Abstract
The RING finger proteins HdmX and Hdm2 share significant structural and functional similarity. Hdm2 is a member of the RING finger family of ubiquitin-protein ligases E3 and targets the tumor suppressor protein p53 for degradation. Although HdmX also binds to p53, HdmX does not induce p53 degradation. Moreover, HdmX has been reported to interfere with p53 degradation in overexpression experiments. To obtain insight into the mechanism by which HdmX interferes with p53 degradation, we studied the effect of HdmX on the E3 activity of Hdm2 in vitro. Surprisingly, this revealed that HdmX stimulates Hdm2-mediated ubiquitination of p53 and that HdmX facilitates ubiquitination of Hdm2 and vice versa. In addition, down-regulation of HdmX expression within cells results in the accumulation of both p53 and Hdm2. Because HdmX alone does not have appreciable E3 activity, these data indicate that HdmX acts as a stimulator, rather than as an inhibitor, of the E3 activity of Hdm2 and that, at least under certain conditions, HdmX is actively involved in the degradation of both p53 and Hdm2.
The RING finger protein HdmX (MdmX in mouse) was originally isolated as a protein that interacts with the tumor suppressor p53 (1). In addition, it shows significant amino acid sequence similarity to the proto-oncoprotein Hdm2 (Mdm2 in mouse). The most conserved regions of HdmX and Hdm2 comprise an N-terminal p53-binding domain, a central acidic domain of yet unknown function, a zinc-binding motif, and a C-terminal RING finger motif (1–3). Genetic analyses in mice demonstrated that both Mdm2- and MdmX-deficient mice are viable only in a p53 null background (4, 5). Although Mdm2 and MdmX null mice die at different stages during embryonic development, this finding clearly indicates that both Mdm2 and MdmX are required to keep the growth-suppressive properties of p53 under control.
RING finger motifs are frequently found in proteins with the property of a ubiquitin-protein ligase E3 or in proteins that are part of an E3 complex (6–8). E3s are commonly assumed to be responsible for the selective recognition of substrate proteins of the ubiquitin-dependent proteolytic pathway. Functional analyses indicate that, in a simplified view, RING finger E3s have a modular structure consisting of at least two functional domains. One domain, which determines the substrate specificity of the respective E3, is required for specific interaction with the respective substrate protein. The other domain, the RING finger motif, is assumed to mediate the interaction with its cognate ubiquitin-conjugating enzyme E2, which in turn catalyzes the covalent attachment of ubiquitin to the substrate protein (6–8). Notably, these domains can be displayed either on a single polypeptide chain (single subunit E3s) or on several distinct proteins that are present within one complex (multisubunit E3s).
It is well established that the growth-suppressive properties of p53 are in part controlled by selective degradation via the ubiquitin-proteasome system and that ubiquitination with subsequent degradation of p53 is mainly facilitated by Hdm2 (3, 9–15). Because Hdm2 contains both a p53-binding domain and a RING finger domain, Hdm2 can be considered as a single subunit E3 (which does not exclude that Hdm2 functions as a dimer, rather than a monomer). In addition, Hdm2 seems to target itself for ubiquitination with subsequent degradation (12, 13). Although HdmX is structurally similar to Hdm2 and both interfere with p53 function, HdmX does not seem to function as an E3 for p53 (1–5). Moreover, overexpression experiments suggested that HdmX interferes with Hdm2-mediated degradation of p53 as well as with degradation of Hdm2 (2, 16–18).
HdmX and Hdm2 form heteromeric complexes via their C-terminal RING finger domains (2, 19). Because the RING finger domain of HdmX is sufficient to inhibit Hdm2-mediated degradation of p53 (2, 17, 18), a possible explanation for the inhibitory activity of HdmX was that binding of HdmX blocks the E3 activity of Hdm2. However, by using in vitro ubiquitination assays and small interfering RNA (siRNA)-mediated down-regulation of HdmX expression within cells, we report here that HdmX functions as a stimulator of Hdm2 E3 activity and that HdmX is required to keep p53 at low levels in cells under normal growth conditions.
Materials and Methods
Cell Lines, Plasmids, and Protein Expression. H1299 cells were grown in RPMI medium 1640, and MCF-7 and U2OS cells were grown in DMEM. Both media were supplemented with 10% (vol/vol) FBS.
Bacterial expression constructs for GST fusion proteins of full-length Hdm2, ΔN101-Hdm2, Hdm2-C464A, full-length HdmX, ΔN101-HdmX, and HdmX-C463A, respectively, were generated by PCR-based approaches (further details will be provided on request). For in vitro translation, cDNAs for Hdm2 and HdmX were cloned into the expression vector pcDNA3.1 (Invitrogen), respectively. For transient transfection experiments, HdmX was expressed with an N-terminal HA-tag from pcDNA3.1. The expression construct for Mdm2 (kindly provided by M. Oren, Weizmann Institute, Rehovot, Israel) has been described (9, 20). The expression constructs for wild-type p53 and ΔN43-p53 have been described (21). Dihydrofolate reductase (DHFR) was expressed as an N-terminally HA-tagged form from pcDNA3.1.
