also required for the optimal binding of another important signalling mediator, the phospholipase C-γ[13].
Although Syk and Zap-70 belong to the same family of PTKs they reveal substantial differences in their mechanisms of regulation [1]. Whereas Zap-70 expresses in T and NK cells, Syk regulates signalling in B cells, mast cells, neutrophils and macrophages. More importantly, the kinase activity of Syk is intrinsically 100-fold higher than that of Zap-70 and its stimulation does not require phosphorylation by kinases of the Src family. In contrast, activation of Zap-70 strictly requires the phosphorylation of tyrosine residues in its inter-SH2 linker region by Src. Moreover, inter-domain B is 23 aminoacid longer in Syk than in Zap-70 likely influencing its association with phoshorylated targets. Structural studies have provided some clues on the structural basis of these functional differences between Syk and Zap-70. The atomic structure of a tandem SH2 domain of Syk bound to a double phosphorylated ITAM peptide shows that each SH2 functions as an independent binding module [4]. In contrast, Zap-70 utilises both SH2 domains as a binding module for just one tyrosine [14]. More recently, the atomic structure of the inhibited conformation of Zap-70 has been solved by X-ray crystallography [15] revealing that the interaction between the tandem SH2 region and the catalytic domain likely reduces the flexibility required for catalysis. Importantly, two tyrosine residues located in the inter-SH2 region interact with the kinase domain to maintain this auto-inhibited conformation. These residues must become phosphorylated by Src kinases in order to activate Zap-70.
An drawback for the full understanding of the mechanism of activation of the Syk protein is the lack of information regarding the three-dimensional (3D) structure of the full-length protein. Indeed, up to know, only 3D structures of fragments of Syk have been characterized using either crystallization or magnetic nuclear resonance (MNR) techniques [4, 16]. We have used single particle electron microscopy (EM) [17] to elucidate the 3D structure of full-length Syk in its non-phosphorylated at low resolution. This approach has revealed a compact conformation shaped by the regulatory and the kinase domains similar to that of Zap-70.
2. Materials and methods
2.1. Purification of GST-Syk and auto-phosphorylation
Rat Syk-GST was cloned as described and purified as a GST fusion [18, 19]. Phosphorylation assays were performed by incubation 1 mg/ml of Syk-GST with 1 mM ATP and 2.5 mM MgCl2 for 60 min at 25 °C.
2.2 Kinase assays
To test the kinase activity of GST-Syk, we used an in vitro kinase reaction assay in the presence of [γ-32P]ATP, as indicated before [20]. As substrates, we used GST proteins fused to either the Vav1 CH-Acidic region [21] or the N-terminal region of Cbl-b [22] that were purified from IPTG-induced E. coli cultures using gluathione-coated beads.
2.3 Electron microscopy and image processing
A few microliters of the purified GST-Syk and the phosphorylated protein were adsorbed to glow discharged carbon coated grids and negatively stained using 2% uranyl acetate. The grids were observed in a JEOL 1230 operated at 100 kV and micrographs were taken at 50000× magnification under low-dose conditions with the specimen holder tilted at 0 and 15 degrees. These were digitized using a Minolta Dimage Scan Multi PRO scanner at 2400 dpi corresponding to a final 2.1 Å/pixel at the specimen. Particles from the micrographs were extracted, normalised, centred and filtered using different commands found in the EMAN software [23]. The data from tilted and untilted micrographs were processed together following a strategy that avoided bias from our previous knowledge of the atomic structures of isolated domains or Zap-70. Images were processed using angular refinement methods in EMAN [23] and using several volumes as starting references. Images were classified into homogeneous groups using reference-free methods (refine2d.py command in EMAN and using the XMIPP platform [24] and the best averages were used to build several reference volumes using common lines (startAny command in EMAN). We also used artificial noisy ellipses and Gaussian blobs as starting models as before [25, 26]. Projections from the initial reference volumes were confronted to the data set using angular refinement methods implemented in EMAN [23]. In all cases and after several iterations, the different strategies used converged to similar solutions. Refinement was performed with an angular coverage of 8 degrees to generate 280 classes, each containing approximately between 4 and 14 particles. The resolution was estimated by Fourier Shell Correlation (eotest command in EMAN) to be 24 Å (using the 0.5 cross-correlation coefficient criteria) for the reconstructions of Syk. The density map and the atomic structures were visualized with UCSF Chimera from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, CA [27]. Thresholds were chosen to account for 100% of the protein mass.
