Abstract
The lack of knowledge of the three-dimensional structure of the trimeric, catalytic (C) subunit of aspartate transcarbamoylase (ATCase) has impeded understanding of the allosteric regulation of this enzyme and left unresolved the mechanism by which the active, unregulated C trimers are inactivated on incorporation into the unliganded (taut or T state) holoenzyme. Surprisingly, the isolated C trimer, based on the 1.9-Å crystal structure reported here, resembles more closely the trimers in the T state enzyme than in the holoenzyme:bisubstrate-analog complex, which has been considered as the active, relaxed (R) state enzyme. Unlike the C trimer in either the T state or bisubstrate-analog-bound holoenzyme, the isolated C trimer lacks 3-fold symmetry, and the active sites are partially disordered. The flexibility of the C trimer, contrasted to the highly constrained T state ATCase, suggests that regulation of the holoenzyme involves modulating the potential for conformational changes essential for catalysis. Large differences in structure between the active C trimer and the holoenzyme:bisubstrate-analog complex call into question the view that this complex represents the activated R state of ATCase.
Integration of metabolic pathways and tight control of protein functions require allosteric regulation, whose hallmark is functional cooperativity. In their pioneering paper aimed at accounting for the properties of allosteric proteins, such as hemoglobin and regulatory enzymes, Monod, Wyman, and Changeux (1) postulated that these oligomeric proteins exist in two (or more) conformational states and that ligands control the equilibrium between a low-affinity or less active, taut (T state) form and a high-affinity or more active, relaxed (R state) quaternary structure. This two-state model has been shown to account for both the homotropic cooperativity and the heterotropic effects (inhibition and activation) exhibited by Escherichia coli aspartate transcarbamoylase (ATCase; aspartate carbamoyltransferase, carbamoyl phosphate:l-aspartate carbamoyltransferase; EC 2.1.3.2), which catalyzes the first committed step in pyrimidine biosynthesis (2–4). In addition, physical chemical measurements of the change in quaternary structure and catalytic activation promoted by substoichiometric amounts of the bisubstrate analog (5), N-(phosphonacetyl)-L-aspartate (PALA), were interpreted readily in terms of the model (2).
In parallel with tests of the two-state model based on enzyme kinetics and solution studies of the ligand-promoted global conformational changes in ATCase, Lipscomb and his colleagues (6–9) conducted detailed crystallographic investigations of the wild-type enzyme and various mutant forms. The extensive data include high-resolution structures of unliganded ATCase (8), PDB ID 6atl, and the PALA-liganded enzyme (9), PDB ID 8atc. Large conformational differences between the free and PALA-bound forms of the holoenzyme were proposed to define the structural differences between the T and R states.
Despite the extensive knowledge of the structure of E. coli ATCase, which is composed of two catalytic (C) trimers and three regulatory dimers, there is no understanding of the conformational changes in the active, nonallosteric C trimers on their incorporation into the holoenzyme. This information is essential for the complete understanding of the allostery of ATCase, because the two-state model includes the assumption that the R state conformation, by virtue of reduced quaternary constraints compared with the T state, resembles the structure of isolated, active subunits. To test this assumption and to understand changes in both the structure and properties associated with assembly of an active subunit into an allosteric enzyme, we determined the high-resolution x-ray crystal structure of the isolated, unregulated C trimer of ATCase. This structural information, in comparison with the results of earlier crystallographic studies of ATCase, provides an explanation for the high activity of the isolated C trimer compared with the virtually inactive, unliganded holoenzyme and leads to crucial questions regarding our understanding of the allosteric transition.
