Abstract
Zinc-finger protein transcription factors (ZFP TFs) can be designed to control the expression of any desired target gene, and thus provide potential therapeutic tools for the study and treatment of disease. Here we report that a ZFP TF can repress target gene expression with single-gene specificity within the human genome. A ZFP TF repressor that binds an 18-bp recognition sequence within the promoter of the endogenous CHK2 gene gives a >10-fold reduction in CHK2 mRNA and protein. This level of repression was sufficient to generate a functional phenotype, as demonstrated by the loss of DNA damage-induced CHK2-dependent p53 phosphorylation. We determined the specificity of repression by using DNA microarrays and found that the ZFP TF repressed a single gene (CHK2) within the monitored genome in two different cell types. These data demonstrate the utility of ZFP TFs as precise tools for target validation, and highlight their potential as clinical therapeutics.
Defects in transcriptional regulation underlie numerous disease states, most notably cancer (1). A major goal of current strategies for correcting such defects is to achieve sufficient specificity of action (2). Designed zinc-finger protein transcription factors (ZFP TFs) emulate natural transcriptional control mechanisms and therefore provide an attractive tool for precisely regulating gene expression. Accurate control of gene expression is important for understanding gene function (target validation) and for developing therapeutics to treat disease (3). We and others have used engineered ZFP TFs to either activate or repress a variety of endogenous gene targets (4–11). For these proteins, or any other gene-regulation technology, to succeed as tools in drug discovery or direct agents in the clinic, their specificity of action within the genome must be precise, a challenging criterion to meet given the size and complexity of the human genome. Recent studies with small interfering RNA (12, 13) and antisense DNA/RNA (14) have illuminated the magnitude of the task of achieving single-gene specificity in regulating the human genome.
We focus here on the use of ZFP TFs in the area of oncology and specifically on the emerging role of checkpoint kinase 2 (CHK2). CHK2 acts as a key integrator of DNA-damage signals regulating cell-cycle progression, DNA repair, and cell death by phosphorylating a variety of substrates, including the p53 tumor suppressor protein (15, 16). Here we show that a designed ZFP TF targeted to a unique 18-bp recognition sequence in the promoter of the CHK2 gene binds the intended site within chromatin and represses CHK2 transcription in vivo. Moreover, repression of CHK2 by this engineered ZFP TF occurs with remarkable specificity, while simultaneously reducing CHK2 protein to levels that functionally ablate the action of this kinase. Finally, we show that constitutive expression of the ZFP TF in telomerase-immortalized, untransformed human fibroblasts provides stable repression of the CHK2 gene and results in loss of DNA-damage-induced CHK2-dependent phosphorylation of p53 on Ser-20. These data demonstrate that ZFP TFs can be exquisitely specific, yet potent repressors of gene expression and, therefore, are potentially powerful reagents for target validation and therapeutic interventions in vivo.
Materials and Methods
Cell Culture. The HEK293 cell line was obtained from the American Type Culture Collection. HEK293 T-REx and U2OS T-REx cell lines were purchased from Invitrogen. Each line was maintained as recommended by the supplier.
Mapping of DNase I-Accessible Chromatin Regions in CHK2 Locus. Human HEK293 nuclei were treated with DNase I as described (17). Genomic DNA isolation, restriction enzyme digestion, and Southern blot analysis were then carried out as described (5, 7) except that the restriction enzymes and probe used were as indicated in Fig. 1.
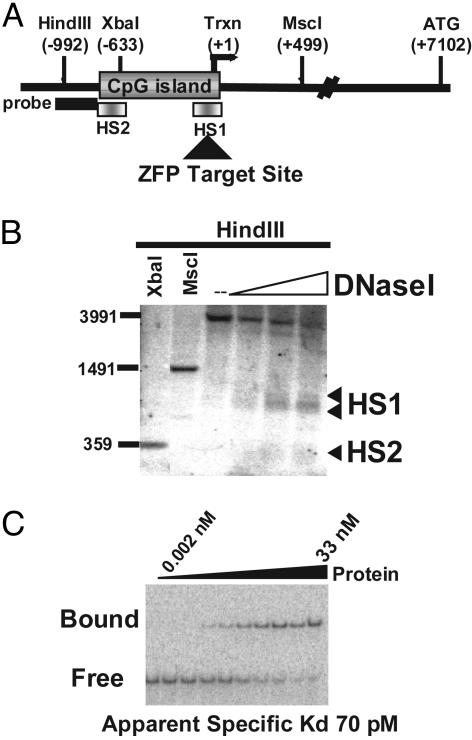
Fig. 1.
Identification of a ZFP TF for the regulation of the human CHK2 gene. (A) A schematic representation of the CHK2 promoter indicating the positions of restriction sites and probe used in the DNase I hypersensitive site mapping and the experimentally determined major start site of transcription (Trxn). (B) DNase I hypersensitivity map of the CHK2 promoter. The XbaI and MscI digests serve as location markers, and the numbers indicated on the left refer to the positions of the size markers run alongside. (C) Electrophoretic mobility-shift assay of ZFP-5475 used to determine the apparent in vitro Kd for this DNA-binding protein. Maltose-binding-protein-purified ZFP protein was titrated by using a 3-fold dilution series as indicated.
Synthesis, Purification, and Gel-Shift Analysis of ZFPs. ZFP TFs targeted to a predetermined region in the CHK2 gene were assembled by linking three two-finger modules as described (18, 19). The resultant ZFP gene was cloned into the pMal-c2 plasmid (New England Biolabs) as a fusion with DNA encoding maltose-binding protein. Maltose-binding protein–ZFP fusions were then expressed and affinity-purified by using an amylose/agarose resin (New England Biolabs). Binding studies were performed as described except that the target site, 5′-ACCCGGGTTCCCCTCGGG-3′, was incorporated into the DNA oligonucleotides (5, 7).
