Abstract
A universal PCR method for the rapid amplification of minimal enediyne polyketide synthase (PKS) genes and the application of this methodology to clone remaining prototypical genes from producers of structurally determined enediynes in both family types are presented. A phylogenetic analysis of the new pool of bona fide enediyne PKS genes, consisting of three from 9-membered producers (neocarzinostatin, C1027, and maduropeptin) and three from 10-membered producers (calicheamicin, dynemicin, and esperamicin), reveals a clear genotypic distinction between the two structural families from which to form a predictive model. The results from this study support the postulation that the minimal enediyne PKS helps define the structural divergence of the enediyne core and provides the key tools for generating enediyne hybrid genes/molecular scaffolds; by using the model, a classification is also provided for the unknown enediyne PKS genes previously identified via genome scanning.
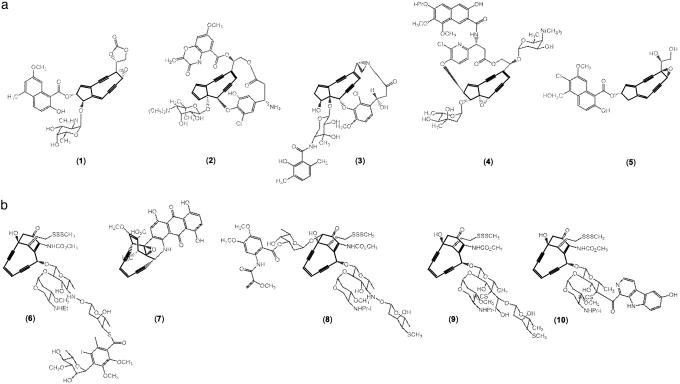
The enediyne family of antibiotics (Fig. 1) is characterized structurally by an enediyne core unit consisting of two acetylenic groups conjugated to a double bond or incipient double bond within a 9- or 10-membered ring (1–3). To date, five unique 9-membered enediynes (1–5) (Fig. 1), also often designated as the chromoprotein enediynes, and five additional distinct naturally occurring 10-membered enediynes (6–10) (Fig. 1) have been elucidated structurally (4–6). In general, these enediynes contain three distinct structural elements: a DNA-recognition unit, which serves to deliver the metabolite to its target DNA; an activation component, which sets the stage for cycloaromatization; and the enediyne “warhead,” which cycloaromatizes to a highly reactive diradical species and, in the presence of DNA, results in oxidative strand scission of the targeted sequence (7–10). In vitro and in vivo studies are consistent with the role of enediynes as DNA-damaging agents and suggest that they may even favor cleavage at certain chromosomal sites and/or tertiary structures (11, 12). Although this extraordinary reactivity invokes incredible potency (some enediynes are >8,000-fold more potent than adriamycin), the enediynes are similar to most cytotoxics in their general lack of tissue specificity. However, targeting via polymer-assisted delivery devices 1-poly(styrene-maleic acid)-conjugated neocarzinostatin or conjugation to tumor-specific monoclonal antibodies (as in the 6-based Mylotarg) has led to clinical success (13, 14).
Fig. 1.
The naturally occurring enediynes. (a) The 9-membered enediynes include neocarzinostatin (1), C1027 (2), maduropeptin (3), kedarcidin (4), and N1999A (5). (b) The 10-membered family includes calicheamicin (6), dynemicin (7), esperamicin (8), namenamicin (9), and shishijimicin (10). The enediyne core structures are highlighted in black.
Although feeding experiments with 13C-labeled precursors unambiguously established that both the 9- and 10-membered enediyne cores were derived (minimally) from eight head-to-tail acetate units (15–17), it remained controversial, until very recently, whether the enediyne cores are assembled by de novo polyketide biosynthesis or degradation from a fatty acid precursor. The elucidation of the biosynthetic loci for prototypical enediynes from each family (1 and 2 as models for 9-membered enediynes and 6 as the model for 10-membered enediynes) led to the remarkable discovery of a highly conserved enediyne polyketide synthase (PKS) gene (ncsE, sgcE, and calE8 for 1, 2, and 6, respectively), which suggested that the biosynthesis of all enediyne warheads may proceed via similar polyunsaturated polyketide intermediates and established a PKS paradigm for the enediyne biosynthesis (18, 19) (the neocarzintostatin cluster was cloned, sequenced, and characterized from Streptomyces carzinostaticus ATCC 15944 by W.L., E.W.-P., and B.S., unpublished data). Guided by this information, recent high-throughput genome scanning also led to the surprising discovery of functional enediyne PKS genes in diverse organisms previously not known as enediyne producers, the presumed enediyne products of which were not elucidated structurally (20).
