Abstract
Little is known about the large ectodomain of MET, the product of the c-met protooncogene and receptor for hepatocyte growth factor/scatter factor (HGF/SF). Here, we establish by deletion mutagenesis that the HGF/SF and heparin-binding sites of MET are contained within a large N-terminal domain spanning the α-chain (amino acids 25–307) and the first 212 amino acids of the β-chain (amino acids 308–519). Neither the cystine-rich domain (amino acids 520–561) nor the C-terminal half of MET (amino acids 562–932) bind HGF/SF or heparin directly. The MET ectodomain, which behaves as a rod-shaped monomer with a large Stokes radius in solution, binds HGF/SF in the absence or presence of heparin, and forms a stable HGF/SF–heparin–MET complex with 1:1:1 stoichiometry. We also show that the ligand-binding domain adopts a β-propeller fold, which is similar to the N-terminal domain of αV integrin, and that the C-terminal half contains four Ig domains (amino acids 563–654, 657–738, 742–836, and 839–924) of the unusual structural E set, which could be modeled on bacterial enzymes. Our studies provide 3D models and a functional map of the MET ectodomain. They have broad implications for structure-function of the MET receptor and the related semaphorin and plexin proteins.
Keywords: Ig domain, sema domain, integrin α-chain, hidden Markov models, semaphorins
Receptor tyrosine kinases (RTKs) mediate intercellular signals, which are essential for the development and maintenance of the cells of multicellular organisms. The minimal domain structure of RTKs consists of an extracellular ligand-binding domain, a single transmembrane helix, and a cytoplasmic kinase domain. This minimal structure, however, is very rare, and, typically, the extracellular moiety of RTKs, the ectodomain, consists of complex and distinctive domain sets, which enable classification of the RTKs in different families (1).
There is a strong preference for certain domains to occur in the ectodomain of RTKs. The fibronectin type-3 (FN-3) domain, for example, is present as two copies in the large Eph receptor family, three copies in the insulin and IGF-1 receptors, and at least seven copies in the rod outer segment receptor (1). Cystine-rich domains of variable length are also commonly found in RTKs.
A large number of RTKs contain Ig domains and the ectodomain of certain families consists solely of Ig domains: the fibroblast growth factor (FGF) receptors contain two or three, depending on RNA splicing, the platelet-derived growth factor (PDGF), colony-stimulating factor 1 (CSF1), KIT, and FLT kinase/serine-threonine kinase 1 (FLK2/STK1) receptors contain five, and the FMS-like (FLT1), FLK1, FLT4, and cholecystokinin 4 (CCK4) receptors contain seven (1). Ig domains can also be present in combination with FN-3, cystine-rich, or other domains (1). Interestingly, most Ig domains present in RTKs and cell adhesion molecules belong to a distinct structural set known as the I set, with architecture intermediate between the V and C1 sets (2).
MET, the RTK encoded by the c-met protooncogene (3, 4), is the receptor for hepatocyte growth factor/scatter factor (HGF/SF) (5), which is a large polypeptide growth factor discovered as a protein causing dispersion of epithelial colonies and cell migration (SF) (6, 7), and as a liver mitogen (HGF) (8–10). HGF/SF and MET are essential for the development of several tissues and organs, including the placenta (11, 12), liver (11), and several groups of skeletal muscle (13). They also play a major role in the abnormal migration of cancer cells as a result of overexpression or MET mutations (14).
In contrast to extensive data on the signal transduction pathways activated by MET (15), little is known about extracellular MET. Here we report a functional map of the MET ectodomain by deletion mutagenesis, sequence analysis, and comparative modeling.
Materials and Methods
Expression and Purification of MET Proteins. Two silent mutations were introduced in codons Q559 and I560 of a full-length human MET cDNA to remove a BglII site. MET deletions lacking the endogenous leader (amino acids 1–24) were generated by PCR as MluI–BglII inserts. For monomeric MET proteins, inserts were cloned in-frame between a 21-aa Ig leader and a hexahistidine sequence. For dimeric proteins, inserts were cloned between the same Ig leader and the hinge, CH2 and CH3 domains of the human γ1 antibody constant region gene, followed by a hexahistidine sequence. These constructs cause MET dimerization through the antibody (Fc) portion. For expression, MET constructs in plasmid pA71d were transfected in the mouse myeloma line NS0 or in Lec 8 cells (16). Stable transfectants were selected in 0.75 mg/ml hygromycin, screened for expression, and positive cultures were cloned and expanded for protein production. Monomeric MET proteins were purified on a nickel-nitrilotriacetic acid (Ni-NTA) agarose column (catalog no. MG3398; Qiagen, Valencia, CA) and eluted with 0.4 M imidazole, followed by further purification on a Mono S column (catalog no. 17–0547-01; Amersham Biosciences). Purification of dimeric MET proteins was carried out on Ni-NTA agarose, followed by chromatography on Protein A Sepharose CL-4B (catalog no. 17–0780-01; Amersham Biosciences).