Also, pairs of micrographs were obtained and processed following the Random Conical Tilt (RCT) method [28] which allows deriving 3D structures from defined views of the specimen. Pairs of micrographs were taken for each area, with and without tilting the specimen holder. Pairs of particles were selected and all the processed was performed using the XMIPP platform [24]. Untitled data was classified and aligned in 2D and distinct 2D averages were selected and their tilted data used to build 3D reconstructions. Since these RCT 3D structures are obtained from defined views, they are not affected by the averaging of views from different orientations that happens in angular refinement methods. The RCT structures were found to preserve the overall structural features of the model derived by angular refinement.
The handedness chosen to render GST-Syk was that providing the best fit with the atomic structure of the homologous Zap-70 [15]. Both possible hands of the reconstruction were tested for fitting and it was found that only one hand provided a reasonable fitting of this atomic structure.
2.4 Fitting of atomic structures
Atomic structures deposited in the Protein Data Bank and used in the fitting experiments were as follows: 1A81 (Syk tandem SH2 domains, but used without considering the bound peptide) [4], 1XBA (Kinase domain) [16] and 2OZO (Zap-70) [15]. Fitting was carried out by a 6-dimensional search using colores from the software SITUS [29, 30] and the ADP_EM platform [31] using both the method of cross-correlation and the Laplacian filter. Fittings were performed using both possible hands.
3. Results
3.1 Purification and Functional assay of Syk
We purified Syk by fusing a glutathione S-transferase (GST) tag at its N-terminus. Previous results indicated that such tagging strategy does not affect the biological functions of Syk or its sites of auto-phosphorylation [32, 33]. Similar findings have been reported for other non-receptor PTKs such as Lck [18]. GST-Syk was expressed in baculovirus-infected insect cells, purified to homogeneity using glutathione-coated beads. The correct functionality of the purified protein was tested first by the analyses of its capacity to auto-phosphorylate using in vitro kinase reactions. We observed an optimal phosphorylation of the kinase, as demonstrated by the shift in the electrophoretic mobility of GST-Syk after the kinase reaction (Fig. 1B). This shift was inhibited when in vitro kinase reactions were carried out either in the absence of ATP or, alternatively, when ATP was combined with an inhibitor of the kinase activity of Syk (piceatannol), indicating that it was a result of the autophosphorylation of the kinase (data not shown). Mass spectrometry analysis also confirmed that the slow migrating band of Syk-GST was indeed phosphorylated in tyrosine residues located within the intedomain B and the C-terminal tail (Fig. 1C) (supplementary Fig. S1 on line). To further support the correct functionaly of the protein, the activity of GST-Syk was demonstrated by its ability to trans-phosphorylate known substrates such as Cbl-b and Vav1 (Fig. 1D).
3.2 Electron microscopy and 3D structure of Syk
Non-phosphorylated GST-Syk molecules were clearly seen in negatively stained electron micrographs (Fig. 2A) and images of single molecules suggested that GST-Syk has the rough shape of a square (Fig. 2B). Micrographs were taking after tilting the specimen holder from 0 up to 15 degrees to help increase the angular sampling of possible orientations of the molecules binding to the support film. A total of 6,299 images, tilted and untilted, were selected, combined and subjected to iterative refinement. Importantly, the structural information of Zap-70 [15] was never used as an input during refinement (see Materials and methods). As a further control, 3D reconstructions were also derived using the random conical tilt method (RCT) on typical square-shaped views [28]. The general structural features of the RCT map supported the reconstruction defined by angular refinement (result not shown).
Fig. 2.
