Abstract
One of the most intriguing disease-related mutations in human prion protein (PrP) is the Tyr to Stop codon substitution at position 145. This mutation results in a Gerstmann–Straussler–Scheinker-like disease with extensive PrP amyloid deposits in the brain. Here, we provide evidence for a spontaneous conversion of the recombinant polypeptide corresponding to the Y145Stop variant (huPrP23–144) from a monomeric unordered state to a fibrillar form. This conversion is characterized by a protein concentration-dependent lag phase and has characteristics of a nucleation-dependent polymerization. Atomic force microscopy shows that huPrP23–144 fibrils are characterized by an apparent periodicity along the long axis, with an average period of 20 nm. Fourier-transform infrared spectra indicate that the conversion is associated with formation of β-sheet structure. However, the infrared bands for huPrP23–144 are quite different from those for a synthetic peptide PrP106–126, suggesting conformational non-equivalence of β-structures in the disease-associated Y145Stop variant and a frequently used short model peptide. To identify the region that is critical for the self-seeded assembly of huPrP23–144 amyloid, experiments were performed by using the recombinant polypeptides corresponding to prion protein fragments 23–114, 23–124, 23–134, 23–137, 23–139, and 23–141. Importantly, none of the fragments ending before residue 139 showed a propensity for conformational conversion to amyloid fibrils, indicating that residues within the 138–141 region are essential for this conversion.
Transmissible spongiform encephalopathies (the prion diseases) are a group of fatal neurodegenerative diseases of animals and humans (1–4). This relatively diverse group includes bovine spongiform encephalopathy in cattle, scrapie in sheep, chronic wasting disease in deer and elk, and kuru, Creutzfeldt–Jakob disease, fatal familial insomnia, and Gerstmann–Straussler–Scheinker (GSS) disease in humans. Clinically, prion diseases can exhibit sporadic, inherited, or infectious presentations. Regardless of the primary etiology, these diseases are associated with a conversion of the cellular prion protein, PrPC, into an abnormal form, termed PrPSc. This conversion, which seems to occur without any covalent modifications, is believed to involve a major conformational change in the prion protein (1–4). In contrast to PrPC, the pathogenic PrPSc isoform is characterized by a relatively high content of β-sheet structure, partial resistance to proteolytic digestion, insolubility in non-ionic detergents, and a propensity to aggregate into amyloid-like fibrils and plaques (1–7). The mechanistic and structural aspects of the PrPC → PrPSc conversion remain, however, poorly understood.
The “protein-only” hypothesis asserts that the transmission of transmissible spongiform encephalopathies does not require nucleic acids, and that PrPSc itself is the infectious prion pathogen (1, 8). This novel pathogen is believed to self-propagate by a mechanism involving binding to cellular prion protein and catalyzing its conversion to the PrPSc state. Although the ultimate proof for the protein-only model (the evidence that the normal form of prion protein alone can indeed be converted to an infectious agent) is still missing, the central role of prion protein in the disease pathogenesis is documented by a wealth of biochemical and genetic data (1–4). Furthermore, the notion that proteins can act as infectious agents that replicate by self-propagating conformational changes is supported by recent studies on prion-like phenomena in yeast and fungi (9–11).
An important argument in favor of the protein-only hypothesis of transmissible spongiform encephalopathies is the link between hereditary prion diseases and mutations in the gene coding for human prion protein. Over 20 mutations in the human PrP gene have been shown to segregate with inherited Creutzfeldt–Jakob disease, GSS disease, or fatal familial insomnia (1, 4, 12). All of these disorders are autosomal, dominantly inherited conditions. The pathogenic process in patients carrying these mutations seems to develop spontaneously. Furthermore, in some cases, the material from brain tissue of these patients was shown to be infectious for experimental animals (12, 13). Therefore, the mutant proteins associated with these diseases provide an invaluable model for studying molecular mechanisms of the PrPC → PrPSc conversion in cell models and in vitro. One of the most intriguing disease-related mutations is the tyrosine to stop codon substitution at position 145. This mutation, resulting in a truncated prion protein huPrP145Stop, has been linked to prion disease with extensive PrP-amyloid deposits in the central nervous system (14, 15). The phenotype of this disease is similar to some other familial GSS disorders (12). However, attempts to transmit this variant of the disease to mice have not yet been successful (13). Here, we demonstrate that, unlike the wild-type prion protein, the recombinant polypeptide corresponding to huPrP145Stop undergoes in vitro a very efficient self-propagating conversion to β-sheet-rich fibrils. Furthermore, we identify a specific polypeptide region that is essential for conformational conversion of the huPrP145 variant and, likely, also of the full-length prion protein.