ZFP Expression Constructs Used for Cell-Culture Studies. For transient transfection studies, the ZFP TF was cloned into a repression-domain construct as described (8). The resulting construct, pTracer-ZFP-KOX1, contains an N-terminal ZFP DNA-binding domain, a nuclear localization signal (PKKKRKV) from simian virus 40 large T antigen, and the KOX1 repression domain. The ZFP itself was assembled from an archive of two-finger modules described in ref. 20, wherein the amino acid residues of the helical regions (from the –1 to +6 positions) responsible for specific DNA binding are F1, RSDHLSR; F2, DNRDRTK; F3, DRKTLIE; F4, TSSGLSR; F5, RSDHLSE; and F6, TSSDRTK.
Cell Culture and Transient Transfections. HEK293 cells were grown in DMEM supplemented with 10% FBS in a 5% CO2/95% air incubator at 37°C. For transfections, HEK293 cells were plated in 12-well plates at a density of 250,000 cells per well and transfected 1 day later by using Lipofectamine 2000 reagent (GIBCO/BRL) according to the manufacturer's recommendations, with 1.75 μl of Lipofectamine 2000 reagent and 0.5 μg of ZFP plasmid DNA per well. The medium was removed and replaced with fresh medium 6–12 h after transfection. Transfection efficiency was assessed in each independent experiment by the use of a GFP expression plasmid control; in all experiments, an apparent transfection efficiency of 80–90% GFP-positive cells was observed.
Retroviral Constructs, Virus Preparation, and Generation of Stable Lines. A self-inactivating retroviral vector containing a tetracycline-inducible ZFP expression cassette was constructed and used for virus generation. In brief, the pSIR vector (Clontech) was modified to contain the cytomegalovirus promoter and the tetracycline operator sequences from pcDNA4-TO (Invitrogen). The coding region of ZFP-5475-KOX1 was inserted downstream of the inducible promoter by cloning into the modified pSIR vector (Clontech). Virus-containing supernatant was generated by transient transfection of the resulting plasmid, pSIR-TO-ZFP-5475-KOX1, into the Phoenix packaging line (G. P. Nolan, Stanford University, Stanford, CA) as described (21). For stable cell-line generation, HEK293 T-REx and U2OS T-REx cells were transduced with supernatant obtained above containing retrovirus encoding ZFP-5475-KOX1 and selected in medium containing 800 μg/ml G418 (Invitrogen). Individual clones were isolated and analyzed for doxycycline (DOX)-dependent expression of ZFP-KOX1 expression and corresponding repression of the endogenous gene target.
Quantitative RT-PCR Analysis of RNA Expression (TaqMan). Cells were lysed and total RNA was prepared by using the high-pure RNA isolation kit (Roche Diagnostics) according to the manufacturer's recommendations. Real-time quantitative RT-PCR analysis using TaqMan chemistry in a 96-well format on an ABI 7700 SDS machine (Perkin–Elmer) was performed as described (5). The primer/probes used are available on request. The results were analyzed by using sds version 1.6.3 software.
Microarray Analysis. Global changes in gene expression were analyzed by using a U133A GeneChip array (Affymetrix, Santa Clara, CA) and a GeneArray scanner (Agilent Technologies, Palo Alto, CA). RNA samples were prepared according to the manufacturer's recommendations. Data analysis to determine differentially expressed genes was carried out by using Affymetrix genechip mas version 5.0 and dmt version 3.0 software. The “Change Call” indicated does not relate to the P value; rather, for a probe set to be called “up” or “down,” criteria of (i) a 2-fold difference in expression level between experiment and control, and (ii) a 100% confidence call were applied. For the HEK293 experiments three independent single-cell-derived clones were analyzed in duplicate with fold change determined by using Affymetrix dmt version 3.0 statistical software and the “low signal log ratio” algorithm. For the U2OS experiments an individual single-cell-derived clone was analyzed in duplicate, and fold change was determined by using Affymetrix dmt version 3.0 statistical software and the signal log ratio algorithm.
Immunoblotting. Western blot analysis of protein expression was performed as described (6), followed by immunoblotting by using antibodies against CHK2 (catalog no. 2391, ProSci, Poway, CA) and TFIIB (sc-225, Santa Cruz Biotechnology).
Chromatin Immunoprecipitation. Chromatin immunoprecipitation was performed by using the chromatin immunoprecipitation assay kit according to the manufacturer's instructions (Upstate Biotechnology, Lake Placid, NY) and as described (6) except that an anti-hemagglutinin epitope tag antibody (sc-7392, Santa Cruz Biotechnology) was used throughout. Plasmids encoding hemagglutinin-tagged constructs were assembled as described (6).
hTERT-Immortalized Human Cell Studies. Human fibroblasts (strain 82-6) were obtained, cultured, and immortalized with an hTERT-expressing retrovirus, as described (22, 23). The ZFP-5475 cDNA was subcloned into the pLXSN retroviral vector, infectious virus was produced, and hTERT-expressing cells were infected and selected as mass cultures. The cells were then plated at clonal densities, and single-cell clones were expanded for analysis. Whole-cell lysates were prepared and analyzed for the indicated proteins by immunoblotting, using commercially available antibodies that recognize CHK2 (Santa Cruz Biotechnology), actin (Chemicon), p53 (Oncogene Research Products, EMD Biosciences, San Diego), and p53-Ser-20 (Cell Signaling Technology, Beverly, MA), as described (22).
