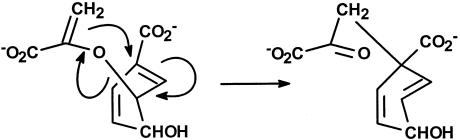
The action of chorismate mutase is a paradigm of enzyme catalysis, because the enzyme is a structurally simple protein that accelerates a straightforward unimolecular reaction: a concerted, intramolecular rearrangement of chorismate to prephenate in which one carbon–carbon bond is formed and one carbon–oxygen bond is broken (Fig. 1). The reaction itself is a vital step in the biosynthesis of aromatic amino acids, lending the enzyme a biological importance equal to its importance in understanding mechanisms of catalysis. In this issue of PNAS, Hur and Bruice (1) have applied the near-attack conformation (NAC) concept, which Bruice and coworkers have been developing for several years (2, 3), to the accelerations produced by several different chorismate mutases, developing thereby what might be called a microhistory of the catalytic process for these enzymes, tracing events between reactant state and transition state. The findings are illuminating and likely to generate considerable surprise.
Fig. 1.
Chorismate rearranges to prephenate: the reaction catalyzed by chorismate mutases. Very roughly and schematically, the structures represent reactive conformers.
Explaining enzyme catalysis has always posed an adequate challenge. Enzymes are powerful catalysts, some accelerating their target reactions by factors that can exceed 1020 (4). Chorismate mutases accelerate their target reaction by ≈106- to 107-fold. The details of how such accelerations are orchestrated in the course of a process in which the enzyme encounters reactant molecules, may, in some cases, shepherd them through various intermediate states to the product molecule(s), and then releases the latter before launching a new catalytic cycle, have been a subject of research, speculation, and debate since the discovery of enzymes.
One of the main conceptual tools for organizing the findings has been Pauling's insight (5) that an enzyme is a biomolecule that specifically binds and thus stabilizes the transition state of the target reaction. When couched in the customary solution-chemist's version of the transition-state theory [H. S. Johnston (6) called it the “ultrasimple” version], the idea that an enzyme achieves catalysis only by means of net stabilization of the transition state has been a comfortable theme in mechanistic enzymology for some time (7). This formulation is one in which reaction rate constants such as the familiar kcat and kcat/KM increase with catalysis only as the catalyst decreases the difference in Gibbs free energy between the transition state and the reactant state. It thus offers as a great advantage the path-independence of thermodynamic state functions. Attention could be focused solely on the transition state and the reactant state, with no consideration of intervening states because their properties could not contribute to catalysis. The microhistory of an enzymic reaction could therefore be deemed of no interest because none of its features could influence the experimentally observable rate constants.
For many, discoveries of the last one or two decades have begun to suggest that this account is inadequate. In particular, the discovery and documentation by Klinman and coworkers of the vital role of quantum tunneling in enzymic reactions involving hydrogen transfer (8), raised the question of exactly how to incorporate these effects into the rate-constant description. Related studies of environmental coupling in hydrogen-transfer catalysis by Benkovic, Hammes-Schiffer, and their coworkers (9) are contributing to this reconsideration. Kuznetsov and Ulstrup (10) and Schwartz and coworkers (11), for example, have proposed formulations alternative to transition-state theoretical ones, whereas Gao and Truhlar (12) have emphasized that modern versions of transition-state theory, which are very far from “ultrasimple,” are extremely robust and versatile. Karplus (13, 14) has reviewed the prospects for incorporating into the description other current, innovative forms of theory, particularly dynamical simulations, to provide a more complete and satisfying picture of the details of enzyme catalysis.
The time is therefore clearly upon us for our understanding of enzymic catalysis to be enhanced by descriptions of the microhistory between reactant state and transition state, and theory is the appropriate tool for the task. Whether the totality of new information will eventually require a conceptual approach distinct even from the powerful modern versions of transition-state theory remains to be seen. Whatever the theoretical framework that will be needed, the basic questions likely to be illuminated seem numerous. In particular, the manner in which active site structures (and perhaps remote and extended networks in the protein structure) act to move reactant molecules across the energy barrier is of high interest. This is particularly true for cases like chorismate mutase, where atoms that move to form and break bonds are all heavy, nonhydrogen atoms. Quantum tunneling is not then an effective option for achieving catalytic acceleration.
In their development of the microhistory of chorismate mutase, Hur and Bruice (1) consider five different examples of protein catalysis as well as the “uncatalyzed” reaction in aqueous solution. They show that a NAC of chorismate that is poised for the critical bond-formation event (having the overall conformation of the transition state and the correct orientation of the reacting orbitals for smooth bond formation) exists in the active site of each catalyst in a population that exactly reflects the catalytic activity of that catalyst. They obtain the standard free energy of formation of the NAC in each environment and then subtract this value from the experimentally determined free energy of activation to find the free energy required to convert NAC to transition state. This value is uniformly 16 kcal/mol for all of the catalysts and for the uncatalyzed reaction in free aqueous solution!
Hur and Bruice have therefore shown that catalyst interactions (electrostatic and hydrophobic interactions) stabilize a transition-state-like conformation that approximates the transition state except that the forming bond is longer by ≈1.5–2 Å than it will be in the transition state. These interactions constitute the entirety of the attractive interactions that stabilize the transition state. As the critical bond formation and bond shortening occur, the stabilizing interactions are maintained quantitatively so that a uniform barrier of 16 kcal/mol is surmounted in every catalytic case and in the uncatalyzed reaction. As the transition state is attained and the new bond is partially formed, the energetic cost of sorting the reactive conformations out of the vast conformational array of free chorismate disappears, the new bond securing the molecule in the correct cyclic form. The entirety of the catalyst–substrate attractive interactions now appears as transition-state stabilization.