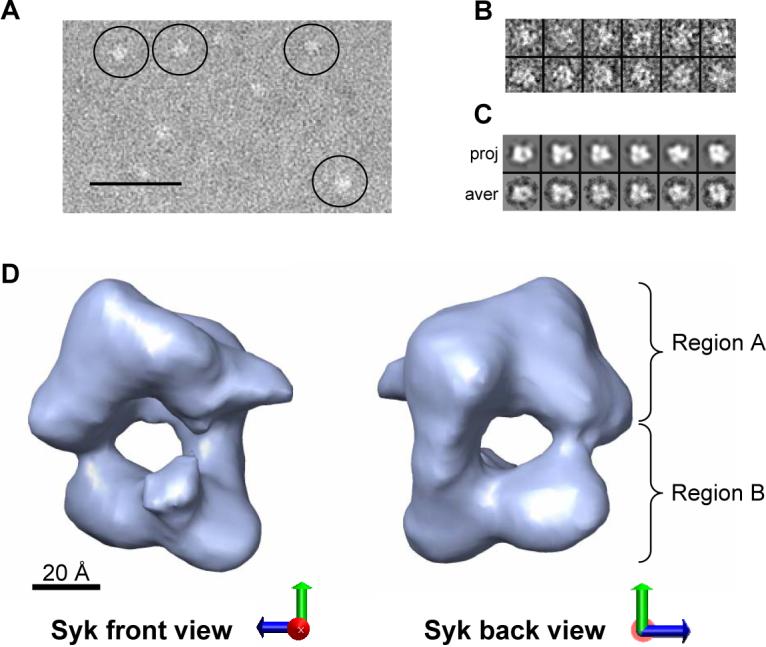
3D Structure of full-length non-phosphorylated GST-Syk. (A) EM fields of negatively stained Syk. Some molecules highlighted within circles. Scale bar corresponds to 50 nm. (B) Gallery containing a representative collection of single molecules. Particles from the micrographs were extracted, normalised, centred and filtered using different commands found in the EMAN software [23]. (C) Panel containing the projections (upper row) and their corresponding class averages (lower row) obtained after angular refinement. (D) Front and back views of the 3D reconstruction of GST-Syk. Scale bar represents 20 Å.
The three-dimensional (3D) reconstruction of GST-Syk at a resolution of 24 Å (Fig. 2D) indicates that this kinase displays a square-shaped conformation with dimensions compatible with those of the atomic structures solved for individual domains. Two major intramolecular regions could be defined: a bulky massive region (Fig. 2D, region A) and a two-lobule domain (Fig. 2D, region B). Such arrangement seemed highly reminiscent of the recently published inhibited conformation of the related Zap-70 protein [15]. In order to define the hand of our 3D reconstruction of Syk and simultaneously analyse the similarities and differences between Syk and Zap-70 [15], we fitted this within the EM map using computational procedures implemented in the SITUS [30] and the ADP_EM platforms [31]. The solutions provided by these automatic methods are less biased and less subjective than a manual fitting. Solutions were obtained where Zap-70 (Fig. 3A, yellow colour) fitted into the reconstruction of Syk (Fig. 3A, blue colour) confirming the structural resemblance between the two proteins. Notably, only one possible handedness in the EM reconstruction allowed an adequate fitting of Zap-70′s x-ray structure. Most importantly, this fitting allowed us to assign the two structural regions in GST-Syk (regions A and B) to specific domains of the protein. Accordingly the two-lobule region B likely corresponded to the two SH2 tandem domains whereas the bulky region A must comprise the kinase domain and the GST tag. The catalytic and regulatory domains in Syk would therefore form a compact protein whereas the tag would be placed at the back of the kinase domain. Despite the similarity between Zap-70 and Syk, the fitting between the two structures is not perfect indicating that conformational differences exist. More specifically, the EM reconstruction of Syk reveals a protrusion in the regulatory region likely corresponding to the inter-SH2 linker whose position is not identical to that of the equivalent region in Zap-70 (Fig. 3A).
Fig. 3.