Results
An Engineered ZFP TF Repressor of the Human CHK2 Gene. Engineered ZFP TFs can regulate the expression of endogenous genes in vivo (see ref. 11 for a recent review). To identify engineered ZFP TFs capable of controlling transcription of the CHK2 gene, we first mapped the chromatin architecture of the human CHK2 promoter to determine regions of “open” or accessible chromatin. Fig. 1A shows a schematic representation of the human CHK2 locus, indicating positions of the probe and restriction enzyme cutting sites used in the DNase I hypersensitive site-mapping experiment. Two accessible or hypersensitive (HS) sites were identified, designated HS1 and HS2 (Fig. 1B). HS1 contained the major start site of transcription, as determined by rapid amplification of cDNA ends (or RACE) (data not shown). The sequence of the HS1 site was therefore used to design a six-finger ZFP TF (ZFP-5475) recognizing the site 5′-ACCCGGGTTCCCCTCGGG-3′ constructed from an archive of zinc-finger DNA-binding modules (20). This ZFP TF consisted of a string of three two-finger units, which was demonstrated to have increased specificity over more conventional poly-zinc-finger peptide units in vitro (18). The in vitro DNA-binding characteristics of this protein are shown in Fig. 1C. ZFP-5475 binds its intended target sequence with an apparent Kd of ≈70 pM, a value that is similar to natural TFs (24). Furthermore, when linked to the Krüppel-associated box A/B repression domain (25) from the N-terminal region of the KOX1 protein (26), this ZFP decreased the level of CHK2 mRNA in a dose-dependent manner, achieving up to 50% repression in transient transfection assays (Fig. 2A).
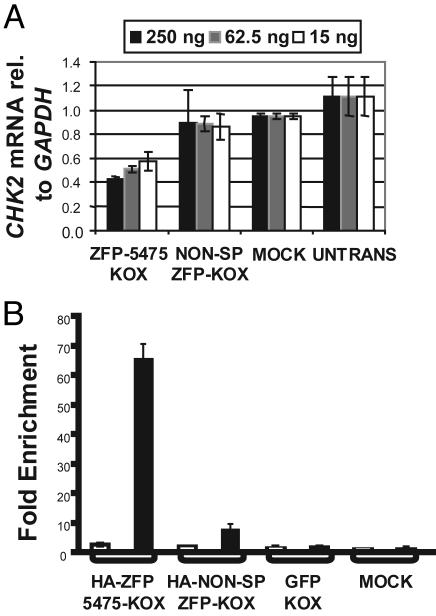
Fig. 2.
ZFP-5475 regulates the expression of the endogenous CHK2 gene. (A) ZFP-5475 represses CHK2 expression in cultured cells. HEK293 cells transfected with the plasmids indicated were assayed for CHK2 mRNA by quantitative RT-PCR (TaqMan) after 72 h. The CHK2 mRNA levels were normalized relative to an internal control of GAPDH mRNA and are expressed as this ratio. Charts represent data from a minimum of two independent experiments, with means and standard deviations shown. Transfection efficiency was assessed in each independent experiment by the use of a GFP expression plasmid control; in all experiments an apparent efficiency of 80–90% GFP-positive cells was observed. (B) ZFP-5475 binds to its intended target site within the CHK2 promoter in vivo. HEK293 cells transfected with the indicated plasmids were assayed for enrichment of the CHK2 promoter by chromatin immunoprecipitation with primers specific for the ZFP-proximal region. Enrichment was quantified by RT-PCR. Results are expressed as the fold increase of the ratio to the GAPDH control relative to the results for nontransfected cells, the value of which is arbitrarily set to 1. The same samples were analyzed with primers specific for the p16 locus as a second internal control (open bars). No enrichment was observed with preimmune serum (data not shown).
The repression of CHK2 mRNA levels depended on the Krüppel-associated box A/B repressor domain because transfection of a construct expressing the DNA-binding domain alone failed to repress CHK2 gene expression. Moreover, when we switched the repressor domain with the p65 activation domain of NF-κB (27), CHK2 mRNA increased (data not shown).
To confirm that CHK2 repression resulted from a direct interaction between ZFP-5475 and its intended target site, we used chromatin immunoprecipitation analysis. An ≈65-fold enrichment of the CHK2 promoter fragment containing the ZFP TF binding site was observed in the presence of ZFP-5475 (HA-ZFP5475-KOX) relative to a control fragment from the GAPDH gene (Fig. 2B). Neither transfection with a plasmid encoding GFP-KOX nor a nonspecific ZFP TF invoked a significant enrichment of the CHK2 promoter fragment. Moreover, no enrichment of a control fragment at the p16 gene was observed. Thus, this ZFP TF is indeed bound to the expected region of the CHK2 promoter in vivo.
We conclude that the engineered TF ZFP-5475 binds to and regulates the expression of the CHK2 gene in vivo.
Regulatable and Reversible Repression of CHK2 in Stable Inducible Cell Lines. To eliminate the contribution of untransfected cells in the transient transfection assays of repression, we constructed stable cell lines in which the T-REx system (Invitrogen) provided inducible expression of the ZFP TF. We created vectors that placed ZFP TF expression under the control of a tetracycline operator-regulated cytomegalovirus promoter, and introduced them into HEK293 (HEK293 T-Rex) and U2OS (U2OS T-Rex) cells by retroviral transduction. We isolated single-cell-derived clones and tested for DOX-dependent repression of the CHK2 gene. The results from 16 HEK293 T-REx clones are shown in Fig. 3A.