However, given the remarkable similarity in both sequence and organization of the enediyne PKS from both loci known to encode structurally characterized 9- or 10-membered enediynes, it remains to be determined what factor(s) predefines the biosynthetic divergence of the enediyne core among these architecturally distinct enediyne families. More specifically, (i) does the enediyne PKS produce a precursor universal to all families, which then is acted on by divergent accessory enzymes/proteins, or (ii) is there an apparent familial distinction between the enediyne PKSs that might invoke an early divergence in biosynthesis? To begin to recruit the tools for addressing this fundamental issue, herein we report a general PCR method for the rapid amplification of genes encoding enediyne PKSs and the application of this methodology to clone remaining prototypical genes from producers of structurally determined enediynes in both family types. A phylogenetic analysis of the new pool of bona fide enediyne PKS genes, consisting of three from 9-membered producers (1–3) and three from 10-membered producers (6–8) reveals a clear genotypic distinction between the two structural families. Contrary to genome-scanning methods, the described method provides very rapid access to the minimal enediyne PKS gene cassettes yet requires little knowledge of the producing organism or pathway a priori. The results from this study support the postulation that the enediyne PKS may play a role in the structural divergence of the enediyne core unit and clearly provides the tools for the generation of hybrid genes to study this unique issue further. By using this model, a classification is also provided for the unknown enediyne PKS genes previously identified via genome scanning.
Materials and Methods
Bacterial Strains, Vectors, and Culture Conditions. Escherichia coli DH5α was used as a general host for routine subcloning. Actinomadura madurae ATCC 39144 (the 3-producing strain), Micromonospora chersina ATCC 53710 (the 7-producing strain), and Actinomadura verrucosospora ATCC 39334 (the 8-producing strain) were obtained from the American Type Culture Collection. The PCR-cloning vector pGEM-T Easy was purchased from Promega. Culturing of spores from enediyne producers followed a four-stage fermentation process. Each stage consisted of a 5-day growth in the appropriate medium and temperature. The cells were subsequently collected by centrifugation, homogenized, and added to a fresh 50-ml culture. Specific conditions for each organism were: A. madurae, incubation at 30°C and 746 × g in R2YE medium; M. chersina, incubation at 26°C and 746 × g in ISP medium 2 (Difco); and A. verrucosospora, incubation at 28°C and 746 × g in ISP medium 4.
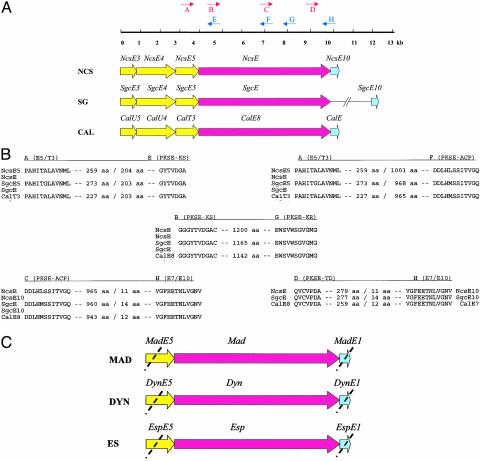
Primer Design and PCR Amplification of Minimal Enediyne PKS Genes. To clone the enediyne PKS gene madE from A. madurae, dynE from M. chersina, and espE from A. verrucosospora, sets of degenerate primers were designed according to various conserved regions of the enediyne PKS genes from the 1 and 2 loci as models for 9-membered enediynes and from the 6 gene cluster as a model for 10-membered enediynes (Fig. 2 and Table 1). Primers A–E and A–F, were designed to amplify ≈1.4 or ≈3.8 kb, respectively, of the N-terminal enediyne PKS gene, whereas primers C–H and D–H were generated to provide ≈2.9 or ≈0.9 kb, respectively, of the enediyne PKS C-terminal sequence. The PCR product resulting from primer pair B–G should present an ≈3.5-kb internal fragment of the enediyne PKS gene, which overlaps with PCR products A–F and C–H, respectively.
Fig. 2.