EXPERIMENTAL PROCEDURES
ATCase, containing a His-6 sequence at the amino terminus of each regulatory chain, was purified as described elsewhere (10), and unmodified C subunits were isolated from the His-tagged holoenzyme by treatment with neohydrin followed by chromatography (11). The expected mass of the catalytic chains, based on their amino acid sequence, was confirmed by mass spectrometry. In contrast to previous crystallization experiments (12) on the C trimer that did not yield detailed information, a high-resolution structure was determined from a different crystal form of the trimer obtained by vapor diffusion at 4°C from 5-μl hanging drops of 0.1 M Tris⋅HCl, pH 7.8/0.04 M Ca(OAc)2/7.5% polyethylene glycol (PEG) 8,000 and 5 μl of C subunit (6.1 mg/ml) in 10 mM Tris⋅HCl, pH 7.5/1 mM 2-mercaptoethanol. The crystals grew in space group P212121, with unit cell dimensions of a = 56.10 Å, b = 82.54 Å, and c = 210.52 Å. Crystals (1.0 × 0.3 × 0.2 mm) were equilibrated to 42 mM Tris⋅Cl, pH 7.5/42 mM Ca(OAc)2/29% PEG 8,000/22% (vol/vol) methylpentanediol and flash cooled on a nylon loop in liquid N2. Data were collected from a single crystal at 100 K by using a Mar345 image plate on Beamline 9-1 at the Stanford Synchrotron Radiation Laboratory (Stanford, CA). The data were integrated and scaled by using mosflm (13) and scala (14) to give Rmerge = 8.7% for 80,183 independent reflections from 20- to 1.88-Å resolution. The data were 99.6% complete overall, and, in the highest resolution shell, data had I/σI values of 11.4 overall and of 2.9 in the 1.95- to 1.88-Å shell. Molecular replacement was carried out with amore (15) with a trimeric search model constructed from the PALA-bound ATCase structure (9). The molecular-replacement solution yielded an initial R factor of 51%, which was reduced to 48% by rigid-body refinement. Iterative cycles of positional and restrained B factor refinement with TNT (16) followed by model building with O (17) yielded the final refined structure. The final model contained 6,810 protein atoms in the three polypeptide chains, 789 water molecules, and two Ca2+ atoms. There were no Ramachandran violations, and the rms deviations from ideal bond lengths and angles were 0.008 and 1.85 Å, respectively.
RESULTS
The crystal structure of the C trimer was determined by molecular replacement by using a search model derived from the holoenzyme:PALA complex (9) from which the bisubstrate inhibitor was removed. This choice was guided by the proposal that the holoenzyme:PALA complex represents the R state enzyme (6, 7, 18) and the evidence that the R state is highly active, although not as active as the isolated C trimer (19, 20). The electron density for most of the protein was readily interpretable (Fig. 1A), and the structure was refined to R and free R values of 0.189 and 0.290, respectively, by using all the data from 20- to 1.88-Å resolution.
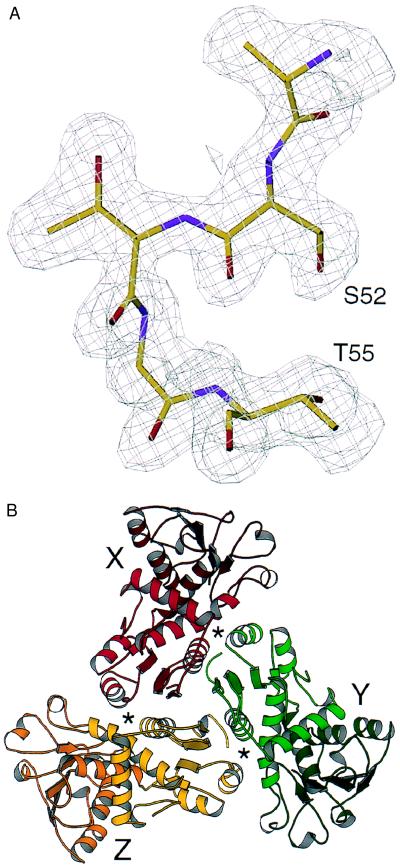
Figure 1.
Structure of the ATCase C trimer. (A) The quality of the electron density maps is illustrated by the 20- to 1.88-Å resolution, 2Fo-Fc map contoured at 1σ, showing residues of the phosphate-binding loop, Ser-52 through Thr-55. (B) Ribbon diagram (21) of the X (red), Y (green), and Z (gold) chains of the C trimer, viewed along the trimerization axis. The active sites, which contain residues from adjacent chains, are designated by asterisks.