Binding Assays. Immulon B 96-well plates were coated with recombinant single-chain (R494E) or two-chain HGF/SF in 50 mM phosphate buffer, pH 6.0. Wells were blocked and incubated with dimeric MET constructs at the concentrations shown in Fig. 2 a and b, and bound MET was detected with horseradish peroxidase-conjugated rabbit anti-human IgG (catalog no. P0214, DAKO). For heparin binding, dimeric MET constructs were loaded on a HiTrap heparin HP column (catalog no. 7–0406-01; Amersham Biosciences) in 50 mM phosphate/150 mM NaCl, pH 7.4, and eluted with a linear gradient of NaCl as shown in Fig. 2 c and d.
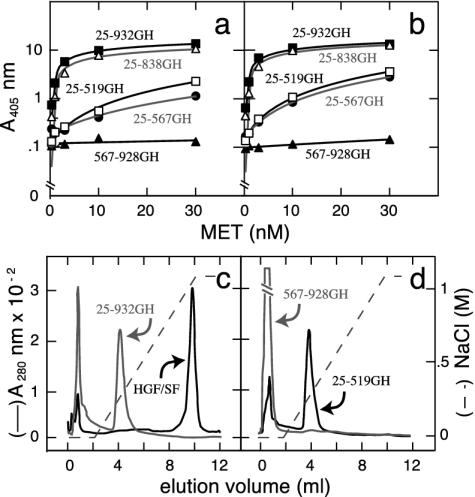
Fig. 2.
Binding of MET deletions to HGF/SF (a and b) or heparin (c and d). (a and b) Binding of MET deletions to single-chain (a) or two-chain (b) HGF/SF, as measured in a solid phase assay. (c and d) Binding of three MET constructs (25–519GH, 25–932GH, and 567–928GH) to immobilized heparin. Both full-length MET (25–932GH) and MET 25–519GH showed binding, whereas MET 567–928GH showed none. The strong heparin binding of mature (two-chain) HGF/SF is shown for comparison in c.
Solution Behavior of HGF/SF, MET, and HGF-SF–MET Complexes by Gel Electrophoresis and Analytical Sedimentation. Monomeric MET proteins were characterized by agarose gel electrophoresis in 10 g/liter agarose gels in 50 mM Mes, pH 6.7, for 4 h at 50 mA. Analytical sedimentation experiments were performed in a Beckman Optima XL-A ultracentrifuge with an An60 Ti rotor. Sedimentation velocity was at 20°C and various speeds (30–52,000 revolutions per min). Only a single cell was used and this was scanned repeatedly. Data were analyzed by the dc/dt method (17, 18) by using the program dc/dt+ (19) with partial specific volumes, and solvent density and viscosity were calculated from their compositions with the program sednterp (D. B. Hayes, T. Laue, and J. Philo from the rasmb software archive, www.bbri.org/RASMB/rasmb.html). Sets of 8–12 scans were analyzed to give plots of g(s*) against s*20,w, where g(s*) is the amount of material (in D280 units) sedimenting between s* and (s* + δs*), where δs* is set as small as the data allow, and s*20,w is the apparent sedimentation coefficient, corrected to water at 20°C. The scans were also fitted with models for either one or two components, calculating s*20,w and molecular mass (from s and a fitted diffusion coefficient, based on the broadening of the boundary). Plots of the residuals between the absorbance calculated from the model, with the fitted parameters against s*20,w were made to allow assessment of the adequacy of the model fit.
3D Models of the Sema and Ig Domains of MET. Fold assignment was carried out by using dynamic statistical profiles (hidden Markov models; HMMs) or two sequence-structure homology recognition and alignment procedures: 3dpssm (20) and fugue (21). For HMM analysis, both profile-sequence and profile-profile searches were carried out. For the latter HMM, profiles of query sequences were generated with the SAM-T99 iterative procedure (22), and searches were performed by using the symmetric profile comparison method (M. Madera and J.G., unpublished data). clustalw alignments (23) were also used to build a model directly. In all cases, the superfamily (24) library of HMMs was used as the subject profile database. For comparative modeling, alignments produced by fugue (21) were formatted with joy (25). Models were constructed with modeller (26), refined by using the sybyl (Tripos, St. Louis) force field, and validated by procheck (27), verify3d (28), and joy (25). The four Ig domains were joined by using the program mnyfit (29).
Results
MET Deletions. Membrane-bound MET is cleaved by furin between R307 and S308 (30) (amino acid numbering of MET is from the translation start site, and is based on Swiss-Prot accession no. P08581), yielding an extracellular α-chain (amino acids 25–307), and a longer β-chain (amino acids 308-1390), of which amino acids 308–932 are outside the membrane (refs. 4 and 31 and Fig. 1a, top line). The β-chain contains a short cystine-rich sequence (amino acids 520–561), which is indicated as a black box in Fig. 1a.
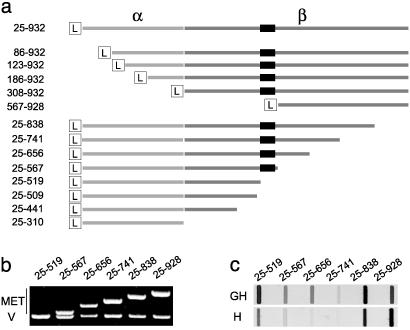
Fig. 1.