(A) Schematic representation of primer design for PCR-based amplification of enediyne PKS (PKSE) fragments. The PKSE and associated genes shown constitute the minimal PKSE cassettes from the 1, 2, and 6 biosynthetic loci, respectively, which are conserved among all enediyne gene clusters known to date. (B) The amino acid motifs used for the design of the degenerate primers A–H, the DNA sequences of which are illustrated in Table 1. The numbers between the motifs indicate the distance in amino acids. The noncoding regions between ORFs, represented by a slash, are <10 nt. (C) Amplified products using the PCR-based approach from genomic DNA isolated from the 3-, 7-, and 8-producing organisms to give madE, dynE, and espE, respectively.
Table 1. Oligonucleotides used for PCR amplification of enediyne PKS genes.
| Primer | Oligonucleotide sequence* |
|---|---|
| Forward | |
| A | 5′-CCCCGCVCACATCACSGSCCTCGCSGTGAACATGCT-3′ |
| B | 5′-GGCGGCGGVTACACSGTSGACGGMGCCTGC-3′ |
| C | 5′-GACGAYCTGCACMTGAGCTCSATCACCGTCGGCCAG-3′ |
| D | 5′-CARGTGTGCGTSCCSGACGCS-3′ |
| Reverse | |
| E | 5′-GCAGGCKCCGTCSACSGTGTABCCGCCGCC-3′ |
| F | 5′-CTGGCCGACGGTGATSGAGCTCAKGTGCAGRTCGTC-3′ |
| G | 5′-CCCATSCCGACSCCGGACCASACSGACCAYTCCA-3′ |
| H | 5′-ACGTTGCCGACSAGRTTSGTYTCCTCGAACCGAC-3′ |
IUB codes for mixed base sites: M = A or C; R = A or G; S = C or G; Y = C or T; K = G or T; V = A, C, or G; B = C, G, or T.
Genomic DNAs of A. madurae, M. chersina, and A. verrucosospora were purified according to the literature protocols (21) except that cells of A. madurae were treated with 2 mg·ml–1 of lysozyme and 1 mg·ml–1 of achromopeptidase to enhance cell lysis. PCRs were performed in 50-μl mixtures (1 μl of diluted template DNA/1× reaction buffer/5% DMSO/1.5 mM MgCl2/0.5 μM of each primer/0.2 mM of each dNTP/1 unit of Taq polymerase) by using the following parameters: 94°C for 5 min; 30 cycles at 94°C for 45 sec, 60–65°C for 1.5 min, and 72°C for 2–5 min; and 72°C for 7 min then cooled to 4°C. Purification of PCR products, subcloning, sequencing, and other required DNA manipulations followed standard methods.
Sequence and Phylogenetic Analysis. The sequences of overlapped PCR fragments from at least five independent clones were assembled into contiguous regions by using the assemble method and inspected by codonpreference, frames, and translate methods available in gcg wisconsin package (Accelrys, San Diego) to identify probable ORFs and restriction sites. The corresponding deduced protein sequences were compared with other known proteins in the databases by available blast methods. Amino acid sequence alignment and phylogenetic analysis were performed by the clustalw method and the drawtree and drawgram methods, respectively, from biology workbench 2.2 software (http://workbench.sdsc.edu).
Results and Discussion
Amplification of the Minimal Enediyne PKS Gene Cassettes. Template genomic DNA for this study included that from A. madurae, M. chersina, and A. verrucosospora, the producing strains for maduropeptin (Fig. 1, 3), dynemicin (Fig. 1, 7), and esperamicin (Fig. 1, 8), respectively. As illustrated in Fig. 2 A and B and Table 1, degenerate PCR primers were designed from conserved regions derived from a comparison among the known minimal enediyne PKS genes from the 1, 2, and 6 biosynthetic loci. Primer sets A–E, A–F, B–G, C–H, and D–H lead to the PCR products, consistent in the predicted sizes of 1.4, 3.8, 2.9, 0.9, and 3.5 kb, respectively. Cloning and sequencing of five individual clones for each representative fragment, followed by assembly and analysis, led to the elucidation of the intact PKS genes for each representative enediyne producer in the study (Figs. 2C and 3). The corresponding DNA sequence for the 3 enediyne PKS gene madE was deposited under the GenBank accession no. AY271660, the 7 dynE under AY162971, and the 8 espE under AY267372. Among the various methods developed for cloning enediyne biosynthetic gene clusters (22), the method presented provides the most rapid route for obtaining the enediyne PKS genes and thus will clearly enhance experiments designed to expedite the understanding and exploitation of enediyne biosynthesis.