For in vitro ubiquitination experiments, the various forms of Hdm2 and HdmX were expressed as GTS fusion proteins in Escherichia coli DH5α. The ubiquitin-activating enzyme E1 and the ubiquitin-conjugating enzyme UbcH5 were expressed in the baculovirus system or in E. coli BL21 by using the pET expression system as described (22).
In Vitro Ubiquitination and Degradation Assays. For in vitro ubiquitination, 1 μl of rabbit reticulocyte lysate-translated 35S-labeled substrate (p53, Hdm2, or HdmX) was incubated in the absence or in the presence of increasing amounts of bacterially expressed Hdm2 (5 ng to 5 μg), 50 ng of E1, 50 ng of UbcH5, and 10 μg of ubiquitin (SIGMA) in the absence or in the presence of increasing amounts of bacterially expressed HdmX (5 ng to 5 μg) in 50-μl volumes. In addition, reactions contained 25 mM Tris·HCl (pH 7.5), 60 mM NaCl, 1 mM DTT, 2–4 mM ATP, and 4–8 mM MgCl2. After incubation at 30°C for 2 h, total reaction mixtures were electrophoresed in 10% SDS-polyacrylamide gels, and the 35S-labeled substrates were detected by fluorography. For degradation of polyubiquitinated Hdm2, 35S-labeled polyubiquitinated Hdm2 was generated in the presence of bacterially expressed HdmX. After 2 h, 5 μl of untreated rabbit reticulocyte lysate (Promega) were added as a source of the 26S proteasome, and the reaction was continued for 2 h at 37°C (23).
Mass Spectrometry. Five micrograms of bacterially expressed Hdm2 or bacterially expressed Hdm2 and HdmX were polyubiquitinated in the presence of E1, UbcH5, and 20 μg of ubiquitin. Total reaction mixtures were electrophoresed on 10% SDS-polyacrylamide gels, and proteins were stained with Coomassie blue. Bands representing polyubiquitinated Hdm2 or Hdm2/HdmX were excised. The samples were digested in gel with trypsin, and prepared for matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (further details will be provided on request). Spectra were acquired on a Bruker Reflex IV time of flight mass spectrometer (Bruker Daltonics) in positive mode.
Transfection, siRNAs, and Immunofluorescence. In transient expression experiments, cells were transfected with the respective constructs in the presence of a reporter construct encoding β-galactosidase by lipofection (DOTAP, Roche). Protein extracts were prepared 20 h after transfection as described (21), and transfection efficiency was determined by measuring β-galactosidase activity. Then, levels of Mdm2, HA-tagged Hdmx, and HA-tagged DHFR were determined by Western blot analysis by using transfection efficiency adjusted protein amounts. HA-tagged HdmX and HA-tagged DHFR were detected by the mouse monoclonal HA.11 (Hiss Diagnostics, Freiburg, Germany). Mdm2 was detected by the mouse monoclonal 4B2 (Santa Cruz Biotechnology).
siRNAs against mRNAs encoding Hdm2, HdmX, and the E6 oncoprotein of human papillomavirus type 16 were obtained from Dharmacon Research (Lafayette, CO). The respective target sequences were as follows: for Hdm2 5′ CAAGAGACCCUGGUUAGAC (si-Hdm2-1) and 5′ CCACCUCACAGAUUCCAGC (si-Hdm2-2); HdmX 5′ GAUUUUGCAUGCAGCA GGU (si-HdmX-1) and 5′ UCAAUCAGGUACGACCAAA (si-HdmX-2); and E6 5′UACAACAAACCGUUGUGUG. For immunofluorescence analysis, exponentially growing cells were transfected with 60–100 pmol of the respective siRNA per well of a 24-well plate by Oligofectamine according to the manufacturer's instructions (Invitrogen). Immunofluoresence analysis was performed 48–72 h after transfection. p53 was detected by the rabbit polyclonal FL393 (Santa Cruz Biotechnology). HdmX was detected by the goat polyclonal D-19 (Santa Cruz Biotechnology), and Hdm2 was detected by the mouse monoclonals SMP14 (Santa Cruz Biotechnology) or 4B2 (Calbiochem). For Western blot analysis, one 6-cm plate of exponentially growing cells was transfected with 0.6–1 nmol of the respective siRNA. The mouse monoclonal 1801 (Calbiochem) was used to detect p53. Hdm2 was detected by a combined immunoprecipitation/Western blot analysis as described (21). HdmX was detected by immunoprecipitation with a rabbit polyclonal antibody (kindly provided by A. G. Jochemsen, Leiden University, Leiden, The Netherlands) (24), followed by Western blot analysis using the goat polyclonal D-19. Enhanced chemiluminescence was performed according to the manufacturer's instructions (Amersham Pharmacia).