Deletion mapping and expression of MET domains. (a) A schematic view and sequence boundaries of N- and C-terminal deletions of the MET ectodomain. The α- and β-chains are shown in different shades of gray. The L indicates a 21-aa Ig leader used for secretion of MET proteins and the black box corresponds to the cystine-rich sequence (amino acids 520–561) of the MET β chain. (b) The cDNAs corresponding to several C-terminal deletions of the MET ectodomain (top bands, MET) are shown along with a vector band (V). (c) Expression of the same MET deletions in supernatants of stable transfectants of the mouse myeloma line NS0. H and GH indicate monomeric and dimeric MET constructs, respectively.
To map the ligand-binding domain, two sets of MET deletions were produced in the mouse myeloma line NS0, either as histidine-tagged (H) monomeric proteins, or as fusions to the Fc region of the human γ1 antibody heavy chain yielding dimeric histidine-tagged MET proteins (GH). Four shorter N-terminal deletions could not be expressed at measurable levels, but a larger one (amino acids 567–928) yielded stable MET proteins (Fig. 1a). A total of 16 C-terminal deletions were generated, several of which are shown in Fig. 1a (sequence boundaries), Fig. 1b (DNA inserts), and Fig. 1c (protein expression). The highest levels of expression were seen either with the amino acid 25–519 constructs or with the larger proteins (amino acids 25–932 and 25–838). Intermediate constructs (amino acids 25–567, 25–656, and 25–741) were expressed at low levels, especially as monomeric proteins (Fig. 1c). Differences in expression levels reflected the properties of the constructs and not bias in selection. Truncations of MET 25–519 (Fig. 1a) could not be expressed at detectable levels.
The First 519 Amino Acids of MET Are Sufficient for Binding HGF/SF and Heparin. Binding of dimeric MET deletions to single-chain (R494E) or two-chain HGF/SF is shown in Fig. 2 a and b. There were no significant differences in MET binding to the two ligands. The strongest binding was observed with the longest constructs, but binding of 25–519GH and 25–567GH was readily measurable. In contrast, construct 567–932GH showed no binding (Fig. 2 a and b). Thus, the N-terminal part of the MET ectodomain (amino acids 25–519) is sufficient for binding HGF/SF, whereas the C-terminal part (amino acids 567–932) has no binding activity, but increases binding to the N-terminal one (amino acids 25–519).
There is evidence that MET binds heparan sulfate proteoglycans (HSPGs) (32–34). Thus, three MET constructs were used to map the region of MET responsible. The full MET ectodomain (construct 25–932GH) bound immobilized heparin, albeit with lower apparent affinity than HGF/SF (Fig. 2c). Binding of 25–519GH was indistinguishable from full-length MET (25–932GH). In contrast, MET567–928GH exhibited no binding (Fig. 2d). Thus, both the HGF/SF- and the HSPG-binding sites are contained in amino acids 25–519 of MET.
HGF/SF–MET Complexes. The availability of soluble, monomeric forms of the MET receptor enabled studies of its solution properties and HGF/SF binding. Fig. 3a shows SDS gel electrophoresis under reducing conditions of MET 25–838H expressed in NS0 (lane 1) or Lec 8 cells (lane 2). The increased mobility of the β- and α-chain bands in MET from Lec 8 cells is due to reduced glycosylation by Lec 8 as a result of a mutation in the UDP-Gal transporter. Lane 3 is MET 25–928H from Lec 8 cells. Binding of full-length, monomeric MET to HGF/SF was studied by gel filtration (data not shown), native gel electrophoresis, and velocity sedimentation. Fig. 3b shows the electrophoretic mobility of HGF/SF, MET, and HGF/SF–MET complexes in the presence or absence of heparin. At pH 6.7, HGF/SF displayed anodic mobility, whereas MET exhibited no mobility. Incubation of HGF/SF or MET with heparin resulted in HGF/SF–heparin or MET–heparin complexes with increased negative charge. Incubation of HGF/SF and MET with or without heparin resulted in distinct HGF/SF–MET or HGF/SF–heparin–MET complexes (Fig. 3b).
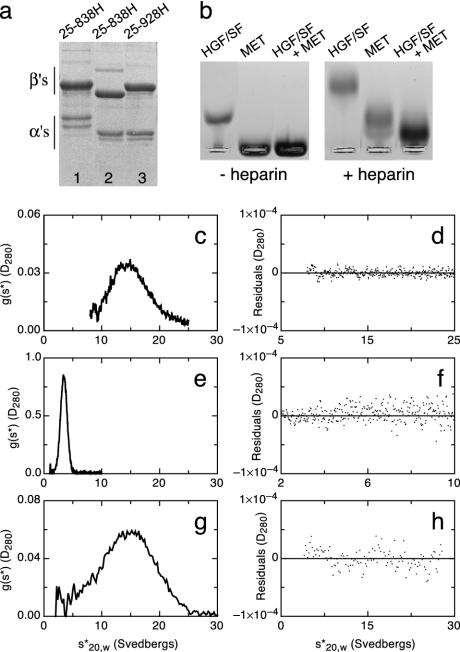
Fig. 3.