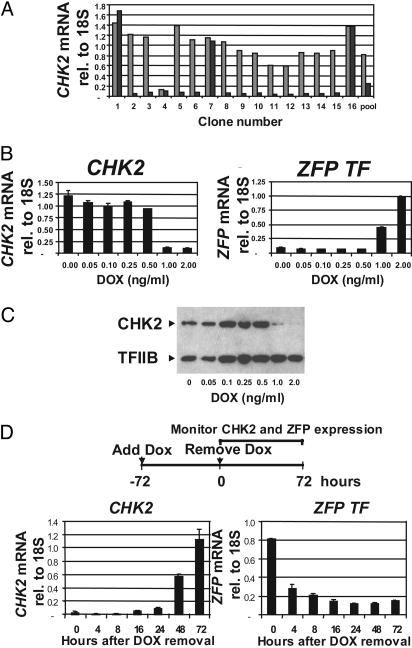
Fig. 3.
Regulatable expression of the ZFP TF drives inducible and reversible repression of CHK2. (A) Isolated single-cell-derived clones show inducible repression of CHK2 mRNA expression. ZFP TF-transduced HEK293 T-REx clones were assayed for CHK2 mRNA by quantitative RT-PCR (TaqMan) after 48 h in the presence (▪) or absence ( ) of 1 ng/ml DOX. mRNA assays were done as in Fig. 2 except that the CHK2 mRNA levels were normalized relative to an internal 18S rRNA control. “Pool” refers to the antibiotically selected ZFP TF-transduced HEK293 T-REx cell population before single-cell cloning. (B) CHK2 repression correlates with ZFP TF expression. An individual, isolated, single-cell-derived clone was assayed for both CHK2 mRNA (Left) and ZFP TF mRNA (Right) over the range of DOX concentrations indicated and normalized as above. (C) CHK2 protein is abolished by expression of the ZFP TF. Whole-cell lysates obtained from the experiment described in B were assayed for the presence of CHK2 by immunoblotting and normalized to the signal from TFIIB. (D) ZFP TF repression of CHK2 is reversible. The experimental strategy is shown diagrammatically above the graphs. After 72 h of DOX treatment at 1 ng/ml, the DOX was removed and both CHK2 mRNA and ZFP TF mRNA were assayed by RT PCR as described in B at the times indicated.
) of 1 ng/ml DOX. mRNA assays were done as in Fig. 2 except that the CHK2 mRNA levels were normalized relative to an internal 18S rRNA control. “Pool” refers to the antibiotically selected ZFP TF-transduced HEK293 T-REx cell population before single-cell cloning. (B) CHK2 repression correlates with ZFP TF expression. An individual, isolated, single-cell-derived clone was assayed for both CHK2 mRNA (Left) and ZFP TF mRNA (Right) over the range of DOX concentrations indicated and normalized as above. (C) CHK2 protein is abolished by expression of the ZFP TF. Whole-cell lysates obtained from the experiment described in B were assayed for the presence of CHK2 by immunoblotting and normalized to the signal from TFIIB. (D) ZFP TF repression of CHK2 is reversible. The experimental strategy is shown diagrammatically above the graphs. After 72 h of DOX treatment at 1 ng/ml, the DOX was removed and both CHK2 mRNA and ZFP TF mRNA were assayed by RT PCR as described in B at the times indicated.
The majority (12 of 16) of clones showed DOX-dependent repression of CHK2 mRNA levels, indicating a high frequency with which inducible repression was obtained. Moreover, most clones showed >10-fold repression, resulting in barely detectable CHK2 transcript levels. Of particular importance for the functional assays described below, mRNA levels of the related checkpoint kinase CHK1 (28) were unaffected by ZFP induction (data not shown, but see Table 1). Target gene repression depended on the ZFP expression level, as the increasing ZFP mRNA levels obtained by increasing DOX concentrations correlated well with the degree of CHK2 repression at both the mRNA and protein levels (Fig. 3 B and C). Essentially identical results were obtained in U2OS T-REx clones, indicating that the results were not specific to a particular cell type (see below).
Table 1.
Genes regulated by the CHK2-specific ZFP TF in HEK293 T-REx stable lines
| No. | Probe set | Fold change (down) | Confidence call | P value | Change call | Gene name |
|---|---|---|---|---|---|---|
| 1 | 210416_s_at | 9.4 | 100 | <0.001 | Down | Homo sapiens protein kinase hChk2 |
| 2 | 208739_x_at | 1.5 | 83 | 0.056 | None | H. sapiens MIF2 suppressor (HSMT3) |
| 3 | 203012_x_at | 1.3 | 75 | 0.085 | None | H. sapiens ribosomal protein L23a (RPL23A) |
| 4 | 201665_x_at | 1.3 | 75 | 0.192 | None | H. sapiens ribosomal protein S17 (RPS17) |
| 5 | 206074_s_at | 2.5 | 66 | 0.009 | None | H. sapiens high-mobility group protein isoforms I and Y (HMGIY) |
| 6 | 200817_x_at | 1.3 | 66 | 0.131 | None | H. sapiens ribosomal protein S10 (RPS10), mRNA |
| 7 | 208909_at | 1.3 | 66 | 0.068 | None | H. sapiens ubiquinol-cytochrome c reductase |
| 8 | 208738_x_at | 1.3 | 66 | 0.230 | None | H. sapiens cDNA highly similar to HSSMT3B H. sapiens mRNA |
| 9 | 211765_x_at | 1.3 | 66 | 0.431 | None | H. sapiens peptidylprolyl isomerase A (cyclophillin A) |
| 10 | 212734_x_at | 1.3 | 66 | 0.010 | None | CLONE=IMAGE:1745177 Hs. 180842 ribosomal protein L13 |
The growth characteristics of induced HEK293 T-Rex and U2OS T-Rex cells were indistinguishable from uninduced cells after ≈2 weeks of culture. Moreover, repression of CHK2 was maintained throughout this period (data not shown, but see Fig. 5). These results indicate that cells tolerated persistent expression of the ZFP TF. Target gene repression required the continuous presence of the ZFP TF, because removing DOX from the culture medium reduced ZFP expression to background levels within ≈24 h followed by recovery of CHK2 gene expression (Fig. 3D). Taken together, these data demonstrate that the repression of target gene expression driven by the ZFP is dramatically effective (>10-fold repression) and is also stable, regulatable, and reversible.