Hur and Bruice identify a number of amino acid residues that stabilize the NAC and transition state in equal measure. They particularly stress the role of arginine residues that interact with the two carboxylate groups of chorismate to stabilize the conformation that brings the two reactive carbon atoms into proximity, and the valine, isoleucine, leucine, and phenylalanine residues that provide a hydrophobic environment for the less polar fragments of the chorismate molecule when it is in the NAC conformation.
The calculations of Hur and Bruice suggest a conceivable explanation for an otherwise somewhat mystifying result from Hilvert and coworkers (15). They created a mutant chorismate mutase wtih an electrically neutral citrulline residue in the place of arginine 90, one of the residues involved in catalysis. Indeed, the catalysis is strongly compromised, but the wild-type enzyme and the mutant enzyme bind a transition-state analog inhibitor with equilibrium constants that are only slightly different (1.2 and 6.8 μM). It might have been expected that the inhibitor, which is structurally constrained to a conformation resembling both transition state and NAC, would have bound to the mutant enzyme substantially less strongly (in the mutant–enzyme active site, only a small minority of the substrate is found in the NAC form, which lies at a free energy above that of the enzyme–substrate complex by 4.1 kcal/mol). Hur and Bruice find that their simulations suggest that the inhibitor–mutant enzyme interactions are altered from those in the wild-type enzyme. Thus, it is possible that the strong binding of the inhibitor by the mutant enzyme may be an artifact of this unusual binding.
A microhistory of enzyme-catalyzed processes traces events between reactant state and transition state.
The microhistory of Hur and Bruice begins with the enzyme–substrate complex, which in the most active catalysts contains a high population of the NAC or reactive conformation. The microhistory can be extended one step earlier by a theoretical study of Guo et al. (16). They considered the question of whether the very small fraction of reactive conformations present among the many other conformations of chorismate in free solution is selected out and specifically bound by the enzyme. The alternative is that the more common unreactive conformations can also be bound and then find their way, perhaps with enzyme assistance, into the reactive form. The answer is that unreactive conformers are indeed bound and that active-site groups assist in bringing them into and maintaining them in the reactive form as the system passes into the NAC and then to the transition state.
The NAC approach does not lack critics, but the new findings should be congenial to some of them. In their recent study of the chorismate mutase case, Štrajbl et al. (17) note that, although they too identify a NAC effect, it should be preferable to center computational attention on the transition state “rather than to evaluate the NAC effect that might or might not help in predicting the catalytic effect.” In many cases, of course, this would be good advice. But in the particular case of chorismate mutase catalysis, Hur and Bruice now find that the free energy of NAC formation precisely predicts the magnitude of the catalytic effect because of the constant 16 kcal/mol barrier that is crossed, beginning at the NAC structure, in the case of all of the catalyzed and uncatalyzed reactions they examined.
Theoretical approaches to enzyme mechanism are thus expanding our vision from the one imposed by the limited possibilities of a previous time, and allowing us to explore events at all points along the pathway from reactant molecules through the transition state.
See companion article on page 12015.
References
- 1.Hur, S. & Bruice, T. C. (2003) Proc. Nat. Acad. Sci. USA 100, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruice, T. C. (2002) Acc. Chem. Res. 35, 139–148. [DOI] [PubMed] [Google Scholar]
- 3.Bruice, T. C. & Kahn, K. (2000) Curr. Opin. Chem. Biol. 4, 540–544. [DOI] [PubMed] [Google Scholar]
- 4.Miller, B. G. & Wolfenden, R. (2002) Annu. Rev. Biochem. 71, 847–885. [DOI] [PubMed] [Google Scholar]
- 5.Pauling, L. (1946) Chem. Eng. News 24, 1375–1377. [Google Scholar]
- 6.Johnston, H. S. (1966) Gas Phase Reaction Rate Theory (Ronald Press, New York), p. 128.
- 7.Gandour, R. D. & Schowen, R. L., eds. (1978) Transition States of Biochemical Processes (Plenum, New York).
- 8.Knapp, M. J. & Klinman, J. P. (2002) Eur. J. Biochem. 269, 3113–3121. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal, P. K., Billeter, S. R., Rajagopalan, P. T., Benkovic, S. J. & Hammes-Schiffer, S. (2002) Proc. Natl. Acad. Sci. USA. 99, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuznetsov, A. M. & Ulstrup, J. (1999) Can. J. Chem. 77, 1085–1096. [Google Scholar]
- 11.Antoniou, D., Caratzoulas, S., Kalyanaraman, C., Mincer, J. S. & Schwartz, S. D. (2002) Eur. J. Biochem. 269, 3103–3112. [DOI] [PubMed] [Google Scholar]
- 12.Gao, J. & Truhlar, D. G. (2002) Annu. Rev. Phys. Chem. 53, 467–505. [DOI] [PubMed] [Google Scholar]
- 13.Karplus, M. (2003) Biopolymers 68, 350–358. [DOI] [PubMed] [Google Scholar]
- 14.Karplus, M. (2002) Acc. Chem. Res. 35, 321–323. [DOI] [PubMed] [Google Scholar]
- 15.Kienhöfer, A., Kast, P. & Hilvert, D. (2003) J. Am. Chem. Soc. 125, 3206–3207. [DOI] [PubMed] [Google Scholar]
- 16.Guo, H., Cui, Q., Lipscomb, W. N. & Karplus, M. (2003) Angew. Chem. Int. Ed. Engl. 42, 1508–1511. [DOI] [PubMed] [Google Scholar]
- 17.Štrajbl, M., Shurki, A., Kato, M. & Warshel, A. (2003) J. Am. Chem. Soc. 125, 10228–10237. [DOI] [PubMed] [Google Scholar]