Materials and Methods
Plasmid Construction and Protein Purification. The plasmid encoding huPrP23–231 with N-terminal linker containing a His-6 tail and a thrombin cleavage site has been described (16). Constructs for the expression of huPrP145Stop and other C-truncated variants of human prion protein were prepared by introducing, at specific positions of the above template plasmid, a Stop codon TAG or TGA. This preparation was accomplished by sitedirected mutagenesis using appropriate primers and a QuikChange kit (Stratagene). The proteins were expressed in Escherichia coli and purified essentially as described by using a nickel-nitrilotriacetic acid agarose resin (16, 17), with the exception that no glutathione was present in the refolding buffer. The N-terminal His-6 tag was cleaved with biotinylated thrombin (Novagen), followed by removal of thrombin by using streptavidin-agarose beads. The free-polyhistidine tag was removed by dialysis against distilled water. The purity of the final products was >98% as judged by SDS/PAGE. The identity of each protein was further confirmed by mass spectrometry. Protein concentration was determined by measuring absorbance at 280 nm.
Thioflavine T (ThT) Assay. The progress of amyloid fibril formation was followed by a fluorimetric ThT assay (18). In the unseeded reaction, protein samples were rapidly transferred from distilled water to a buffer (typically 50 mM potassium phosphate, pH 6.5) and incubated at 25°C. Small aliquots of each sample were withdrawn at different time points and diluted to a final concentration of 4 μM in 50 mM phosphate buffer, pH 6.5, containing 10 μM ThT. After 2 min incubation at room temperature, the fluorescence of ThT dye was measured at 482 nm by using the excitation wavelength of 450 nm. In the seeded reaction, a small amount (1% wt/wt) of preformed fibrillar aggregates was added to protein solution in buffer, and the kinetics of fibril formation were followed as described above. To break up very large aggregates, the fibrillar protein used for seeding was briefly sonicated in a bath-type sonicator.
Fourier-Transform Infrared (FTIR) Spectroscopy. To prepare samples for FTIR spectroscopy, proteins were first dialyzed against distilled water and then lyophilized. The lyophilized material was dissolved at a concentration of 400 μM in a buffer prepared in D2O (50 mM potassium phosphate, pD 6.5) and placed between two calcium fluoride windows separated by a 50-μm Teflon spacer. After incubation at 25°C for appropriate time periods, the spectra were acquired at room temperature on a Bruker IFS 66 FTIR spectrometer. For each spectrum, 256 interferograms were co-added and Fourier-transformed to give a resolution of 2 cm-1. The spectra in the 1,500–1,800 cm-1 region were corrected for the weak absorption of the buffer. Overlapping infrared bands were partially resolved by Fourier self-deconvolution procedure (19).
Electron Microscopy. After incubation at 25°C for appropriate periods of time, protein samples showing positive response in the fluorimetric ThT assay were analyzed by electron microscopy. A drop of the incubation mixture was placed on a carbon-coated 600-mesh copper grid and negatively stained with 2% aqueous uranyl acetate. Grid preparations were visualized by using a Jeol 1200 transmission electron microscope operating at 80 kV.
Atomic Force Microscopy (AFM). For AFM, ThT-positive samples were diluted at least 10 times and then applied to a freshly cleaved muscovite mica substrate. After a 60-s incubation, the substrate was rinsed with two 50-μl aliquots of ultra-pure water to remove salts and unbound protein. The preparation was then dried in a stream of nitrogen and mounted on the microscope scanner. The images were acquired in a tapping mode on a Multimode atomic force microscope (Digital Instruments, Santa Barbara, CA) equipped with a Nanoscope IV controller and a type E scanner. All images were obtained in air by using single-beam silicon cantilever probes.