Monomeric full-length MET and HGF/SF-MET complexes. (a) SDS/PAGE under reducing conditions of MET 25–838H from NS0 (lane 1) or Lec 8 cells (lane 2) and MET 25–928H from Lec 8 cells. (b) Gel electrophoresis under native conditions of HGF/SF, MET, and HGF/SF-MET complexes, in the absence or presence of heparin (c–h) Velocity sedimentation analysis of HGF/SF (c and d), MET (e and f), and the HGF/SF–heparin–MET complex (g and h). (c, e, and g) Plots of g(s*) against s*20,w. (d, f, and h) Plots of the residuals, from fitting models to the data, against s*20,w. Experiments shown in b, e, and g were carried out with equimolar concentrations of HGF/SF and MET 25–928H derived from Lec 8 cells (4 × 10–6 M) and a 2.5-fold excess of heparin.
Fig. 3 c–h shows the behavior of HGF/SF (c and d), MET (e and f), and the HGF/SF–heparin–MET complex (g and h) analyzed by ultracentrifugation, with plots of g(s*) against s*20,w on the left, and plots of the residuals, from fitting models to the data, against s*20,w on the right. Velocity sedimentation of HGF/SF alone showed a wide peak, which could not be fitted satisfactorily by a model with a single component, but required two components, of roughly similar optical density, with an s*20,w of 14.4 and 17.7 S and a molecular mass of 74 and 21 kDa, respectively. The presence of two species of different s*20,w values may reflect an equilibrium between structurally distinct forms, as seen with the homologue plasminogen, which exists in open and closed conformations (35).
MET alone (Fig. 3 e and f) showed a single, symmetric peak, with the data well fitted by a model for a single component with s*20,w = 3.5 S and M = 109 kDa (a value intermediate to calculated masses of 102.6 and 117.0 kDa without and with core N-linked carbohydrates). This is a low sedimentation coefficient for the molecular mass and, together with a Stokes radius of 56 Å from gel filtration experiments (data not shown), indicates a nonglobular, rod shape for the MET ectodomain.
Velocity sedimentation analysis of the HGF/SF–MET complex showed a complex boundary, which required at least two components in the model to produce a reasonable fit, implying that the binary HGF/SF–MET complex is unstable in solution under the conditions used. In contrast, the HGF/SF–heparin–MET complex yielded a symmetrical peak (Fig. 3g), which was well fitted by a model with a single component with s*20,w = 15.4 S and Mr = 179 kDa (Fig. 3h). Whereas this molecular mass is somewhat lower than that calculated for a 1:1:1 HGF/SF–heparin–MET complex (≈205 kDa), it is compatible only with such a complex and not with complexes of higher stoichiometries (2:1:2 or 2:2:2).
The Ligand-Binding Domain of MET Has a β-Propeller Fold. Amino acids 25–519 of MET exhibit weak homology (7.9–12.2% identity, 50.7–52.5% similarity) to a sequence present in the semaphorins, a large protein family involved in axon guidance, and in their plexin receptors (10.1–14.7% identity, 54.0–56.5% similarity) (36). This so-called sema domain was initially described as a cystine-rich sequence of ≈550 amino acids. The cystine-rich region, however, is confined to the last ≈50 amino acids, and may be related to the one present in MET (amino acids 520–561; Fig. 1), and in the N-terminal domain of the β-chain of integrins (37).
Database searches with dynamic statistical profiles (HMMs) of the sequence of the sema domain of MET and several semaphorins produced marginal but consistent hits to six-, seven-, or eight-bladed β-propellers. Hits to other folds were never consistent. Sequence-structure homology recognition using sequence profiles in combination with secondary structure and solvent accessibility (3dpssm) (20) or environment-specific substitution tables and a database of structural profiles (fugue) (21), yielded strong predictions for a β-propeller fold for the sema domain (E values of 5.04e × 10–2 and 4.61e × 10–2 for 3dpssm and Z scores of 4.40 and 3.77 for fugue for SEMA4A and SEM7A) with lower scores for MET (1.09e 10–1 and 2.84). In the majority of cases, the top predictions returned seven-bladed β-propeller domains; some returned six- or eight-bladed propeller domains.
Fig. 4 a and b show HMMs of a seven-blade β-propeller [amino acids 1–438 of the integrin αV chain, Protein Data Bank (PDB) ID code 1L5G] (ref. 38 and Fig. 4a) and amino acids 1–519 of MET (Fig. 4b). Hydrophilic amino acids are blue and hydrophobic amino acids are red. Blue lines underneath the HMM of 1L5G correspond to the clusters of four β-strands, which pack against each other to produce the seven blades (38). Hydrophobicity peaks with individual blades and generally with the inner β-strands (Fig. 4a). Blue and yellow lines underneath the HMM of amino acids 1–519 of MET correspond to segments predicted by jpred (39) as β-strands (24) or α-helices (4). Alternating stretches of hydrophobicity in the MET HMM correspond to segments where many of the predicted β-strands cluster (Fig. 4b). These features define the putative boundaries of six blades (blades 1–4, 6, and 7). The MET sequence included between blades 4 and 6 (amino acids 297–436) contains the three predicted α-helices of mature MET (amino acids 311–318, 325–330, and 365–372) interspersed with two β-strands (amino acids 339–345 and 358–362), followed by a region with little secondary structure (Fig. 4b). We considered the possibility that this sequence may correspond to a domain inserted in a six-blade β-propeller, but were unable to assign a distinct fold to amino acids 297–436 of MET. Thus, we conclude that this sequence contains two long connecting loops and an additional blade, and we propose a seven-blade β-propeller fold for the sema domain of MET and semaphorins.