Fig. 3.
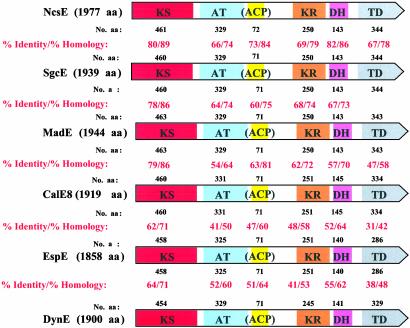
Domain organization and sequence comparison between bona fide PKSs for 9-membered (1–3) and 10-membered (6–8) enediyne core structures. Domain designations: KS, ketosynthase; AT, acyltransferase; ACP, putative acyl carrier protein; KR, ketoreductase; DH, dehydratase; TD, terminal domain (possibly containing a phosphopantetheinyl transferase).
Consistent with previously identified enediyne PKSs, the translated products MadE, DynE, and EspE share the common overall enediyne PKS domain organization (KS, AT, ACP, KR, DH, and TD) illustrated in Fig. 3 (refs. 18–20 and W.L., E.W.-P., and B.S., unpublished data) and exhibit significant head-to-tail sequence homology. The substantial functional domain similarities among all identified enediyne PKSs including MadE, DynE, and EspE support the notion that the enediyne cores of both 9- and 10-membered enediynes share a common polyketide origin. The high sequence similarity to the known minimal enediyne PKS gene cassettes from SgcE, NcsE, and CalE8 strongly imply that the PCR-derived MadE, DynE, and EspE are involved in the biosynthesis of 3, 7, and 8, respectively. As additional support, madE was found recently to restore the production of 1 in S. globishporus SB1005, (a ΔsgcE mutant strain incapable of 1 production as a result of inactivation of the critical enediyne PKS gene sgcE) (W.L., E.W.-P., and B.S., unpublished data).
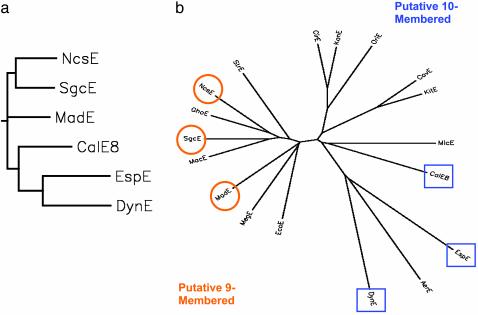
Phylogenetic Analysis and Construction of a Familial Model. In conjunction with the previously elucidated 1 NcsE, 2 SgcE, and 6 CalE, the combined set of bona fide enediyne PKS genes now is comprised of three known members of the 9-membered family (1–3) and three known members of 10-membered origin (6–8). Phylogenetic analysis of the entire set of cloned bona fide enediyne PKSs revealed the separation of this set into two main branches, the segregation of which coincides with the distinction of the core enediyne ring size (Fig. 4a). The enediyne PKSs therefore must have evolved from the same ancestor, divergent evolution from which gives rise to the 9- and 10-member-specific enediyne enzymes. This uniquely divergent relationship provides a basis for a predictive model from which the fundamental structure of unknown enediyne core structures can be predicted directly based on a cloned enediyne pksE gene, which in turn can be accessed rapidly via PCR. Thus, future rapid PCR-based enediyne “genotyping” of new enediyne-producing isolates could assist in focusing efforts on unique structural leads. This categorical genotyping might be enhanced further by PCR-based screens for the highly conserved, 9-membered-exclusive chromoproteins (23). In comparison, PCR-based screens for homologs of the calicheamicin-resistance gene calC possibly could also be used as an additional secondary 10-membered enediyne marker (24, 25).
Fig. 4.
A phylogenetic comparison of bona fide enediyne minimal PKSs (a) and all putative enediyne PKSs identified to date (b). PDB ID codes: Lechevalieria aerocolonigenes AerE, AAO25864; Micromonospora echinospora CalE8, AAM94794; Streptomyces cavourensis subsp. washingtonensis CavE, AAO25858; Streptomyces citricolor CirE, AAO25874; M. chersina DynE, AAN79725; Streptomyces sp. 100/Eco52 EcoE, AAO25879; A. verrucosospora EspE, AY267372; Streptomyces ghanaensis GhaE, AAO25844; Streptomyces kaniharaensis KanE, AAO25869; Kitasatospora sp. CECT 4991 KitE, AAO25848; Streptomyces macromomyceticus MacE, AAO25894; A. madurae MadE, AY271660; Micromonospora megalomicea subsp. nigra MegE, AAO25852; Micromonospora sp. 046/Eco11 MicE, AAO25884; S. carzinostaticus NcsE, unpublished; Amycolatopsis orientalis OriE, AAO25836; Streptomyces globisporus SgcE, AY048920; Streptomyces sp. 171/Eco105 StrE, AAO25889.