Results
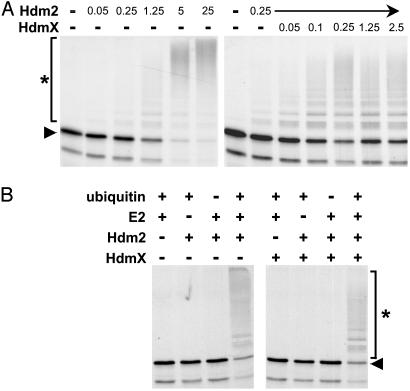
HdmX Stimulates Hdm2-Mediated Ubiquitination of p53. To obtain insight into the mechanism by which HdmX interferes with Hdm2-mediated degradation of p53, the effect of HdmX on Hdm2-mediated ubiquitination of p53 was tested in a recently established in vitro ubiquitination system (25). In this system, ubiquitination of in vitro translated p53 is dependent on the amount of Hdm2 used (Fig. 1A). Unexpectedly, addition of bacterially expressed recombinant HdmX stimulated the efficiency of Hdm2-mediated p53 ubiquitination at all Hdm2 concentrations tested (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.). The most significant effect of HdmX (≈10-fold stimulation) was observed at conditions where the amount of Hdm2 used was rate-limiting with respect to both the percentage of p53 ubiquitinated and the number of ubiquitin moieties attached to individual p53 molecules. Importantly, HdmX was not able to induce ubiquitination of p53 in the absence of Hdm2 (Fig. 1B), demonstrating that the effect of HdmX depends on the additional presence of Hdm2.
Fig. 1.
HdmX stimulates Hdm2-mediated ubiquitination of p53. (A) Increasing amounts of bacterially expressed GST fusion proteins of Hdm2 and HdmX, respectively, were incubated with in vitro translated radiolabeled p53 under standard ubiquitination conditions for 2 h. The reaction products were analyzed by SDS/PAGE followed by fluorography. Amounts of Hdm2 and HdmX used are indicated in 10–1 μg. (B) Determination of the requirements for ubiquitination of in vitro translated p53 by bacterially expressed Hdm2 (250 ng) and by Hdm2/HdmX (25 ng each), respectively. E2, UbcH5. The running positions of the nonmodified form and of the ubiquitinated forms of p53 are indicated by an arrowhead and an asterisk, respectively.
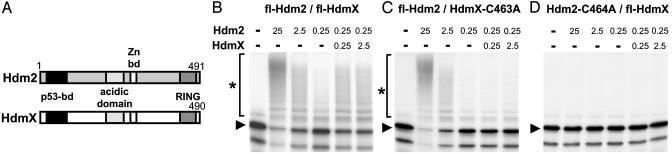
Hdm2 and HdmX have been reported to form heteromeric complexes via their RING finger domains (2, 19). Mutational analysis of Hdm2 and HdmX showed that the presence of an intact RING finger motif on both proteins is required for the stimulating effect of HdmX (Fig. 2 B–D). The N-terminal p53-binding domain of HdmX was dispensable for the stimulating effect (Fig. 7C, which is published as supporting information on the PNAS web site), indicating that HdmX stimulates p53 ubiquitination via heterocomplex formation with Hdm2. It should be noted, however, that ubiquitination of p53 in vitro was also observed by an Hdm2 mutant, in which the N-terminal p53-binding domain was deleted (ΔN101-Hdm2), in the presence and in the absence of HdmX (Fig. 7D). This result indicates that, in vitro, Hdm2 and possibly HdmX can interact with p53 independent of the known p53 binding domain. Nonetheless, the reaction was specific insofar as a p53 mutant, in which the N-terminal 43 aa containing the binding site for Hdm2 and HdmX were deleted and which is not degraded by Hdm2 in vivo, did also not serve as a substrate for Hdm2 or the Hdm2/HdmX complex in vitro (Fig. 7E).
Fig. 2.
An intact RING finger motif of Hdm2 and HdmX is required for HdmX to stimulate p53 ubiquitination. (A) Schematic representation of the structure of Hdm2 and HdmX. bd, binding domain. (B–D) In vitro ubiquitination of p53 in the presence of different amounts of the various Hdm2 and HdmX proteins as indicated. Amounts of Hdm2 and HdmX used are indicated in 10–1 μg. fl, full-length protein. C463A and C464A, the respective cysteine residues of HdmX and Hdm2, were replaced by alanine. The running positions of the nonmodified form of p53 and of the ubiquitinated forms of p53 are indicated by an arrowhead and an asterisk, respectively.