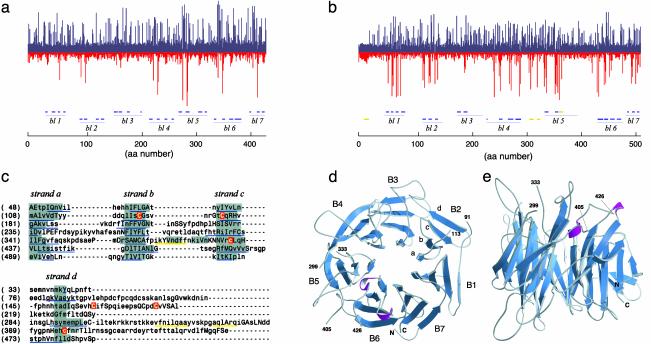
Fig. 4.
Predicted structure of the ligand-binding domain of MET. (a and b) HMMs of amino acids 1–438 of the α-chain of the β-propeller domain of integrin αV (1L5G) (a) and amino acids 1–519 of MET. bl, blade. (b) Each column plots the probabilistic distribution of hydrophilic (blue) and hydrophobic (red) amino acids at each position. Column height is proportional to the distance from the generic null distribution. Blue lines below the 1L5G HMM are β-strands in the crystal structure of 1L5G (38). Blue or yellow lines below the MET HMM (and in c below) are β-strands or α-helices predicted with jpred (39). (c) Alignment of the seven-blade β-propeller 3D model of the MET ligand-binding domain. Regions boxed in gray are assigned as β-strands in the model, and residues in small or capital letters are solvent accessible or solvent inaccessible, respectively. The six cysteine residues shown in white on a red background are predicted to form three disulfide bonds between strands b and c of blade 2, in the d-a loop between blades 2 and 3, and between strands c and d of blade 5. (d and e) Ribbon diagrams of the β-propeller model of the ligand-binding domain of MET (amino acids 33–516) viewed from the top and side.
A 3D model of the MET ligand binding domain was constructed by using the atomic coordinates of 1L5G (Fig. 4c). The two sequences exhibit 8.1% identity and 49.2% similarity. Regions boxed in gray correspond to β-strands in the 3D model, whereas sequences underlined in blue and yellow correspond to segments predicted as β-strands or α-helices by jpred (39). The model has good stereochemistry with 84.2% of the residues in the most favored conformations, 14.7% in allowed conformations, and only 1.1% in disallowed conformations in the Ramachandran plot. Fig. 4 d and e show ribbon diagrams of the model of the sema domain of MET, as viewed from the top or the side with three segments omitted (amino acids 92–112, 300–332, and 406–425), because they were predicted to form long and flexible loops connecting blades 1 and 2, 4 and 5, and 5 and 6 respectively.
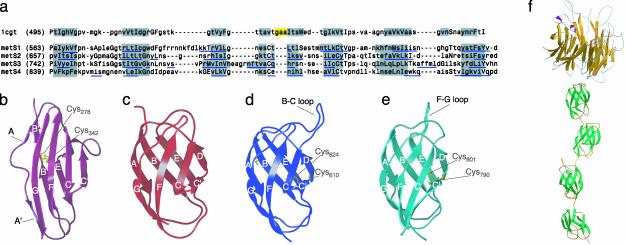
The C-Terminal Region of Extracellular MET Contains Four E Set Ig Domains. The C-terminal region of the MET ectodomain (amino acids 561–932) contains four proline- and glycine-rich repeats of 22–23 amino acids in length spaced by sequences 84–97 amino acids in length which, when aligned from the first proline, showed additional sequence conservation (Fig. 5a). Secondary structure prediction of the four putative domains (herewith defined as S1, S2, S3, and S4) yielded a firm prediction for all β-proteins and a conserved pattern of β-strands (underlined in blue in Fig. 5a). superfamily (24) and 3dpssm (20) returned a prediction for an Ig fold for domains S1, S2, and S3 with decreasing statistical scores. fugue (21) returned a prediction for an Ig fold for all four domains with the highest statistical scores for the first two.
Fig. 5.
Sequence and predicted structure of the stalk region of the MET ectodomain. (a) Sequence alignment of domain E of B. stearothermophilus cyclodextrin. Glucanotransferase (1CGT) with four segments of the MET sequence (amino acids 563–656, 657–741, 742–838, and 839–928, which are referred as domains S1, S2, S3d and S4). Regions boxed in gray correspond to β-strands in 1CGT or predicted to form β-strands in the Ig models of MET. Regions boxed in yellow correspond to α-helices in 1CGT. Residues in small or capital letters are solvent accessible or solvent inaccessible, respectively, and segments of the MET sequence predicted as β-strands or α-helices by jpred (39) are underlined in blue. (b and c) Ribbon representations of typical I set (domain 3 of FGF receptor 2, 1e0) (b) and E set (domain E of 1CYG (c) domains. (d and e) Ribbon representations of 3D models of the first (S1) and third (S3) Ig domains of the stalk region of MET. The image shows the long B-C and F-G loops of these domains and the cysteine residues involved in potential intradomain disulfides. (f) Overall view of the MET ectodomain, based on the results of this article. A small cysteine-rich domain located between the β-propeller and Ig domains (amino acids 520–561) is not shown.