The C trimer model (Fig. 1B) contains 878 of 930 amino acids and 791 solvent molecules. As seen previously in structures of the holoenzyme (8, 9), each catalytic chain in the isolated C trimer contains two domains bordering a large, substrate-binding cleft (Fig. 2A). Trimerization is mediated primarily by the amino-terminal, carbamoyl-phosphate-binding domain (residues 1–134). The carboxyl-terminal, aspartate-binding domain (residues 150–284) is connected to the amino-terminal domain by two α–helices (residues 135–149 and 285–305), which comprise a hinge between domains.
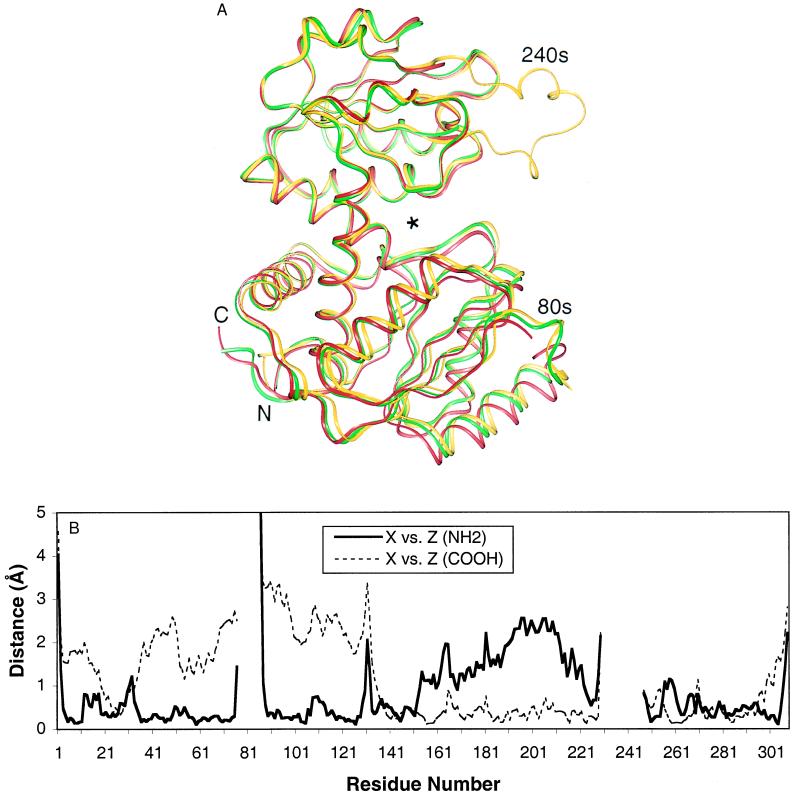
Figure 2.
Structural differences among the catalytic chains in the C trimer. (A) Ribbon representation of the catalytic chains superimposed (22) by using the carboxyl-terminal domains (residues 150–229 and 252–284) of chains X (red), Y (green), and Z (gold). An asterisk marks the active site. Differences in the interdomain hinge angle in the three chains are apparent from the displacement of the nonsuperimposed, amino-terminal domains. The superimposed, carboxyl-terminal domains show close correspondence. Two flexible loops (80s and 240s) containing active-site residues differ among the chains and exhibit disordered regions, shown as breaks in the ribbons. (B) Shifts in Cα positions of chains X and Z superimposed by the backbone atoms of the amino-terminal (residues 1–73 and 90–134; thick, solid line) or carboxyl-terminal (thin, dashed line) domain, plotted as a function of residue number. Gaps result from disordered regions in one or both chains. The individual domains of the C trimer closely resemble each other (<0.3-Å shifts) in tertiary structure, except for the loop regions. Large shifts in the nonsuperimposed domains reflect the hinge-angle variation.
In contrast to the 3-fold crystallographic symmetry in the holoenzyme, the individual chains in the isolated C trimer show substantial differences in local conformation and interdomain hinge angle (Fig. 2A). Superpositions of either domain of each chain indicate backbone shifts generally <0.3 Å within that domain and some localized changes of 1–3 Å (Fig. 2B). Larger main-chain differences are apparent at the boundaries of the disordered 80s loop, which contributes to the active site shared with the adjacent chain. In contrast to the local differences in the superimposed domains, the nonsuperimposed domains show larger global shifts, averaging 1–4 Å (Fig. 2B). The hinge angles between domains of the X, Y, and Z chains of the C trimer differ by up to 4.8° (Figs. 2 and 3A), indicating that the hinge angles of the individual chains are not tightly coupled. Such global variations between crystallographically independent chains have been interpreted generally to indicate conformational flexibility in solution (23).