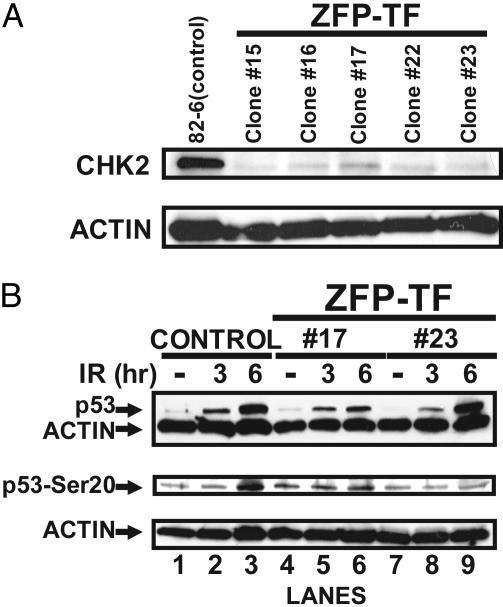
Fig. 5.
ZFP TF repression of CHK2 prevents the DNA-damage-dependent phosphorylation of p53 at Ser-20. (A) Constitutive expression of the ZFP TF in isolated single-cell-derived clones of hTERT immortalized, untransformed human fibroblasts results in repression of CHK2 at the protein level. Whole-cell lysates from five different single-cell-derived clones were assayed by immunoblotting for CHK2 expression. As a loading control the blot was reprobed with an antibody recognizing actin. (B) ZFP TF-driven repression of CHK2 ablates the DNA-damage-dependent phosphorylation of p53 at Ser-20. Control cells (transduced with an insertless vector) and two ZFP TF single-cell-derived clones shown above to repress CHK2 expression by Western blotting were challenged with 10 Gy of x-irradiation. Whole-cell lysates were obtained at the indicated times and analyzed by immunoblotting with the indicated antibodies. Actin was used as a loading control as above.
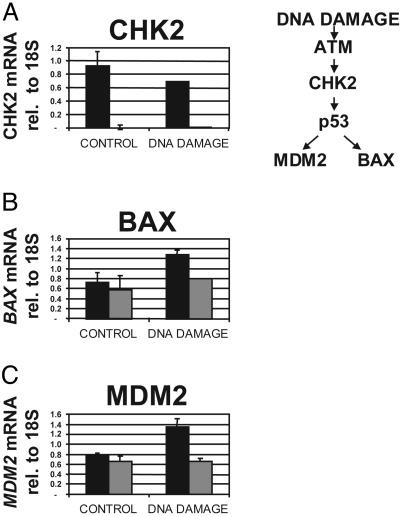
CHK2-Dependent p53 Function Is Abolished After ZFP-Driven Repression. CHK2 is a cell-cycle checkpoint kinase that phosphorylates several key regulators of cell proliferation in response to DNA damage, most notably p53 (see refs. 28–30). One consequence of CHK2-dependent p53 phosphorylation is an increase in p53 transactivation activity (31). This increased activity is manifest by elevated expression of p53 target genes, such as MDM2, BAX, and p21. Indeed, cells from Chk2–/– mice fail to induce expression of these p53 targets after DNA damage by ionizing radiation (31). To confirm that the repression of CHK2 by the ZFP TF functionally abolished CHK2 activity, U2OS T-REx cells were challenged by a DNA-damaging agent (camptothecin) in the presence or absence of the ZFP (i.e., in the presence or absence of DOX). Camptothecin is a topoisomerase I inhibitor, and was previously reported to stimulate a CHK2-dependent DNA-damage response in cultured cells (32). As shown in Fig. 4, in uninduced cells lacking ZFP expression, camptothecin activated the p53-dependent DNA-damage pathway, resulting in increased expression of BAX and MDM2. In contrast, induced cells, in which the ZFP repressed transcription of the CHK2 gene (Fig. 4A), failed to show a camptothecin-dependent increase in BAX and MDM2 expression 8 h after treatment (Fig. 4 B and C). After 24 h of camptothecin treatment, however, activation of both MDM2 and BAX1 was observed in the ZFP TF cell line but did not reach the levels observed for this line in the absence of DOX (data not shown). These results are very similar to the results of similar experiments performed with Chk2–/– mouse thymocytes (31). These data indicate that the CHK2-specific ZFP TF causes a functional CHK2 knock-down phenotype similar to that originally obtained through genetic ablation of the CHK2 gene.
Fig. 4.
ZFP-driven repression of CHK2 functionally eliminates the DNA-damage-dependent transactivation of p53. An isolated single-cell-derived clone of U20S T-REx transduced with a vector encoding inducible expression of the ZFP TF was cultured in the presence ( ) or absence (▪) of DOX. DNA damage was induced by addition of 10 μM camptothecin 72 h after the addition of DOX (DNA DAMAGE) or not (CONTROL). The levels of CHK2 mRNA (A), BAX mRNA (B), and MDM2 mRNA (C) were assayed 8 h after drug addition and are shown relative to 18S rRNA levels as described in the legend of Fig. 3. ATM, ataxia-telangiectasia-mutated kinase.