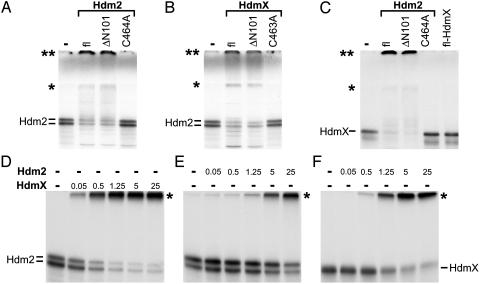
HdmX Stimulates Hdm2 Ubiquitination and Vice Versa. To test whether HdmX affects autoubiquitination of Hdm2 (12, 13), in vitro-translated radiolabeled Hdm2 was incubated in the presence of recombinant ubiquitin-activating enzyme E1 and recombinant UbcH5, an ubiquitin-conjugating enzyme E2 known to support Hdm2-mediated ubiquitination of p53 in vitro (see Fig. 1). Under the conditions used, autoubiquitination of in vitro-translated Hdm2 was observed only in the additional presence of bacterially expressed Hdm2 (Figs. 3 A and E). This result indicates that the concentration of Hdm2 needs to reach a threshold level for autoubiquitination to occur, suggesting that autoubiquitination of Hdm2 proceeds via homo(di)mer formation. Similar to the addition of recombinant Hdm2, addition of bacterially expressed HdmX also induced ubiquitination of Hdm2 (Figs. 3 B and D). Moreover, the amount of recombinant HdmX required to induce Hdm2 ubiquitination was significantly lower than the amount of recombinant Hdm2 (compare Fig. 3 D with E), which is consistent with previously published data indicating that HdmX has a higher affinity for Hdm2 than Hdm2 for itself (19). In addition, HdmX was efficiently ubiquitinated by Hdm2 (Fig. 3 C and F). Because HdmX was not able to induce its own ubiquitination (Fig. 3C; see also Fig. 8, which is published as supporting information on the PNAS web site), this result further indicates that, under the conditions used, HdmX does not function as an E3 on its own but is only active in complex with Hdm2.
Fig. 3.
HdmX stimulates ubiquitination of Hdm2 and vice versa. In vitro-translated, radiolabeled Hdm2 (A and B) or HdmX (C) were incubated with various forms of bacterially expressed GST fusion proteins of Hdm2 (1 μg) and HdmX (1 μg) under standard in vitro ubiquitination conditions as indicated. fl, full-length. ΔN101 represents an Hdm2 or HdmX mutant, of which the N-terminal 101 aa of Hdm2 or HdmX were deleted; C464A and C463A, the respective cysteine residues of HdmX and Hdm2 were replaced by alanine. Note that, in this particular experiment, the ubiquitinated forms of both Hdm2 (A and B) and HdmX (C) run at two positions indicated by an asterisk and by a double asterisk. The position indicated by a single asterisk coincides with the top of the separating gel, and the position of the double asterisk coincides with the top of the stacking gel. The reason for the different migration behavior of the ubiquitinated forms (top of separating gel vs. top of stacking gel) is unknown, but note that analysis of the different forms by mass spectrometry revealed no differences with respect to the lysine residues of ubiquitin used for chain formation (see also legend to Fig. 4). (D–F) Titration analysis of HdmX-mediated ubiquitination of Hdm2 (D) and of Hdm2-mediated ubiquitination of Hdm2 (E) and HdmX (F). Amounts of Hdm2 and HdmX used are indicated in 10–1 μg. The running positions of ubiquitinated forms of Hdm2 and HdmX are indicated by an asterisk.
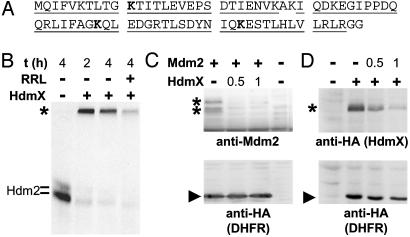
HdmX Stimulates Degradation of Hdm2. In many cases, modification of proteins by polyubiquitination targets these proteins for proteasome-mediated degradation. In recent years, however, it has become clear that ubiquitination of proteins also serves nonproteolytic functions, analogous to other protein modifications including phosphorylation (6–8, 26). Notably, the eventual fate of a given ubiquitinated protein seems to be dependent on the identity of the lysine residue of ubiquitin used for polyubiquitin chain formation. Ubiquitin chains linked via lysine residue 48 of ubiquitin target modified proteins to the proteasome (27). Similarly, lysine 29-linked chains have also been associated with proteasome-mediated degradation, whereas lysine 63-linked chains seem to change the biochemical properties of a modified protein rather than to target it to the proteasome (6–8, 26, 28, 29). A suitable, and nonbiased, approach to determine lysine residue(s) of ubiquitin used for chain formation is provided by mass spectrometry analysis. In vitro ubiquitination of Hdm2 and HdmX is an efficient process insofar that amounts of polyubiquitinated Hdm2 and/or HdmX sufficient for mass spectrometry analysis can be readily obtained (in the μg-mg range; data not shown). Mass spectrometry analysis of polyubiquitin chains formed by Hdm2 in the absence and in the presence of HdmX revealed that, in both cases, three lysine residues are used for polyubiquitin chain formation, namely lysine 48, lysine 63, and lysine 11 (Fig. 4A; Table 1, which is published as supporting information on the PNAS web site), suggesting that the presence of HdmX does not change the identity of ubiquitin chains formed in the presence of Hdm2. These results are supported by a recent study using ubiquitin mutants (30). A similar analysis for p53 ubiquitinated in the presence of Hdm2 or of Hdm2/HdmX was not possible because this reaction is rather inefficient (i.e., the amount of ubiquitinated p53 obtained is not sufficient to allow analysis by mass spectrometry).