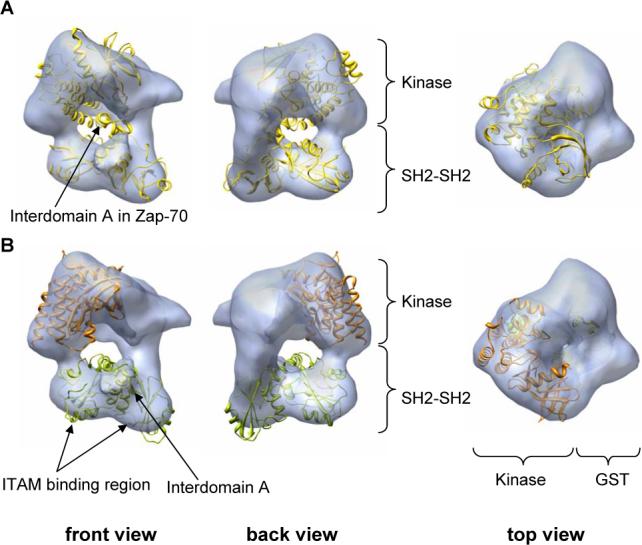
Fitting of atomic structures into the EM map of GST-Syk. (A) Views of the fitting of the atomic structure of Zap-70 (yellow colour) [15] into the EM map of Syk (blue density shown as transparency). The handedness chosen to represent GST-Syk was that providing the best fit with Zap-70. Both possible hands of the reconstruction were tested for fitting and it was found that only one hand provided a reasonable fitting of this atomic structure. (B) Fitting of the atomic structures of SH2-interdomainA-SH2 domain (green colour) and kinase domain (orange colour) of Syk into the EM map (blue density displayed as transparency). Atomic structures deposited in the Protein Data Bank and used in the fitting experiments were as follows: 1A81 (Syk tandem SH2 domains, but used without considering the bound peptide) [4] and 1XBA (Kinase domain) [16].
Since atomic structures of both regulatory and catalytic domains of Syk are known [4, 16], we fitted these within the EM map in order to better assign areas of the 3D reconstruction to those specific domains. We considered the assignment of regulatory and catalytic domains revealed previously by the fitting of Zap-70 to the whole protein, and the regulatory and kinase domain of Syk were then fitted within their corresponding region in the map (Fig. 3B). We fitted the N-terminal SH2-interdomain A-SH2 region (Fig. 3B, green colour) within the bottom part of the map (Fig. 3B, blue density). A sufficiently precise orientation of these domains within region B could be defined where the helical interdomain A fitted the protrusion between the two lobules (Fig. 3B). This fitting suggested that the phosphotyrosine binding pockets of each SH2 domain are not occluded by other regions in the structure and, therefore, are fully accessible for binding to either phosphorylated ITAM sequences or downstream partners. In conclusion, the bottom part of our map could be quite positively assigned to the SH2-SH2 cassette, leaving the remaining of the 3D reconstruction (region A) to account for the kinase domain and the GST tag. Accordingly, fitting experiments revealed that indeed kinase (Fig. 3B, orange colour) and GST atomic structures could be nicely placed within that portion of the map. The combined density of the kinase domain and the GST tag fully occupied region A (fitting for the GST tag not shown in the figure).
Taken together, the 3D structure of Syk is consistent with a compact conformation highly reminiscent of that described for the inhibited version of Zap-70, though the precise relative disposition of regulatory and catalytic regions is slightly different (see discussion). As in Zap-70, the phosphotyrosine binding pockets of the two Syk SH2 domains are located in non occluded areas of the molecule and therefore are fully accessible for recognition and binding to the phosphorylated ITAM sequences (Fig. 3B).
4. Discussion
The analysis of the Syk EM structures reported here provides a first model of the structural organisation of full-length Syk protein. In its inhibited state, Syk displays a compact conformation likely sustained by interactions between the catalytic and regulatory regions. This compact arrangement is similar to those described for other non-receptor PTKs of the Src family [3], and very closely reminiscent of that shown by Zap-70 [15], the second member of the Syk family of kinases. In apo-Syk the orientation of the SH2 domains is such that the ITAM binding pockets seem to be facing outwards accessible to bind ITAMs in an activated receptor. Importantly such disposition contrasts the general organisation of Src where the binding pockets of its SH2-SH3 domains face inwards towards the back side of the kinase domain [2]. These propositions for Syk are further sustained by the recently solved structure of Zap-70 [15]. Interactions between the inter-SH2 linker (interdomain A) and the SH2-kinase linker (interdomain B) with the kinase domain in Zap-70 were found to maintain the inactive conformation.