Application of the Model to Unknown Enediynes. A recent report of high-throughput genome scanning revealed a set of 12 functional enediyne PKS genes found dispersed among genomes of widely diverse organisms previously not known as enediyne producers (20). Although the optimized fermentation products of these organisms were not characterized structurally, extracts were found to be positive in a cell-based enediyne-specific assay used to detect DNA damage, consistent with the mode of enediyne action. Application of our newly devised familial model (Fig. 4b) suggests that five of the unknown potential enediyne PKSs are expected to produce 9-membered products (macE, ghoE, strE, megE, and ecoE), whereas the remaining seven are genotypically predictive of 10-membered core formation (micE, kitE, cavE, aerE, kanE, cirE, and oriE). Although future fermentation product isolation and structural characterization will help definitively support or refute this postulation, this exercise illustrates the exceptional speed and potential power of enediyne-producer familial classification.
Impact on Understanding Enediyne Biosynthesis. A comparison of Figs. 3 and 4 reveals that, although there is an identical functional domain similarity among all enediyne PKSs (and, thus, likely a common early transient intermediate), a clear 9-membered versus 10-membered familial distinction also emerges after phylogenetic analysis. We propose that this distinction is critical to the alternate folding and early modification steps that lead to the divergence of the 9- and 10-membered enediynes and may be a result of unique protein–protein interactions with the other postulated enediyne biosynthesis-associated proteins of the minimal enediyne PKS cassette. Consistent with this model, flanking the enediyne PKS gene are a group of 5–10 genes, presumed to encode enediyne accessory enzymes (unusual oxidoreductases or proteins of unknown function) that are unique to enediyne loci and organizationally conserved among all enediyne gene clusters characterized to date. Cumulatively, this model suggests that all enediyne PKSs are capable of generating an early common intermediate, which then advances toward divergent pathways as dictated by associate proteins (Fig. 5) (22, 26). Based on this model, the key to many of the unique stages of enediyne biosynthesis such as folding and/or triple-bond construction may reside, in part, with these unique associate proteins and their interaction with the core enediyne PKS.
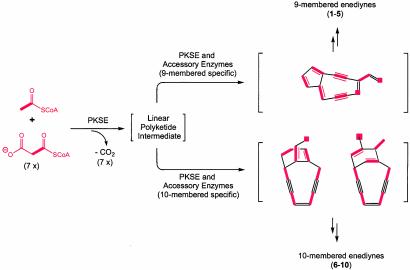
Fig. 5.
A unified scheme for enediyne core biosynthesis implicating the enediyne PKS (PKSE) as responsible for a common polyketide intermediate and enediyne PKS accessory enzymes, in conjunction with the PKSE, to diverge into the 9- or 10-membered enediyne core pathways. Atoms that were incorporated intact from the acyl CoA precursors to the enediyne cores 1, 7, and 8 (15–17) are shown in bold red.
Acknowledgments
This contribution was supported in part by grants from the National Institutes of Health to J.S.T. (GM58196, CA84374, and AI52218) and B.S. (CA78747). B.S. is the recipient of National Science Foundation CAREER Award MCB9733938 and National Institutes of Health Independent Scientist Award AI51687. J.S.T. is an Alfred P. Sloan Research Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PKS, polyketide synthase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY271660 (3 enediyne PKS gene madE), AY162971 (7 dynE), and AY267372 (8 espE)].