A remarkable feature of these outputs was the assignment of the Ig domains of MET, not to the I set, but to the E set, which is a structural set of the Ig domain found earlier in bacterial enzymes (such as galactose oxidases, glycosyltransferases, amylases, sialidases, and cellulases), as well as in transcription factors of the NF-κB/REL/dorsal family such as NFAT, jun, fos, and NF-κB (40). Fig. 5b shows a typical I set domain, namely domain 3 of FGF receptor 2 (41). The domain shows a buried, intersheet, disulfide bond between strands B and F, which is in common with the V and C1 sets, and a split A strand (A/A′), which switches from the ABE sheet to the GFCC′ (2). Fig. 5c shows a typical E set domain, namely domain E of Bacillus (Geobacillus) stearothermophilus cyclodextrin glucanotransferase (1CYG). The domain lacks the intersheet disulfide bond between strands B and F, has a continuous A strand hydrogen-bonded to the B strand, and has a 4 + 4 (ABED + GFCC′) strand architecture.
We modeled the S1, S2, S3, and S4 domains of MET by using the atomic coordinates of the E domains of 1CGT, 1CYT, 1CYG, and 1QHP (S1); 1CGT and 1QHP (S2); 1CIU and 1D3C (S3), and 1QHP (S4) as templates, details of which can be obtained from the Protein Data Bank (www.rcsb.org/pdb). All models showed excellent stereochemistry (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Ribbon diagrams of domains S1 and S3 (Fig. 5 d and e, respectively) show a conserved pair of Cys residues located between the C′and D strands and near the end of the E strand (Cys-610 and Cys-624 in S1 and Cys-790 and Cys-801 in S2), respectively. The distance between the sulfur atoms of these pairs of Cys is 6.0, 3.9, 4.6 and 4.7 Å in S1, S2, S3, and S4, respectively. Thus, the Ig domains of MET may contain an alternative type of intersheet disulfide, which is different from the one between the B-F strands of the V, C1, and I set domains and the intrasheet disulfide observed in domain 5 of the tropomyosin receptor kinase A (42). This potential disulfide lends support to the geometry and stereochemistry of the 3D models of the MET Ig domains.
Discussion
Early cross-linking experiments indicated that HGF/SF binds to the MET β-chain (5). This article establishes that the first 519 amino acids of MET are required for HGF/SF binding, which include the first 212 amino acids of the β-chain. It is thus possible that the HGF/SF binding site is contained within this sequence. A heparin-binding site also maps to the same region of MET (Fig. 2 c and d) and presumably mediates interactions with membrane-bound HSPGs.
A complex formed by HGF/SF, heparin, and the whole MET ectodomain has a 1:1:1 stoichiometry in solution (Fig. 3 g and h). If, as with other growth factor-receptor systems, dimerization is a prerequisite for receptor activation, how does MET dimerize on cell surface? Different crystal structures have shown receptor dimers complexed with bivalent ligand, (GH-GH receptor) (43) or dimeric ligand, (NGF-Trk) (42). Recent structures of truncated forms of the EGF receptor ectodomain complexed with EGF (44) or TGFα (45), however, have shown 2:2 complexes resulting from interactions between the two receptors and imply conversion from an inactive EGF receptor dimer into an active one. In essence, although dimerization or oligomerization may be a general prerequisite for activation of RTKs, a variety of structural mechanisms appear to be at work, and the early suggestion that RTKs are activated by ligand-induced receptordimerization (46) may only apply to a subset of ligand-receptor pairs.
There are several mechanisms for MET dimerization, which are compatible with the data reported here. Dimerization may depend on weak dimerization sequences within the ectodomain that operate at higher concentrations of ligand and receptor. Alternatively, it may rely on sequences within the transor juxtamembrane, as in the neu receptor (47). Finally, it may depend on interactions with additional protein(s). There have been reports of selective association between MET and α6β4 integrin (48) and plexin B1 (49) and, whereas a critical role of α6β4 integrin for MET signaling is not supported by genetic studies in the mouse (50–51), the possibility remains for plexin B1.
We assigned folds and produced atomic models of the N-terminal (ligand binding) and C-terminal halves of MET. The proposed β-propeller fold for the ligand binding domain (Fig. 4) is based on three lines of evidence: (i) lack of expression of constructs carrying deletions within amino acid 25–519 suggesting a single domain, (ii) the results of secondary structure prediction, sequence-sequence, and sequence-structure analysis and, (iii) the robust stereochemistry of the three-dimensional model of the MET ligand built on the 7-blade β 3D-propeller of an integrin α-chain (38) (see Results and Table 1).
The use of the integrin β-propeller as a template for comparative modeling of the MET ligand-binding-domain model does not necessarily imply homology. The level of sequence conservation is below the twilight zone; the exon-intron structure of the two genes differ, and the widespread distribution and functions of the β-propeller fold clearly reflect convergent evolution of this protein fold (52). Equally, our analysis does not exclude homology, and we note a striking conservation of two disulfide bonds (between strands b and c and within the d-a loop) in blade 2 of 1L5G and the MET model (Fig. 4c).