Fig. 4.
HdmX stimulates Hdm2/Mdm2 degradation. (A) Amino acid sequence of ubiquitin. The amino acid residues identified by mass spectrometry in three independent experiments are underlined. The lysine residues used for chain formation are indicated in bold. Note that mass spectrometric analysis of samples taken from the top of the separating gel and from the top of the stacking gel (see Fig. 3) yielded identical results. (B) In vitro-translated radio-labeled Hdm2 was incubated with bacterially expressed HdmX (250 ng) at standard ubiquitination conditions. After 2 h, additional rabbit reticulocyte lysate (RRL) was added as a source of the 26S proteasome, where indicated, and the reactions were incubated for an additional 2 h. Note that the disappearance of the polyubiquitinated forms of Hdm2 depended on the presence of ATP, indicating that their disappearance was not due to the action of ubiquitin-specific isopeptidases that may be present in the reticulocyte lysate used (data not shown). The running position of ubiquitinated Hdm2 is indicated with an asterisk. (C and D) Expression constructs encoding HA-tagged HdmX, Mdm2, and HA-tagged DHFR were cotransfected into H1299 cells. Expression levels of the respective proteins were determined by Western blot analysis with the antibodies indicated. The running positions of HdmX and Mdm2, respectively, are indicated with an asterisk, and the running position of DHFR is indicated with an arrowhead. +, 0.5 μg of the respective expression plasmid was cotransfected with the indicated amount of the other construct.
Although it is not possible for technical reasons to determine whether Hdm2 can form mixed polyubiquitin chains (i.e., lysine 48 and lysine 63 are used for linkage within one chain) or whether only one lysine residue is used within one particular chain, the mass spectrometry data indicate that HdmX-facilitated polyubiquitination labels Hdm2 at least in part for proteasome-mediated degradation. To test this hypothesis directly, an in vitro degradation assay was performed (23). In vitro-translated radiolabeled Hdm2 was ubiquitinated in the presence of HdmX. After 2 h, rabbit reticulocyte lysate was added as a source of the 26S proteasome, and the reaction was incubated for an additional 2 h. This procedure resulted in the disappearance of ≈60–70% of the polyubiquitinated forms of Hdm2, strongly indicating that HdmX targets Hdm2 for proteasome-mediated degradation (Fig. 4B). A similar assay was performed for p53 (data not shown). However, degradation of p53 was not observed, regardless of whether p53 was ubiquitinated by Hdm2 or by Hdm2/HdmX. This result is consistent with recently published data indicating that, in vitro, p53 is modified by the attachment of single ubiquitin moieties to several lysine residues of p53 (monoubiquitination) rather than by the attachment of polyubiquitin chains (31). Alternatively, factor(s) might be missing in the in vitro system that are required for p53 degradation but not for Hdm2 degradation.
To obtain evidence that HdmX can also induce degradation of Hdm2 within cells, cotransfection assays were performed. These assays showed that, under the conditions used, coexpression of HdmX results in significantly decreased Mdm2 levels (Fig. 4C) (note that, in these experiments, an expression construct encoding mouse Mdm2 was used for detection reasons). Similarly, coexpression of Mdm2 resulted in significantly decreased levels of HdmX (Fig. 4D). Because in the same experiment HdmX or Mdm2 had no, or only a minor, effect on the expression levels of cotransfected DHFR, these data indicate that HdmX can induce degradation of Mdm2 and vice versa. The effect of HdmX on p53 stability was also studied in cotransfection experiments in the presence or absence of coexpressed Mdm2. At conditions where p53 was efficiently degraded by Mdm2, coexpression of HdmX had either no effect on p53 levels or resulted in an increase of p53 levels (data not shown), in agreement with previously published results (16–18). Under rate-limiting conditions (i.e., Mdm2 levels are too low to induce p53 degradation), HdmX had either no effect on p53 levels or, in rare cases (one of six to eight experiments), HdmX coexpression resulted in decreased p53 levels (data not shown). However, due to the low reproducibility, it is not possible to conclude whether, under conditions of overexpression, HdmX can cooperate with Hdm2 in p53 degradation.