References
- 1.Jones, G. B. & Fouad, F. S. (2002) Curr. Pharm. Des. 8, 2415–2440. [DOI] [PubMed] [Google Scholar]
- 2.Danishefsky, S. J. & Shair, M. D. (1996) J. Org. Chem. 61, 16–64. [Google Scholar]
- 3.Smith, A. L. & Nicolaou, K. C. (1996) J. Med. Chem. 39, 2103–2117. [DOI] [PubMed] [Google Scholar]
- 4.Thorson, J. S., Shen, B., Whitwam, R. E., Liu, W. & Li, Y. (1999) Bioorg. Chem. 27, 172–188. [Google Scholar]
- 5.Thorson, J. S., Sievers, E. L., Ahlert, J., Shepard, E., Whitwam, R. E., Onwueme, K. C. & Ruppen, M. (2000) Curr. Pharm. Des. 6, 1841–1879. [DOI] [PubMed] [Google Scholar]
- 6.Oku, N., Matsunaga, S. & Fusetani, N. (2003) J. Am. Chem. Soc. 125, 2044–2045. [DOI] [PubMed] [Google Scholar]
- 7.Zein, N., Sinha, A. M., McGaharen, W. J. & Ellestad, G. A. (1988) Science 240, 1198–1201. [DOI] [PubMed] [Google Scholar]
- 8.Myers, A. G., Cohen, S. B. & Kwon, B. M. (1994) J. Am. Chem. Soc. 116, 1255–1271. [Google Scholar]
- 9.DeVoss, J. J., Hangeland, J. J. & Townsend, C. A. (1990) J. Am. Chem. Soc. 112, 4554–4556. [Google Scholar]
- 10.Kappen, L. S. & Goldberg, I. H. (1993) Science 261, 1319–1321. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe, C. M. H., Supekova, L. & Schultz, P. G. (2002) Chem. Biol. 9, 245–251. [DOI] [PubMed] [Google Scholar]
- 12.Stassinopoulos, A., Ji, J., Gao, S. & Goldberg, I. H. (1996) Science 272, 1943–1946. [DOI] [PubMed] [Google Scholar]
- 13.Sievers, E. L. & Linenberger, M. (2001) Curr. Opin. Oncol. 13, 522–527. [DOI] [PubMed] [Google Scholar]
- 14.Abe, S. & Otsuki, M. (2002) Curr. Med. Chem. 2, 715–726. [DOI] [PubMed] [Google Scholar]
- 15.Hensens, O. D., Giner, J.-L. & Goldberg, I. H. (1989) J. Am. Chem. Soc. 111, 3295–3299. [Google Scholar]
- 16.Tokiwa, Y., Miyoshi-Saitoh, M., Kobayashi, H., Sunaga, R., Konishi, M., Oki, T. & Iwasaki, S. (1992) J. Am. Chem. Soc. 114, 4107–4110. [Google Scholar]
- 17.Lam, K. S., Veitch, J. A., Golik, J., Krishnan, B., Klohr, S. E., Volk, K. J., Forenza, S. & Doyle, T. W. (1993) J. Am. Chem. Soc. 115, 12340–12345. [Google Scholar]
- 18.Liu, W., Christenson, S. D., Standage, S. & Shen, B. (2002) Science 297, 1170–1173. [DOI] [PubMed] [Google Scholar]
- 19.Ahlert, J., Shepard, E., Lomovskaya, N., Zazopoulos, E., Staffa, A., Bachmann, B. O., Huang, K., Fonstein, L., Czisny, A., Whitwam, R. E., et al. (2002) Science 297, 1173–1176. [DOI] [PubMed] [Google Scholar]
- 20.Zazopoulos, E., Huang, K., Staffa, A., Liu, W., Bachmann, B. O., Nonaka, K., Ahlert, J., Thorson, J. S., Shen, B. & Farnet, C. M. (2003) Nat. Biotechnol. 21, 187–190. [DOI] [PubMed] [Google Scholar]
- 21.Pospiech, A. & Neumann, B. (1995) Trends Genet. 11, 217–218. [DOI] [PubMed] [Google Scholar]
- 22.Shen, B., Liu, W. & Nonaka, K. (2003) Curr. Med. Chem. 10, in press. [DOI] [PubMed]
- 23.Adjadj, E., Quiniou, E., Mispelter, J., Favaudon, V. & Lhoste, J. M. (1992) Biochimie 74, 853–858. [DOI] [PubMed] [Google Scholar]
- 24.Whitwam, R. E., Ahlert, J., Holman, T. R., Ruppen, M. & Thorson, J. S. (2000) J. Am. Chem. Soc. 122, 1556–1557. [Google Scholar]
- 25.Biggins, J. B., Onwueme, K. C. & Thorson, J. S. (2003) Science 301, 1537–1541. [DOI] [PubMed] [Google Scholar]
- 26.Shen, B. (2003) Curr. Opin. Chem. Biol. 7, 285–295. [DOI] [PubMed] [Google Scholar]