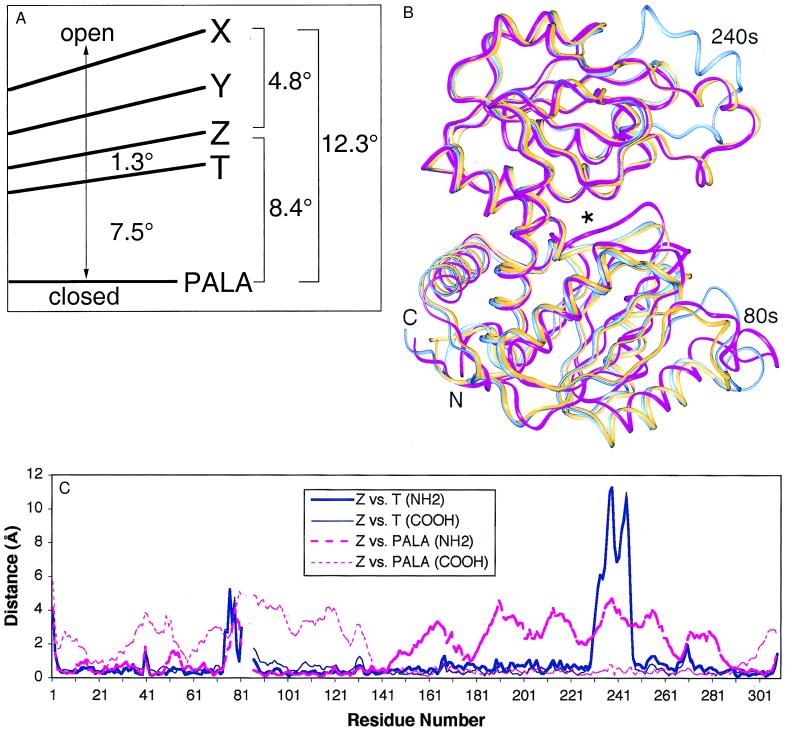
Figure 3.
The C trimer is globally more similar to the T state holoenzyme. (A) Differences in interdomain hinge angles in the catalytic chains of the isolated C trimer, the T state, and the PALA complex of the holoenzyme. The differences of up to 4.8° in hinge angle of the independent X, Y, and Z chains illustrate the asymmetry of the isolated C trimer. In contrast, the C trimers in the holoenzyme structures show exact 3-fold symmetry, and the upper and lower trimers differ in hinge angle by only 0.9° for the T state enzyme and 0.2° for the PALA complex (8, 9). The chains of the isolated C trimer are 1.3–6.0° more open than the catalytic chains of the T state holoenzyme and 8.4–12.3° more open than those in the PALA-liganded holoenzyme structure. (B) Ribbon diagram of chain Z (gold) and catalytic chains from the T state (blue) and PALA-complexed holoenzyme (magenta) superimposed by using the Cα atoms of the carboxyl-terminal domain. An asterisk marks the active site. Interdomain hinge angles are similar for chain Z and the catalytic chain in the T state holoenzyme, and the hinge is more closed in the PALA complex. (C) Shifts in Cα positions between the Z chain of the isolated C trimer and the T state (blue) and PALA complex (magenta) of the holoenzyme. The chains were superimposed by using the amino-terminal (thick lines) or carboxy-terminal (thin lines) domains. With the exception of the 240s loop, which makes a contact between trimers in the crystal, the main chain of the C trimer is more similar to the T state holoenzyme.