) or absence (▪) of DOX. DNA damage was induced by addition of 10 μM camptothecin 72 h after the addition of DOX (DNA DAMAGE) or not (CONTROL). The levels of CHK2 mRNA (A), BAX mRNA (B), and MDM2 mRNA (C) were assayed 8 h after drug addition and are shown relative to 18S rRNA levels as described in the legend of Fig. 3. ATM, ataxia-telangiectasia-mutated kinase.
CHK2 Is the Only Gene Repressed by ZFP-5475. The CHK2-specific ZFP TF repressor protein recognizes an 18-bp sequence that, theoretically, is sufficient to provide a unique address within the human genome. We therefore asked whether the designed ZFP TF indeed regulated just a single gene. In this regard, the CHK2 gene target is an attractive test system for determining the genomewide specificity of ZFP TF because (i) the site to which the ZFP TF binds is indeed unique within the human genome (data not shown), and (ii) CHK2 must be phosphorylated by ATM to become an active kinase capable of phosphorylating substrates such as p53 (33–35). Thus, in undamaged cells, CHK2 remains unphosphorylated and, to a first approximation, inert, thereby eliminating possible downstream or secondary effects that might confound genomewide analyses of the specificity of ZFP TFs.
Three different HEK293 T-REx clones, each demonstrating DOX-inducible CHK2 repression, were analyzed for changes in gene expression in the presence (plus ZFP) or absence (minus ZFP) of DOX. Gene expression changes were determined by using the Affymetrix U133A array, which provides information on 22,225 probe sets or ≈16,000 genes. The results were analyzed by using Affymetrix genechip mas5.0 and dmt3.0 software. For a probe set to be called “up” or “down” (Change Call) criteria of (i) a 2-fold difference in expression level between experiment and control and (ii) a 100% confidence call were applied. The results of this analysis are shown in Table 1.
The only gene that was identified as showing a “down change” (i.e., repression) in this analysis was the intended target, the human CHK2 gene. CHK2 mRNA was repressed ≈10-fold, with a 100% confidence call and P value of <0.001. No other gene on the array was identified by the software as an up or down change. To determine whether this result was peculiar to HEK293 T-Rex cells, we repeated the experiment by using U2OS T-REx cells. Comparison of the genomewide expression profiles of uninduced HEK293 T-REx and U2OS T-REx cells indicated that, of all of the genes that were expressed (scored as “present” by the analysis software), ≈30% were exclusive to one or the other cell line. Despite this difference in uninduced gene expression, the expression of the ZFP TF in U2OS T-REx cells effected repression of only the CHK2 gene (Table 2). Taken together, these data demonstrate that ZFP TFs can regulate target gene expression with singular specificity. Moreover, this specificity is obtained in two different human cell types.
Table 2. Genes regulated by the CHK2-specific ZFP TF in the U20S T-Rex stable line.
| No. | Probe set | Fold change (down) | P value | Change call | Gene name |
|---|---|---|---|---|---|
| 1 | 210416_s_at | 7.1 | 0.003 | Down | H. sapiens protein kinase hChk2 mRNA |
| 2 | 205010_at | 1.9 | 0.058 | None | H. sapiens hypothetical protein (FLJ10613) |
| 3 | 201085_s_at | 1.9 | 0.230 | None | Consensus includes SON DNA-binding protein |
| 4 | 206074_s_at | 1.6 | 0.053 | None | H. sapiens high-mobility group protein isoforms I and Y (HMGIY) |
| 5 | 211767_at | 1.6 | 0.100 | None | H. sapiens similar to RIKEN cDNA 4933405K01 mRNA |
| 6 | 208993_s_at | 1.5 | 0.276 | None | Consensus includes peptidylprolyl isomerase G (cyclophilin G) |
| 7 | 201108_s_at | 1.5 | 0.092 | None | Consensus includes thrombospondin 1 |
| 8 | 208739_x_at | 1.4 | 0.006 | None | H. sapiens MIF2 suppressor (HSMT3) |
| 9 | 215529_x_at | 1.4 | 0.842 | None | Consensus includes H. sapiens mRNA DKFZp434G0572 |
| 10 | 205443_at | 1.3 | 0.076 | None | H. sapiens small nuclear RNA-activating complex mRNA |
All ten genes shown gave 100% confidence calls.