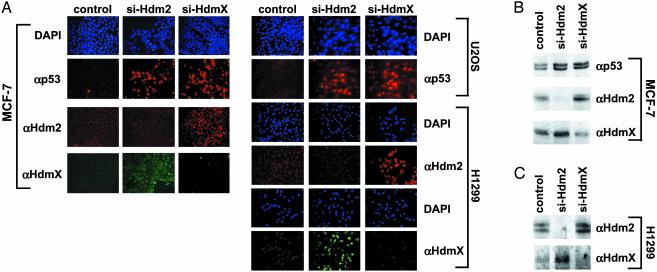
siRNA-Mediated Knockdown of Endogenous HdmX Expression Elevates p53 and Hdm2 Levels. Whereas results obtained in overexpression studies can serve as first indication whether a protein is involved in the degradation of another protein, they may be misleading. For example, overexpression of HdmX may result in a cellular localization of Hdm2 that is inappropriate for p53 degradation or may titrate out other factors involved in degradation of p53 and/or Hdm2 resulting in their unscheduled degradation or stabilization. Therefore, to obtain evidence that endogenous HdmX is involved in degradation of p53 and/or Hdm2, two different siRNAs (32) directed against mRNAs encoding HdmX or Hdm2, respectively, were introduced into cell lines expressing either wild-type p53 (MCF-7, U2OS) or no p53 (H1299) (Fig. 5; see also Fig. 9, which is published as supporting information on the PNAS web site). Immunofluoresence and Western blot analysis showed that transfection of siRNAs directed against Hdm2 mRNA results in a decrease in Hdm2 levels whereas p53 levels increased as expected. (Densitometric analysis of the p53 Western blot shows that p53 levels are increased by 2- to 3-fold in the presence of siRNAs against Hdm2 because, at any given time of analysis, only 40–60% of cells showed p53 staining by immunofluorescence; the actual increase in p53 levels is 4- to 5-fold.) In addition, knockdown of Hdm2 expression results in a similar increase of HdmX levels, indicating that endogenous Hdm2 is involved not only in p53 degradation but also in HdmX degradation. Similarly, knockdown of HdmX expression by siRNA resulted in an increase of both p53 and Hdm2 (by a factor of 4–5; see above) indicating that, in the absence of HdmX, Hdm2 and p53 have a decreased turnover rate. Thus, at normal growth conditions, endogenous HdmX seems to be intrinsically involved in both p53 and Hdm2 degradation.
Fig. 5.
Knockdown of HdmX expression results in increased p53 and Hdm2 levels. siRNAs specific for HdmX and Hdm2 were transfected into the cell lines indicated. As a control, siRNA against the E6 oncoprotein of human papillomavirus type 16, which is not expressed in the cell lines used, was used. (A) The levels of p53, Hdm2, and HdmX were determined 3 days after transfection by immunofluorescence. (B and C) The levels of p53, Hdm2, and HdmX were determined 2 days after transfection by Western blot analysis. MCF-7 cells and U2OS cells express endogenous wild-type p53, whereas p53 is not expressed in H1299 cells. Note that similar results were obtained with two different siRNAs against HdmX (si-HdmX-1, si-HdmX-2; see Materials and Methods) and Hdm2 (si-Hdm2-1, si-Hdm2-2).
Discussion
The data presented in this study indicate that HdmX acts as a positive effector of the E3 activity of Hdm2 in vitro and, at least under certain conditions, plays a positive role in the degradation of p53 and Hdm2 within cells. Furthermore, the data suggest that, in the absence of HdmX, increased levels of Hdm2 are required for efficient degradation of both Hdm2 and p53. Such a situation is achieved under both experimental (i.e., transient transfection experiments) and physiologically relevant [e.g., up-regulation of Hdm2 levels in a p53-dependent manner on DNA damage (3, 14, 15)] conditions.