In addition to global flexibility, the C trimer shows disorder in two functionally important loops containing residues Ser-80, Lys-84, and Gln-231. Little electron density is apparent for residues 77–81 of chain X, 82–85 of chain Y, and 82–86 of chain Z, which comprise the 80s loop. The 240s loop (residues 230–246) shows no interpretable electron density in chains X and Y, but this region is ordered in chain Z, where it makes contact with another trimer in the crystal. Chymotrypsin preferentially and rapidly cleaves the isolated C trimer at Tyr-240 (24), supporting the interpretation that the 240s loop is flexible in solution, and the ordered conformation in chain Z is trapped by crystallization. In the holoenzyme, residues in the 80s and 240s loops make contacts with other chains, indicating that assembly imposes a specific conformation that is similar in each catalytic chain. The conformational constraints in the unliganded holoenzyme result in loss of conformational freedom, catalytic activity, and substrate affinity compared with the isolated C trimer (20).
Surprisingly, the chains in the isolated C trimer are more similar to those in the unliganded, relatively inactive, T state holoenzyme (0.5- to 1.0-Å backbone rms deviations) than to those in the holoenzyme:PALA complex (1.1- to 1.6-Å backbone rms deviations). These results, based on superpositions involving residues 1–73, 90–229, and 252–310, were unexpected, because the structure of the holoenzyme:PALA complex is considered to represent the activated R state enzyme (6, 7, 18), even though all six active sites are occupied by the inhibitor. The hinge angles of the chains in the isolated C trimer, however, are 8.4–12.3° more open than in the holoenzyme:PALA complex and only 1.3–6.0° more open than in the T state holoenzyme (Fig. 3). The conformations of the chains in the E. coli C trimer differ significantly from those in Bacillus subtilis ATCase, a nonallosteric trimeric enzyme with about 35% homology to the catalytic chains in E. coli ATCase (25).
The ordered regions of the active sites in the C trimer also show characteristic similarities to the T state holoenzyme and differences from the holoenzyme:PALA complex. In particular, residues 50–55 in the phosphate-binding loop, which undergo a 2-Å main-chain shift when PALA binds to the holoenzyme, more closely match the T state conformation (Figs. 3C and 4B). The ion pair between Glu-50 and Arg-105 (Fig. 4B), found only in the T state holoenzyme, is present in all three chains of the C trimer.
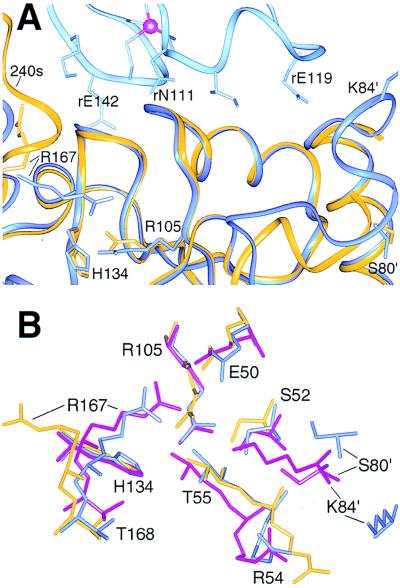
Figure 4.
Similar local conformations near the active sites of the isolated C trimer and the T state holoenzyme. Residues are labeled in the one-letter code. (A) Catalytic (dark blue) and regulatory (r, light blue) chains from the T state holoenzyme superimposed by the amino-terminal domain of the catalytic chains on chain Z (gold) of the C trimer. The 80s loop on the right is flexible in the isolated trimer, and the 240s loop on the left is ordered in chain Z by a crystal contact. (B) Superposition of the active site regions of chain Z (gold), a catalytic chain from the T state (blue), and PALA-liganded (magenta) holoenzyme structures. For clarity, PALA was omitted from the liganded structure. The ordered regions of the active sites in the isolated C trimer more closely resemble the T state holoenzyme. Diagnostic similarities are apparent for Thr-168 and the phosphate binding loop (Ser-52, Arg-54, and Thr-55). Because the isolated C trimer defines an active conformation, the distinct tertiary structure of the holoenzyme:PALA complex may result largely from ligand binding rather than the allosteric transition alone.