Results
Self-Seeded Conversion of the Recombinant huPrP145Stop. The recombinant polypeptide encompassing residues 23–144 of human prion protein (huPrP23–144) was expressed in E. coli and purified to homogeneity. The covalent structure of this polypeptide is identical to that of a disease-associated huPrP145Stop variant (posttranslational modifications such as glycosylation and lipidation affect the C-terminal part of prion protein, beyond residue 144). Freshly purified protein was dialyzed against distilled water. Under these conditions, the protein remained in a monomeric form even after prolonged periods of storage at room temperature. The monomeric state of the protein was verified by dynamic light scattering and size exclusion chromatography. However, on transfer to the phosphate or Tris buffers (pH 6.5–7.5), there was a pronounced time-dependent increase in the viscosity of the solution, indicating self-association of the protein into oligomeric structures.
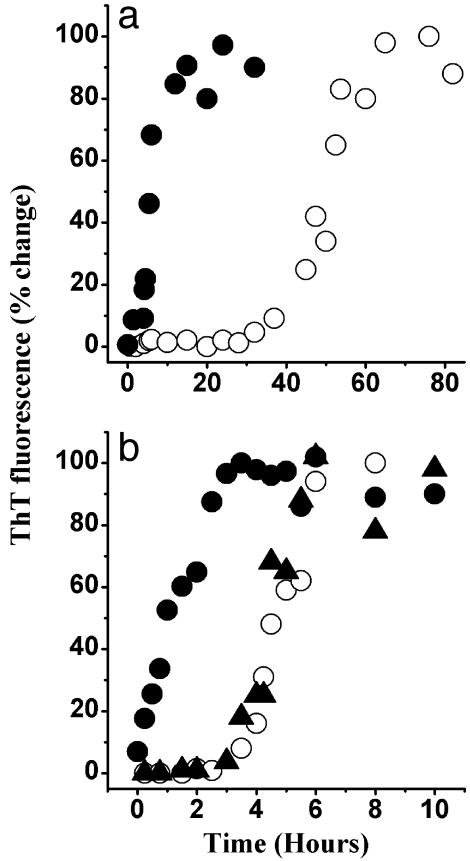
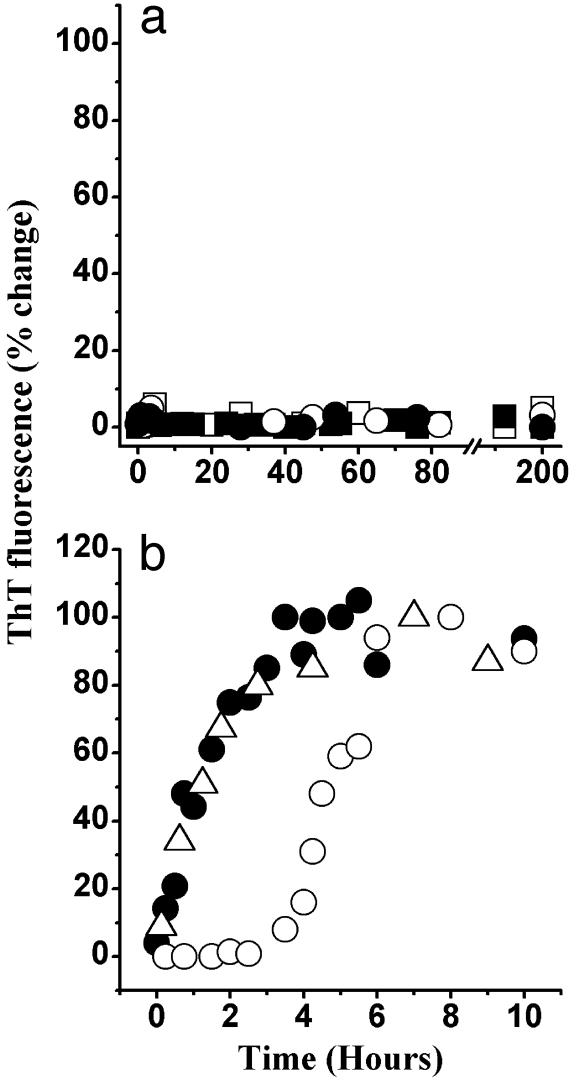
To probe the nature of this self-association, we have used the ThT assay. ThT is believed to specifically bind to amyloid fibrils (18). The increase in dye fluorescence on binding may be used to follow the kinetics of amyloid formation. As shown in Fig. 1, on incubation in the phosphate buffer (pH 6.5), huPrP23–144 acquired the ability to bind ThT. This result strongly suggests formation of amyloid-like structures. The kinetic curves for the assembly of these structures were characterized by a lag phase that was protein concentration-dependent. For example, at a huPrP23–144 concentration of 80 μM, the lag phase was ≈40 h (Fig. 1a) whereas at 400 μM it was reduced to ≈3 h (Fig. 1b). Addition of a small amount (1% wt/wt) of