Collectively, the 3D reconstructions presented in this work indicate that the overall structural organization of Syk is similar to that of Zap-70 [15], the other member of the Syk familiy of kinases. The interactions between the tandem SH2 regulatory region and the kinase domain in Zap-70 are proposed to inhibit the protein by either reducing the flexibility needed to transit to the active conformation and/or precluding the motions required in the kinase domain for the catalytic activity. The fact that we observe a similar compact conformation for Syk suggests that similar regulatory mechanisms must be taking place. Nevertheless, the 3D reconstruction of inhibited Syk reveals a slightly more open conformation than Zap-70. We can not rule out that such conformational difference is just raised by the methodology we have used and non significant given the level of resolution of our EM map. Still, it is also possible that a small conformational difference between Syk and Zap-70 could contribute for their striking differences in activity. In our EM reconstruction, both regulatory and catalytic regions are to some extent rotated slightly outwards when compared to Zap-70 where the inter-domain A is found intimately contacting the kinase domain. Importantly, the interaction between the SH2-SH2 linker of Zap-70 with its catalytic domain defines the structural basis of the inhibited conformation, and the phosphorylation of this region seems to be the hallmark in the activation of the protein. Such structural difference between Syk and Zap-70 would suggest that the inhibition of the kinase activity in Syk could be less stringent than in Zap-70, maybe facilitating the transition from the inactive to the active conformation. Significantly, the activity Syk is intrinsically 100-fold higher than that of Zap-70 and its does not require the phosphorylation of inter-domain A by Src kinases [1]. Syk can be activated directly by auto-phosphorylation [2] and the structural differences we observe between Syk and Zap-70 might therefore contribute to the permissiveness for Syk activation both in vivo and in vitro compared to other protein tyrosine kinases including Zap-70 [1, 2].
Acknowledgements
OL's work is supported by grants SAF2005-00775 (OL) and GEN2003-20239-C06-06 from the Spanish Ministry of Education and Science (MES). XRB's work is supported by grants from the US National Cancer Institute (5R01-CA73735-10), the MES (SAF2006-01789 and GEN2003-20239-C06-01), and the Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/0001, Spanish Ministry of Health). All Spanish funding is co-sponsored by the European FEDER program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- 1.Latour S, Veillette A. Proximal protein tyrosine kinases in immunoreceptor signaling. Curr Opin Immunol. 2001;13:299–306. doi: 10.1016/s0952-7915(00)00219-3. [DOI] [PubMed] [Google Scholar]
- 2.Sada K, Takano T, Yanagi S, Yamamura H. Structure and function of Syk protein-tyrosine kinase. J Biochem (Tokyo) 2001;130:177–186. doi: 10.1093/oxfordjournals.jbchem.a002970. [DOI] [PubMed] [Google Scholar]
- 3.Roskoski R., Jr. Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 4.Futterer K, Wong J, Grucza RA, Chan AC, Waksman G. Structural basis for Syk tyrosine kinase ubiquity in signal transduction pathways revealed by the crystal structure of its regulatory SH2 domains bound to a dually phosphorylated ITAM peptide. J Mol Biol. 1998;281:523–537. doi: 10.1006/jmbi.1998.1964. [DOI] [PubMed] [Google Scholar]
- 5.Latour S, Zhang J, Siraganian RP, Veillette A. A unique insert in the linker domain of Syk is necessary for its function in immunoreceptor signalling. Embo J. 1998;17:2584–2595. doi: 10.1093/emboj/17.9.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catalina MI, Fischer MJ, Dekker FJ, Liskamp RM, Heck AJ. Binding of a diphosphorylated-ITAM peptide to spleen tyrosine kinase (Syk) induces distal conformational changes: a hydrogen exchange mass spectrometry study. J Am Soc Mass Spectrom. 2005;16:1039–1051. doi: 10.1016/j.jasms.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Kimura T, Sakamoto H, Appella E, Siraganian RP. Conformational changes induced in the protein tyrosine kinase p72syk by tyrosine phosphorylation or by binding of phosphorylated immunoreceptor tyrosine-based activation motif peptides. Mol Cell Biol. 1996;16:1471–1478. doi: 10.1128/mcb.16.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visco C, Magistrelli G, Bosotti R, Perego R, Rusconi L, Toma S, Zamai M, Acuto O, Isacchi A. Activation of Zap-70 tyrosine kinase due to a structural rearrangement induced by tyrosine phosphorylation and/or ITAM binding. Biochemistry. 2000;39:2784–2791. doi: 10.1021/bi991840x. [DOI] [PubMed] [Google Scholar]