An intact RING finger domain is required for HdmX to stimulate Hdm2-mediated ubiquitination of p53 and ubiquitination of Hdm2. Because HdmX and Hdm2 can bind to each other via their RING finger domains (2, 19), this finding indicates that complex formation with Hdm2 is required for the stimulating effect of HdmX. This hypothesis is further supported by the notion that, in vitro, a region comprising the RING finger motif of HdmX (amino acid residues 418–490) is sufficient to stimulate Hdm2 activity (data not shown). Why should an HdmX/ Hdm2 complex be more active as an E3 than Hdm2 alone? It is assumed that the RING finger domain of an E3 mediates the interaction with its cognate E2, thereby bringing E2s into close proximity to substrate proteins for ubiquitin conjugation (6–8). An intriguing possibility is that the interaction of the Hdm2/HdmX complex with UbcH5 is more efficient than that of UbcH5 with Hdm2 alone. However, stable interaction between Hdm2 and UbcH5 has not been reported and has also not been observed for the Hdm2/HdmX complex in vitro (unpublished observation). An alternative but not mutually exclusive possibility is that Hdm2 is not active as a monomer but as a dimer or oligomer (as suggested by the observation that Hdm2 has to reach a certain threshold level for autoubiquitination). Data obtained in a yeast two-hybrid system indicate that Hdm2/HdmX heterodimers are more stable than the respective homodimers (19). Thus, lower levels of Hdm2 would be required for E3 activity in the presence of HdmX than in its absence. In this context, it should be noted that it was recently reported that HdmX alone has E3 activity and is capable of ubiquitinating p53, although with very low efficiency (30). In addition, when compared with Hdm2, very high levels of HdmX had to be used to detect p53 ubiquitination. Nonetheless, the inability of HdmX to efficiently function as an E3 in the absence of Hdm2 may be explained by the notion that the RING finger of HdmX alone does only poorly interact with UbcH5.
In contrast to our data (Fig. 4C), HdmX has been reported to stabilize coexpressed Hdm2 in transient transfection experiments (2, 17, 18). Although the actual reason for this difference is not known, it may be explained by differences in the expression levels of the individual proteins achieved in the different experimental systems. Our in vitro data indicate that, at high levels of Hdm2, HdmX has no significant influence on the ubiquitination efficiency of Hdm2 whereas, at low levels, HdmX stimulates Hdm2 ubiquitination. Thus, it seems conceivable that, in vivo, overexpression of HdmX may result in a decrease in the turnover rate of Hdm2 when Hdm2 is highly expressed (e.g., other components of the ubiquitin-conjugation system may be rate-limiting under such conditions). This scenario is similar to what has recently been published for the effect of HdmX on p53 stability in cotransfection assays (33). Regardless of the reason(s) accounting for the different results obtained in overexpression studies, the results obtained by siRNA analysis clearly indicate that endogenous HdmX plays a positive role in the degradation of both p53 and Hdm2, most likely by forming heterodimers with Hdm2.
The notion that the HdmX/Hdm2 complex has higher E3 activity than Hdm2 alone is reminiscent of the interaction of the two RING finger proteins BRCA1 and BARD1 (34–38). Similar to HdmX, BARD1 is a RING finger protein with no appreciable E3 activity. However, it stimulates the E3 activity of BRCA1 via heterodimerization. Thus, activation by specific interaction with other RING finger proteins may be a common mechanism to regulate the activity of RING finger E3 proteins. Another similarity between these two E3 complexes is that both can use lysine 48 and lysine 63 of ubiquitin for polyubiquitin chain formation in vitro (30, 38). In contrast to lysine 48-linked chains, modification of proteins with lysine 63-linked polyubiquitin chains does not seem to target these proteins for degradation but somehow changes their biochemical properties (25, 28, 29). Therefore, although the lysine residues used for ubiquitin chain formation in vivo are not known, it is intriguing to speculate that Hdm2 and Hdm2/HdmX-mediated ubiquitination serves proteolytic as well as not yet characterized nonproteolytic functions.
Supplementary Material
Acknowledgments
We thank N. J. Whitaker, A. G. Jochemsen, and S. Glockzin for comments on the manuscript. The work was supported by grants from the German-Israeli Foundation for Scientific Research and Development (to A.C. and M.S.), the European Community (QLG1-CT-2001-02026 to M.S.), Köln Fortune (to M.S.), and the Fonds der Chemischen Industrie (to M.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DHFR, dihydrofolate reductase; siRNA, small interfering RNA.