Consistent with the enhanced flexibility, the structure of the active C trimer shows previously undetected conformations of the catalytic chains. Despite the similarities to the active sites in the T state enzyme, some residues adopt different conformations in the three active sites of the C trimer. These include the Glu-50/Arg-105 ion pair, which shifts >4 Å in chain X, and Arg-54, which adopts a different conformation in all three chains. Side-chain rearrangements in the C trimer give rise to positional differences up to 14.3 Å and 8.7 Å, respectively, compared with the free and PALA-bound holoenzymes. The conformational heterogeneity of the C trimer contrasts with the 3-fold symmetry observed in the free and ligand-bound states of the holoenzyme (8, 9).
DISCUSSION
In the model of Monod et al. (1), the two alternative conformations, T and R, were defined in reference to functional states. Based on that model and an analysis of data from enzyme kinetics and physical chemical experiments, it was estimated (2) that the T ↔ R equilibrium for unliganded ATCase favored the T state by about 250:1. Hence, the structure of the unliganded holoenzyme (8) generally is considered to represent the T state. There is no structural information, however, regarding unliganded catalytic chains in the R state holoenzyme, leaving open the question whether these chains have the conformation observed in PALA-liganded ATCase.
We have shown here that the structure of the active, isolated C trimer unexpectedly resembles the relatively inactive T state holoenzyme, except that the isolated trimer displays greater flexibility. Evidence for increased flexibility of the isolated C trimer includes differences in the hinge angles of the individual chains (Figs. 2A and 3A) and the lack of interpretable electron density for the 80s and 240s loops. The disorder of the 80s loop is particularly interesting, because this region contributes to the shared active sites in the trimer. Important structural similarities between the isolated C trimer and the holoenzyme T state include the interdomain hinge angles (Fig. 3) and the 50s loop (Fig. 4B), which undergo large changes on PALA binding to the holoenzyme. The structural similarities, together with the attenuation of flexibility and activity, suggest that the T state holoenzyme may be relatively inactive, not because of the conformation of the catalytic chains, but because quaternary constraints hinder localized conformational changes occurring in the catalytic cycle. Thus, the allosteric mechanism of ATCase may depend on modulation of flexibility, with the catalytic chains in the T state restricted to a smaller set of conformations than in the R state.
The large differences in structure and flexibility between the catalytic chains in the C trimer and those in the holoenzyme:PALA complex bring into question the view that the structure of this complex represents the R state enzyme. Binding of substoichiometric amounts of PALA to ATCase leads to a large increase in enzyme activity, despite the occupancy of a large fraction of the active sites by the inhibitor (5, 20). This activation, attributed to the conversion of the unliganded sites from the T to the R conformation, is linked to the global expansion of the ATCase dodecamer, even with approximately one-third of the active sites unoccupied (26). In addition, studies of the titration of the holoenzyme with PALA showed that spectroscopic changes accompanying the global T → R transition are distinct from the spectral perturbations caused directly by PALA binding at unoccupied active sites in the R state enzyme (27).
Preliminary crystallographic experiments on the complex between the isolated C trimers and PALA show large changes in the conformation of the chains caused by PALA binding (J.A.E., P.T.B., T.A., and H.K.S., unpublished results). The tertiary structure of the chains in the C trimer:PALA complex is very similar to that of the catalytic chains in the holoenzyme:PALA complex. The similarity in the structures of the liganded catalytic chains in the two PALA complexes indicate that the specific tertiary structural changes in the fully liganded holoenzyme may arise from local effects caused by PALA binding and not necessarily from the global changes represented by the conversion to the R state enzyme.
The similarity of the chains in the unliganded C trimer and those in the T state holoenzyme is consistent with the view that hinge closure and other tertiary changes observed in the PALA-liganded holoenzyme result from localized effects of PALA binding. Rather than activating the enzyme, stable closure of the hinge in unliganded chains would be expected to block the entrance of substrates to the active sites and thereby inhibit catalysis. These considerations, in conjunction with the dramatic differences in structure between the catalytic chains in the active C trimer and those in the holoenzyme:PALA complex, suggest that the detailed structure of the activated, R state holoenzyme remains a central open question.