ZFP-5475 Functionally Abolishes CHK2 Expression in Telomerase-Immortalized Untransformed Human Fibroblasts. To provide further functional validation of the CHK2-specific ZFP TF repressor, we used retroviruses to constitutively express ZFP-5475 in untransformed human fibroblasts that were immortalized by hTERT, the catalytic subunit of telomerase (23). Several independent single-cell-derived clones were obtained in which ZFP TF-driven CHK2 repression was evident by immunoblot analysis (Fig. 5A). These immortalized, untransformed human cell clones are wild type with respect to p53 function, and thus provide the opportunity to examine the downstream consequences of CHK2 repression in an untransformed human cell. Specifically, DNA damage induces p53 phosphorylation at both Ser-15 and Ser-20 (see ref. 16), and data from Chk2–/– mice identified CHK2 as the kinase responsible for Ser-20 phosphorylation. To determine whether human CHK2 similarly phosphorylates p53 on Ser-20, we x-irradiated control (infected with an insertless retrovirus) and ZFP TF-transduced human cells with 10 Gy of ionizing radiation. We prepared whole-cell extracts 0, 3, and 6 h after ionizing radiation and analyzed them by immunoblotting. In response to ionizing radiation, the cells stabilize p53 protein, as expected of cells with a normal DNA-damage response (29) (Fig. 5B Top, lanes 1–3). Moreover, p53 became phosphorylated on Ser-20 6 h after irradiation (Fig. 5B Middle, lane 3), as expected (30). In the ZFP TF-expressing cells, the Ser-20 phosphorylation signal was abolished (Fig. 5B Middle, compare lanes 3 and 6 or lanes 3 and 9). This result indicates that human CHK2 is necessary for the DNA-damage-dependent phosphorylation of p53 at Ser-20. Ablation of p53 Ser-20 phosphorylation by CHK2 repression did not prevent damage-induced stabilization of p53 protein (Fig. 5B Top, compare lanes 3 and 6 or lanes 3 and 9). ATM phosphorylates p53 at Ser-15 in response to DNA damage (36), and this phosphorylation partially blocks the interaction between p53 and MDM2, which promotes p53 degradation (37). Thus, ATM-dependent phosphorylation may stabilize p53 in the absence of CHK2, a result observed in Chk2–/– mouse cells (31). Whatever the case, our results indicate that, in human fibroblasts, absence of CHK2 specifically abolishes phosphorylation of p53 at Ser-20, but not p53 stabilization (Fig. 5). Taken together, these data show that ZFP TFs can functionally repress target genes in untransformed human cells.
Discussion
We show here that designed ZFP TFs can knock down the mRNA expression of a predetermined target gene, while providing singlegene specificity when ≈16,000 human genes were analyzed. Moreover, the extent of repression achieved by this highly specific engineered TF was sufficient to abolish CHK2 function in two different assays and cell types. This degree of repression is all of the more impressive given that the target gene, CHK2, encodes an enzyme (protein kinase), for which the activity of even minimal residual protein might be expected to functionally compensate for incomplete repression. Indeed, recent data using RNA interference or small interfering RNA targeted to CHK2 in human cells reduced CHK2 protein by only ≈60–75% (38). ZFP TFs are thus shown to be a potent and highly specific alternative to small interfering RNA-based approaches.
We show that the previously described in vitro improvements in ZFP TF architecture (18) (i.e., a series of three two-finger units) can lead to a remarkable biochemical specificity in vivo, even when challenged with the complexity of a 3-billion-bp genome. This compares favorably with recent specificity and genomewide array data using small interfering RNA (13).
The potential therapeutic utility of ZFP TFs stems in part from the exquisite specificity of the ZFP DNA-binding domain, an example of which is shown here, and is supported by a preliminary report documenting in vivo plant studies (39). This specificity, when combined with the potent yet reversible effects of the functional domain, will be key to the success of these reagents in the clinic. As shown in this work, a single ZFP TF can be initially validated by using transient transfection assays, and the same reagent can then be carried forward to more stringent tests of efficacy by using stable, inducible cell lines and untransformed human cells. Indeed, we have shown elsewhere that ZFP TFs can be used successfully in animals (40) before clinical studies. Finally, our data show that ZFP TFs can be constitutively expressed, thus providing stable, long-term target gene regulation. Taken together, these data demonstrate that ZFP TFs can be used across species from cell lines through animal model settings for advanced experimental validation of therapeutic utility.
As our knowledge and understanding of the key genetic determinants of disease improves, singularly specific reagents, such as ZFP TFs, that can activate or repress the expression of any gene are poised to emerge as direct therapeutic interventions with potential application to the entire spectrum of disease-related gene targets.
Acknowledgments
We thank Drs. Fyodor Urnov, Elizabeth Wolffe, and Sean Brennan for careful reading of the manuscript, and Angela Smith for technical assistance. We also thank Edward Lanphier for encouragement and support. This work was supported by National Institutes of Health Grants AG11658 (to J.C.) and AG00266 (to A.D.) and Small Business Innovation Research Grant 9 R44 CA94795-02 (to C.O.P.).
Abbreviations: ZFP, zinc-finger protein; TF, transcription factor; CHK2, checkpoint kinase 2; DOX, doxycycline.