References
- 1.Shvarts, A., Steegenga, W. T., Riteco, N., van Laar, T., Dekker, P., Bazuine, M., van Ham, R. C., van der Houven van Oordt, W., Hateboer, G., van der Eb, A. J. & Jochemsen, A. G. (1996) EMBO J. 15, 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp, D. A., Kratowicz, S. A., Sank, M. J. & George, D. L. (1999) J. Biol. Chem. 274, 38189–38196. [DOI] [PubMed] [Google Scholar]
- 3.Michael, D. & Oren, M. (2002) Curr. Opin. Genet. Dev. 12, 53–59. [DOI] [PubMed] [Google Scholar]
- 4.Montes de Oca Luna, R., Wagner, D. S. & Lozano, G. (1995) Nature 378, 203–206. [DOI] [PubMed] [Google Scholar]
- 5.Parant, J., Chavez-Reyes, A., Little, N. A., Yan, W., Reinke, V., Jochemsen, A. G. & Lozano, G. (2001) Nat. Genet. 29, 92–95. [DOI] [PubMed] [Google Scholar]
- 6.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- 7.Weissman, A. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- 8.Glickman, M. H. & Ciechanover, A. (2002) Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- 9.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. (1997) Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 11.Honda, R., Tanaka, H. & Yasuda, H. (1997) FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- 12.Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H. & Weissman, A. M. (2000) J. Biol. Chem. 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- 13.Honda, R. & Yasuda, H. (2000) Oncogene 19, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 14.Vousden, K. H. & Lu, X. (2002) Nat. Rev. Cancer 2, 594–604. [DOI] [PubMed] [Google Scholar]
- 15.Alarcon-Vargas, D. & Ronai, Z. (2002) Carcinogenesis 23, 541–547. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, M. W. & Berberich, S. J. (2000) Mol. Cell. Biol. 20, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stad, R., Ramos, Y. F., Little, N., Grivell, S., Attema, J., van Der Eb, A. J. & Jochemsen, A. G. (2000) J. Biol. Chem. 275, 28039–28044. [DOI] [PubMed] [Google Scholar]
- 18.Stad, R., Little, N. A., Xirodimas, D. P., Frenk, R., van der Eb, A. J., Lane, D. P., Saville, M. K. & Jochemsen, A. G. (2001) EMBO Rep. 2, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanimura, S., Ohtsuka, S., Mitsui, K., Shirouzu, K., Yoshimura, A. & Ohtsubo, M. (1999) FEBS Lett. 447, 5–9. [DOI] [PubMed] [Google Scholar]
- 20.Haupt, Y., Barak, Y. & Oren, M. (1996) EMBO J. 15, 1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 21.Hengstermann, A., Whitaker, N. J., Zimmer, D., Zentgraf, H. & Scheffner, M. (1998) Oncogene 17, 2933–2941. [DOI] [PubMed] [Google Scholar]
- 22.Nuber, U., Schwarz, S., Kaiser, P., Schneider, R. & Scheffner, M. (1996) J. Biol. Chem. 271, 2795–2800. [DOI] [PubMed] [Google Scholar]
- 23.Glockzin, S., von Knethen, A., Scheffner, M. & Brune, B. (1999) J. Biol. Chem. 274, 19581–19586. [DOI] [PubMed] [Google Scholar]
- 24.Ramos, Y. F., Stad, R., Attema, J., Peltenburg, L. T., van der Eb, A. J. & Jochemsen, A. G. (2001) Cancer Res. 61, 1839–1842. [PubMed] [Google Scholar]
- 25.Hengstermann, A., Linares, L. K., Ciechanover, A., Whitaker, N. J. & Scheffner, M. (2001) Proc. Natl. Acad. Sci. USA 98, 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickart, C. M. (2000) Trends Biochem. Sci. 25, 544–548. [DOI] [PubMed] [Google Scholar]
- 27.Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. (2000) EMBO J. 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spence, J., Gali, R. R., Dittmar, G., Sherman, F., Karin, M. & Finley, D. (2000) Cell 102, 67–76. [DOI] [PubMed] [Google Scholar]
- 29.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J. & Chen, Z. J. (2001) Nature 412, 346–351. [DOI] [PubMed] [Google Scholar]
- 30.Badciong, J. C. & Haas, A. L. (2002) J. Biol. Chem. 277, 49668–49675. [DOI] [PubMed] [Google Scholar]
- 31.Lai, Z., Ferry, K. V., Diamond, M. A., Wee, K. E., Kim, Y. B., Ma, J., Yang, T., Benfield, P. A., Copeland, R. A. & Auger, K. R. (2001) J. Biol. Chem. 276, 31357–31367. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 33.Gu, J., Kawai, H., Nie, L., Kitao, H., Wiederschain, D., Jochemsen, A. G., Parant, J., Lozano, G. & Yuan, Z. M. (2002) J. Biol. Chem. 277, 19251–19254. [DOI] [PubMed] [Google Scholar]
- 34.Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. (2001) Nat. Struct. Biol. 8, 833–837. [DOI] [PubMed] [Google Scholar]
- 35.Hashizume, R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., Ogata, H. & Ohta, T. (2001) J. Biol. Chem. 276, 14537–14540. [DOI] [PubMed] [Google Scholar]
- 36.Baer, R. & Ludwig, T. (2002) Curr. Opin. Genet. Dev. 12, 86–91. [DOI] [PubMed] [Google Scholar]
- 37.Chen, A., Kleiman, F. E., Manley, J. L., Ouchi, T. & Pan, Z. Q. (2002) J. Biol. Chem. 277, 22085–22092. [DOI] [PubMed] [Google Scholar]
- 38.Xia, Y., Pao, G., Chen, H. W., Verma, I. M. & Hunter, T. (2002) J. Biol. Chem. 278, 5255–5263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.