Acknowledgments
We thank S. Doyle, J. F. Kirsch, E. Eisenstein, and R. Cohen for helpful discussions and suggestions on the manuscript and D. King for mass spectra. X-ray data were collected at the Stanford Synchrotron Radiation Laboratory, which is supported by the Department of Energy and the National Institutes of Health. This research was supported by National Institutes of Health/National Institute of General Medical Sciences Research Grant GM 12159 to H.K.S., Research Grant GM 54793 to T.A., and National Research Service Award 19014 to P.T.B.
ABBREVIATIONS
- ATCase
aspartate transcarbamoylase
- C trimer or subunit
catalytic trimer or subunit
- PALA
N-(phosphonacetyl)-l-aspartate
- T state
taut state
- R state
relaxed state
Footnotes
Data deposition: The C trimer atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 3CSU).
References
- 1.Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Howlett G J, Blackburn M N, Compton J G, Schachman H K. Biochemistry. 1977;16:5091–5099. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- 3.Schachman H K. J Biol Chem. 1988;263:18583–18586. [PubMed] [Google Scholar]
- 4.Eisenstein E, Markby D W, Schachman H K. Biochemistry. 1990;29:3724–3731. doi: 10.1021/bi00467a019. [DOI] [PubMed] [Google Scholar]
- 5.Collins K D, Stark G R. J Biol Chem. 1971;246:6599–6605. [PubMed] [Google Scholar]
- 6.Lipscomb W N. Proceedings of the Robert A. Welch Foundation Conference on Chemical Research. XXXVI. Regulation of Proteins by Ligands. Houston: The Robert A. Welch Foundation; 1992. pp. 103–143. [Google Scholar]
- 7.Lipscomb W N. Adv Enzymol Relat Mol Biol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 8.Stevens R C, Gouaux J E, Lipscomb W N. Biochemistry. 1990;29:7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- 9.Ke H, Lipscomb W N, Cho Y, Honzatko R B. J Mol Biol. 1988;204:725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- 10.Graf R, Schachman H K. Proc Natl Acad Sci USA. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y R, Kirschner M W, Schachman H K. Methods Enzymol. 1978;51:35–41. doi: 10.1016/s0076-6879(78)51007-0. [DOI] [PubMed] [Google Scholar]
- 12.Foote A M, Winkler F K, Moody M F. J Mol Biol. 1981;146:389–391. doi: 10.1016/0022-2836(81)90395-8. [DOI] [PubMed] [Google Scholar]
- 13.Leslie A G W, Brick P, Wonacott A T. Daresbury Laboratory Information Quarterly for Protein Crystallography. Vol. 18. Warrington, United Kingdom: Sci. Eng. Res. Counc. Daresbury Lab.; 1986. pp. 33–39. [Google Scholar]
- 14.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 15.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 16.Tronrud D E, Ten Eyck L F, Matthews B W. Acta Crystallogr A. 1987;43:489–501. [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Stevens R C, Chook Y M, Cho C Y, Lipscomb W N, Kantrowitz E R. Protein Eng. 1991;4:391–408. doi: 10.1093/protein/4.4.391. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y R, Syvanen J M, Nagel G M, Schachman H K. Proc Natl Acad Sci USA. 1974;71:918–922. doi: 10.1073/pnas.71.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote J, Schachman H K. J Mol Biol. 1985;186:175–184. doi: 10.1016/0022-2836(85)90267-0. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Fauman E B, Rutenber E E, Maley F, Stroud R M. Biochemistry. 1994;33:1502–1511. doi: 10.1021/bi00172a029. [DOI] [PubMed] [Google Scholar]
- 23.Faber H R, Matthews B W. Nature (London) 1990;348:263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- 24.Powers V M, Yang Y R, Fogli M J, Schachman H K. Protein Sci. 1993;2:1001–1012. doi: 10.1002/pro.5560020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens R C, Reinisch K M, Lipscomb W N. Proc Natl Acad Sci USA. 1991;88:6087–6091. doi: 10.1073/pnas.88.14.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howlett G J, Schachman H K. Biochemistry. 1977;16:5077–5083. doi: 10.1021/bi00642a021. [DOI] [PubMed] [Google Scholar]
- 27.Hu C Y, Howlett G J, Schachman H K. J Biol Chem. 1981;256:4998–5004. [PubMed] [Google Scholar]