In contrast to the globular head of the N-terminal half of the MET ectodomain, the C-terminal region appears to form a stalk structure, which confers an elongated and asymmetric shape to the MET ectodomain (Fig. 5f), which is in agreement with the large Stokes radius and the low sedimentation coefficient experimentally determined (Fig. 3 e and f). The unexpected similarity between the Ig domains of the MET and E set domains raises further questions about the evolution of the MET receptor family. A large semaphorin subfamily contains a single Ig domain, but preliminary analysis suggests features typical of the I domain, and not the E set domain (E.G., unpublished work). This finding implies that the homology between the MET receptor and the semaphorins may be confined to the N-terminal and cystine-rich domain. Crystal structures of the MET receptor, semaphorins, and plexins will be ultimately required to address structural conservation among these protein families, as well as integrins. Meanwhile, the 3D models described here offer the basis for mutagenesis experiments addressing the mechanism of ligand binding and MET activation.
Supplementary Material
Acknowledgments
We thank George Vande Woude for critical reading of the manuscript, Max Paoli for discussions, and Peggy MacCuish for secretarial support. This work was supported by Medical Research Council Program Grant G9704528 (to E.G.). T.L.B. acknowledges the Wellcome Trust Program (Grant 046073) and the Biotechnology and Biological Sciences Research Council Structural Biology Initiative for support. J.G. is supported by a Burroughs-Welcome Fellowship from the Program in Mathematics and Molecular Biology.
Abbreviations: RTK, receptor tyrosine kinase; MET, the RTK encoded by the c-met protooncogene; HGF, hepatocyte growth factor; SF, scatter factor; HMM, hidden Markov model.
References
- 1.Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355–365. [DOI] [PubMed] [Google Scholar]
- 2.Harpaz, Y. & Chothia, C. (1994) J. Mol. Biol. 238, 528–539. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, C. S., Blair, D. G., Oskarsson, M. K., Tainsky, M. A., Eader, L. A. & Vande Woude, G. F. (1984) Cancer Res. 44, 1–10. [PubMed] [Google Scholar]
- 4.Park, M., Dean, M., Kaul, K., Braun, M. J., Gonda, M. A. & Vande Woude, G. (1987) Proc. Natl. Acad. Sci. USA 84, 6379–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottaro, D. P., Rubin, J. S., Faletto, D. L., Chan, A. M., Kmiecik, T. E., Vande Woude, G. F. & Aaronson, S. A. (1991) Science 251, 802–804. [DOI] [PubMed] [Google Scholar]
- 6.Stoker, M., Gherardi, E., Perryman, M. & Gray, J. (1987) Nature 327, 239–242. [DOI] [PubMed] [Google Scholar]
- 7.Gherardi, E., Gray, J., Stoker, M., Perryman, M. & Furlong, R. (1989) Proc. Natl. Acad. Sci. USA 86, 5844–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., Tashiro, K. & Shimizu, S. (1989) Nature 342, 440–443. [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa, K., Tsubouchi, H., Naka, D., Takahashi, K., Okigaki, M., Arakaki, N., Nakayama, H., Hirono, S., Sakiyama, O., Takahashi, K., et al. (1989) Biochem. Biophys. Res. Commun. 163, 967–973. [DOI] [PubMed] [Google Scholar]
- 10.Zarnegar, R. & Michalopoulos, G. (1989) Cancer Res. 49, 3314–3320. [PubMed] [Google Scholar]
- 11.Schmidt, C., Bladt, F., Goedecke, S., Brinkmann, V., Zschiesche, W., Sharpe, M., Gherardi, E. & Birchmeier, C. (1995) Nature 373, 699–702. [DOI] [PubMed] [Google Scholar]
- 12.Uehara, Y., Minowa, O., Mori, C., Shiota, K., Kuno, J., Noda, T. & Kitamura, N. (1995) Nature 373, 702–705. [DOI] [PubMed] [Google Scholar]
- 13.Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A. & Birchmeier, C. (1995) Nature 376, 768–771. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt, L., Duh, F. M., Chen, F., Kishida, T., Glenn, G., Choyke, P., Scherer, S. W., Zhuang, Z., Lubensky, I., Dean, M., et al. (1997) Nat. Genet. 16, 68–73. [DOI] [PubMed] [Google Scholar]
- 15.Comoglio, P. M. (2001) Nat. Cell Biol. 3, E161–E162. [DOI] [PubMed] [Google Scholar]
- 16.Oelmann, S., Stanley, P. & Gerardy-Schahn, R. (2001) J. Biol. Chem. 276, 26291–26300. [DOI] [PubMed] [Google Scholar]
- 17.Stafford, W. F., III (1992) Anal. Biochem. 203, 295–301. [DOI] [PubMed] [Google Scholar]