- 9.Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E, Blancato JK, Vezza PR, McLeskey SW, Mangeat PH, Mueller SC. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–747. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 10.Coopman PJ, Mueller SC. The Syk tyrosine kinase: a new negative regulator in tumor growth and progression. Cancer Lett. 2006;241:159–173. doi: 10.1016/j.canlet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Brdicka T, Kadlecek TA, Roose JP, Pastuszak AW, Weiss A. Intramolecular regulatory switch in ZAP-70: analogy with receptor tyrosine kinases. Mol Cell Biol. 2005;25:4924–4933. doi: 10.1128/MCB.25.12.4924-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert M, Tartare-Deckert S, Couture C, Mustelin T, Altman A. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity. 1996;5:591–604. doi: 10.1016/s1074-7613(00)80273-3. [DOI] [PubMed] [Google Scholar]
- 13.Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J Mol Biol. 2006;356:1222–1236. doi: 10.1016/j.jmb.2005.11.095. [DOI] [PubMed] [Google Scholar]
- 14.Folmer RH, Geschwindner S, Xue Y. Crystal structure and NMR studies of the apo SH2 domains of ZAP-70: two bikes rather than a tandem. Biochemistry. 2002;41:14176–14184. doi: 10.1021/bi026465e. [DOI] [PubMed] [Google Scholar]
- 15.Deindl S, Kadlecek TA, Brdicka T, Cao X, Weiss A, Kuriyan J. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Atwell S, Adams JM, Badger J, Buchanan MD, Feil IK, Froning KJ, Gao X, Hendle J, Keegan K, Leon BC, Muller-Dieckmann HJ, Nienaber VL, Noland BW, Post K, Rajashankar KR, Ramos A, Russell M, Burley SK, Buchanan SG. A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J Biol Chem. 2004;279:55827–55832. doi: 10.1074/jbc.M409792200. [DOI] [PubMed] [Google Scholar]
- 17.Llorca O. Introduction to 3D reconstruction of macromolecules using single particle electron microscopy. Acta Pharmacol Sin. 2005;26:1153–1164. doi: 10.1111/j.1745-7254.2005.00203.x. [DOI] [PubMed] [Google Scholar]
- 18.Spana C, O'Rourke EC, Bolen JB, Fargnoli J. Analysis of the tyrosine protein kinase p56lck expressed as a glutathione S-transferase fusion protein in Spodoptera frugiperda cells. Protein Expr Purif. 1993;4:390–397. doi: 10.1006/prep.1993.1051. [DOI] [PubMed] [Google Scholar]
- 19.Rowley RB, Bolen JB, Fargnoli J. Molecular cloning of rodent p72Syk. Evidence of alternative mRNA splicing. J Biol Chem. 1995;270:12659–12664. doi: 10.1074/jbc.270.21.12659. [DOI] [PubMed] [Google Scholar]
- 20.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Lago M, Lee H, Cruz C, Movilla N, Bustelo XR. Tyrosine phosphorylation mediates both activation and downmodulation of the biological activity of Vav. Mol Cell Biol. 2000;20:1678–1691. doi: 10.1128/mcb.20.5.1678-1691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustelo XR, Crespo P, Lopez-Barahona M, Gutkind JS, Barbacid M. Cbl-b, a member of the Sli-1/c-Cbl protein family, inhibits Vav-mediated c-Jun N-terminal kinase activation. Oncogene. 1997;15:2511–2520. doi: 10.1038/sj.onc.1201430. [DOI] [PubMed] [Google Scholar]
- 23.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 24.Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 29.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 30.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 31.Garzon JI, Kovacs J, Abagyan R, Chacon P. ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics. 2006 doi: 10.1093/bioinformatics/btl625. [DOI] [PubMed] [Google Scholar]
- 32.Furlong MT, Mahrenholz AM, Kim KH, Ashendel CL, Harrison ML, Geahlen RL. Identification of the major sites of autophosphorylation of the murine protein-tyrosine kinase Syk. Biochim Biophys Acta. 1997;1355:177–190. doi: 10.1016/s0167-4889(96)00131-0. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto N, Hasegawa H, Seki H, Ziegelbauer K, Yasuda T. Development of a high-throughput fluoroimmunoassay for Syk kinase and Syk kinase inhibitors. Anal Biochem. 2003;315:256–261. doi: 10.1016/s0003-2697(03)00026-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