References
- 1.Nebert, D. W. (2002) Toxicology 181–182, 131–141. [DOI] [PubMed] [Google Scholar]
- 2.Reid, G. K., Besterman, J. M. & MacLeod, A. R. (2002) Curr. Opin. Mol. Ther. 4, 130–137. [PubMed] [Google Scholar]
- 3.Urnov, F. D. & Rebar, E. J. (2002) Biochem. Pharmacol. 64, 919–923. [DOI] [PubMed] [Google Scholar]
- 4.Beerli, R. R., Dreier, B. & Barbas, C. F., III (2000) Proc. Natl. Acad. Sci. USA 97, 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, L., Spratt, S. K., Liu, Q., Johnstone, B., Qi, H., Raschke, E. E., Jamieson, A. C., Rebar, E. J., Wolffe, A. P. & Case, C. C. (2000) J. Biol. Chem. 275, 33850–33860. [DOI] [PubMed] [Google Scholar]
- 6.Snowden, A. W., Gregory, P. D., Case, C. C. & Pabo, C. O. (2002) Curr. Biol. 12, 2159–2166. [DOI] [PubMed] [Google Scholar]
- 7.Liu, P. Q., Rebar, E. J., Zhang, L., Liu, Q., Jamieson, A. C., Liang, Y., Qi, H., Li, P. X., Chen, B., Mendel, M. C., et al. (2001) J. Biol. Chem. 276, 11323–11334. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds, L., Ullman, C., Moore, M., Isalan, M., West, M. J., Clapham, P., Klug, A. & Choo, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartsevich, V. V. & Juliano, R. L. (2000) Mol. Pharmacol. 58, 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Ren, D., Collingwood, T. N., Rebar, E. J., Wolffe, A. P. & Camp, H. S. (2002) Genes Dev. 16, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamieson, A. C., Miller, J. C. & Pabo, C. O. (2003) Nat. Rev. Drug Discov. 2, 361–368. [DOI] [PubMed] [Google Scholar]
- 12.Doench, J. G., Petersen, C. P. & Sharp, P. A. (2003) Genes Dev. 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G. & Linsley, P. S. (2003) Nat. Biotechnol. 18, 18. [DOI] [PubMed] [Google Scholar]
- 14.Cho, Y. S., Kim, M. K., Cheadle, C., Neary, C., Becker, K. G. & Cho-Chung, Y. S. (2001) Proc. Natl. Acad. Sci. USA 98, 9819–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGowan, C. H. (2002) BioEssays 24, 502–511. [DOI] [PubMed] [Google Scholar]
- 16.Bartek, J., Falck, J. & Lukas, J. (2001) Nat. Rev. Mol. Cell Biol. 2, 877–886. [DOI] [PubMed] [Google Scholar]
- 17.Liang, Y., Li, X. Y., Rebar, E. J., Li, P., Zhou, Y., Chen, B., Wolffe, A. P. & Case, C. C. (2002) J. Biol. Chem. 277, 20087–20094. [DOI] [PubMed] [Google Scholar]
- 18.Moore, M., Klug, A. & Choo, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, M., Choo, Y. & Klug, A. (2001) Proc. Natl. Acad. Sci. USA 98, 1432–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isalan, M., Klug, A. & Choo, Y. (2001) Nat. Biotechnol. 19, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pear, W. S., Nolan, G. P., Scott, M. L. & Baltimore, D. (1993) Proc. Natl. Acad. Sci. USA 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S. H., Kaminker, P. & Campisi, J. (1999) Nat. Genet. 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubio, M. A., Kim, S. H. & Campisi, J. (2002) J. Biol. Chem. 277, 28609–28617. [DOI] [PubMed] [Google Scholar]
- 24.Greisman, H. A. & Pabo, C. O. (1997) Science 275, 657–661. [DOI] [PubMed] [Google Scholar]
- 25.Margolin, J. F., Friedman, J. R., Meyer, W. K., Vissing, H., Thiesen, H. J. & Rauscher, F. J., III (1994) Proc. Natl. Acad. Sci. USA 91, 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vissing, H., Meyer, W. K., Aagaard, L., Tommerup, N. & Thiesen, H. J. (1995) FEBS Lett. 369, 153–157. [DOI] [PubMed] [Google Scholar]
- 27.Ballard, D. W., Dixon, E. P., Peffer, N. J., Bogerd, H., Doerre, S., Stein, B. & Greene, W. C. (1992) Proc. Natl. Acad. Sci. USA 89, 1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shieh, S. Y., Ahn, J., Tamai, K., Taya, Y. & Prives, C. (2000) Genes Dev. 14, 289–300. [PMC free article] [PubMed] [Google Scholar]
- 29.Chehab, N. H., Malikzay, A., Appel, M. & Halazonetis, T. D. (2000) Genes Dev. 14, 278–288. [PMC free article] [PubMed] [Google Scholar]
- 30.Hirao, A., Kong, Y. Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S. J. & Mak, T. W. (2000) Science 287, 1824–1827. [DOI] [PubMed] [Google Scholar]
- 31.Takai, H., Naka, K., Okada, Y., Watanabe, M., Harada, N., Saito, S., Anderson, C. W., Appella, E., Nakanishi, M., Suzuki, H., et al. (2002) EMBO J. 21, 5195–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, Q., Rose, J. H., Zhang, H. & Pommier, Y. (2001) FEBS Lett. 505, 7–12. [DOI] [PubMed] [Google Scholar]
- 33.Matsuoka, S., Rotman, G., Ogawa, A., Shiloh, Y., Tamai, K. & Elledge, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 10389–10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melchionna, R., Chen, X. B., Blasina, A. & McGowan, C. H. (2000) Nat. Cell Biol. 2, 762–765. [DOI] [PubMed] [Google Scholar]
- 35.Ahn, J. Y., Schwarz, J. K., Piwnica-Worms, H. & Canman, C. E. (2000) Cancer Res. 60, 5934–5936. [PubMed] [Google Scholar]
- 36.Khanna, K. K., Keating, K. E., Kozlov, S., Scott, S., Gatei, M., Hobson, K., Taya, Y., Gabrielli, B., Chan, D., Lees-Miller, S. P. & Lavin, M. F. (1998) Nat. Genet. 20, 398–400. [DOI] [PubMed] [Google Scholar]
- 37.Shieh, S. Y., Ikeda, M., Taya, Y. & Prives, C. (1997) Cell 91, 325–334. [DOI] [PubMed] [Google Scholar]
- 38.Ahn, J., Urist, M. & Prives, C. (2003) J. Biol. Chem. 278, 20480–20489. [DOI] [PubMed] [Google Scholar]
- 39.Guan, X., Stege, J., Kim, M., Dahmani, Z., Fan, N., Heifetz, P., Barbas, C. F., III, & Briggs, S. P. (2002) Proc. Natl. Acad. Sci. USA 99, 13296–132301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebar, E. J., Huang, Y., Hickey, R., Nath, A. K., Meoli, D., Nath, S., Chen, B., Xu, L., Liang, Y., Jamieson, A. C., et al. (2002) Nat. Med. 8, 1427–1432. [DOI] [PubMed] [Google Scholar]