- 18.Stafford, W. F., III (1994) Methods Enzymol. 240, 478–501. [DOI] [PubMed] [Google Scholar]
- 19.Philo, J. S. (2000) Anal. Biochem. 279, 151–163. [DOI] [PubMed] [Google Scholar]
- 20.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 21.Shi, J., Blundell, T. L. & Mizuguchi, K. (2001) J. Mol. Biol. 310, 243–257. [DOI] [PubMed] [Google Scholar]
- 22.Karplus, K., Barrett, C. & Hughey, R. (1998) Bioinformatics 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001) J. Mol. Biol. 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 25.Mizuguchi, K., Deane, C. M., Blundell, T. L. & Overington, J. P. (1998) Protein Sci. 7, 2469–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 28.Luthy, R., Bowie, J. U. & Eisenberg, D. (1992) Nature 356, 83–85. [DOI] [PubMed] [Google Scholar]
- 29.Sutcliffe, M. J., Hayes, F. R. & Blundell, T. L. (1987) Protein Eng. 1, 385–392. [DOI] [PubMed] [Google Scholar]
- 30.Komada, M., Hatsuzawa, K., Shibamoto, S., Ito, F., Nakayama, K. & Kitamura, N. (1993) FEBS Lett. 328, 25–29. [DOI] [PubMed] [Google Scholar]
- 31.Giordano, S., Ponzetto, C., Di Renzo, M. F., Cooper, C. S. & Comoglio, P. M. (1989) Nature 339, 155–156. [DOI] [PubMed] [Google Scholar]
- 32.Rubin, J. S., Day, R. M., Breckenridge, D., Atabey, N., Taylor, W. G., Stahl, S. J., Wingfield, P. T., Kaufman, J. D., Schwall, R. & Bottaro, D. P. (2001) J. Biol. Chem. 276, 32977–32983. [DOI] [PubMed] [Google Scholar]
- 33.Lietha, D., Chirgadze, D. Y., Mulloy, B., Blundell, T. L. & Gherardi, E. (2001) EMBO J. 20, 5543–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyon, M., Deakin, J. A. & Gallagher, J. T. (2002) J. Biol. Chem. 277, 1040–1046. [DOI] [PubMed] [Google Scholar]
- 35.Mangel, W. F., Lin, B. H. & Ramakrishnan, V. (1990) Science 248, 69–73. [DOI] [PubMed] [Google Scholar]
- 36.Winberg, M. L., Noordermeer, J. N., Tamagnone, L., Comoglio, P. M., Spriggs, M. K., Tessier-Lavigne, M. & Goodman, C. S. (1998) Cell 95, 903–916. [DOI] [PubMed] [Google Scholar]
- 37.Bork, P., Doerks, T., Springer, T. A. & Snel, B. (1999) Trends Biochem. Sci. 24, 261–263. [DOI] [PubMed] [Google Scholar]
- 38.Xiong, J. P., Stehle, T., Diefenbach, B., Zhang, R., Dunker, R., Scott, D. L., Joachimiak, A., Goodman, S. L. & Arnaout, M. A. (2001) Science 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuff, J. A. & Barton, G. J. (2000) Proteins 40, 502–511. [DOI] [PubMed] [Google Scholar]
- 40.Lo Conte, L., Ailey, B., Hubbard, T. J., Brenner, S. E., Murzin, A. G. & Chothia, C. (2000) Nucleic Acids Res. 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B. & Blundell, T. L. (2000) Nature 407, 1029–1034. [DOI] [PubMed] [Google Scholar]
- 42.Wiesmann, C., Ultsch, M. H., Bass, S. H. & de Vos, A. M. (1999) Nature 401, 184–188. [DOI] [PubMed] [Google Scholar]
- 43.de Vos, A. M., Ultsch, M. & Kossiakoff, A. A. (1992) Science 255, 306–312. [DOI] [PubMed] [Google Scholar]
- 44.Ogiso, H., Ishitani, R., Nureki, O., Fukai, S., Yamanaka, M., Kim, J. H., Saito, K., Sakamoto, A., Inoue, M., Shirouzu, M. & Yokoyama, S. (2002) Cell 110, 775–787. [DOI] [PubMed] [Google Scholar]
- 45.Garrett, T. P., McKern, N. M., Lou, M., Elleman, T. C., Adams, T. E., Lovrecz, G. O., Zhu, H. J., Walker, F., Frenkel, M. J., Hoyne, P. A., et al. (2002) Cell 110, 763–773. [DOI] [PubMed] [Google Scholar]
- 46.Ullrich, A. & Schlessinger, J. (1990) Cell 61, 203–212. [DOI] [PubMed] [Google Scholar]
- 47.Bargmann, C. I., Hung, M. C. & Weinberg, R. A. (1986) Cell 45, 649–657. [DOI] [PubMed] [Google Scholar]
- 48.Trusolino, L., Bertotti, A. & Comoglio, P. M. (2001) Cell 107, 643–654. [DOI] [PubMed] [Google Scholar]
- 49.Giordano, S., Corso, S., Conrotto, P., Artigiani, S., Gilestro, G., Barberis, D., Tamagnone, L. & Comoglio, P. M. (2002) Nat. Cell Biol. 4, 720–724. [DOI] [PubMed] [Google Scholar]
- 50.Georges-Labouesse, E., Messaddeq, N., Yehia, G., Cadalbert, L., Dierich, A. & Le Meur, M. (1996) Nat. Genet. 13, 370–373. [DOI] [PubMed] [Google Scholar]
- 51.van der Neut, R., Krimpenfort, P., Calafat, J., Niessen, C. M. & Sonnenberg, A. (1996) Nat. Genet. 13, 366–369. [DOI] [PubMed] [Google Scholar]
- 52.Jawad, Z. & Paoli, M. (2002) Structure (London) 10, 447–454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.