the preformed huPrP23–144 amyloid to the incubation mixture resulted in elimination of the lag phase. Similar self-assembly curves, although with somewhat different kinetic parameters, were observed for huPrP23–144 at pH 7.5 (data not shown). When the protein was incubated with constant rotation, at a given protein concentration, the lag phase was reduced at least ≈10 times as compared with the unperturbed reaction. As in the absence of rotation, the lag phase could be completely eliminated by addition of fibrillar seeds. However, rotation or stirring promoted clumping of individual fibrils into very large aggregates that had a tendency to precipitate and adhere to the walls of the test tubes. These effects precluded detailed kinetic characterization of the rotated reactions. Overall, the above data indicate that the conformational conversion of huPrP23–144 is a self-seeded reaction that requires oligomeric nuclei.
Fig. 1.
Time course of huPrP23–144 fibrillization monitored by ThT fluorescence. The protein was incubated in the absence (○) or presence (•) of preformed fibrillar seeds (1% wt/wt). (a) Protein incubated at a concentration of 80 μM. (b) Protein incubated at a concentration of 400 μM. Triangles (▴)in b represent the fibrillization kinetics of huPrP23–144 incubated in the presence of fibrillar seeds of synthetic PrP106–126 peptide (5% wt/wt). All samples were in 50 mM potassium phosphate buffer, pH 6.5, at a temperature of 25°C.
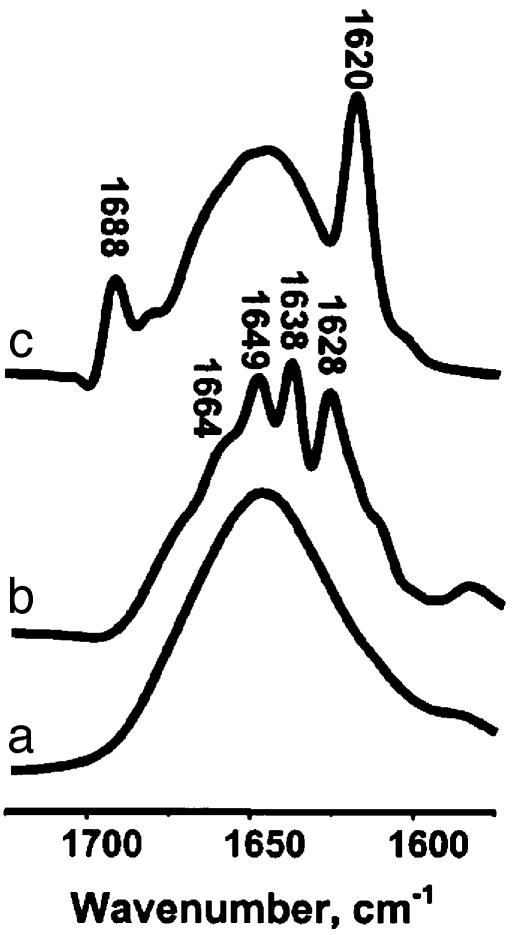
Changes in the Secondary Structure. The changes in protein secondary structure during the transition of huPrP23–144 monomer to the oligomeric, ThT-binding form were probed by FTIR spectroscopy. The conformation-sensitive amide I mode for the monomeric protein is strongly dominated by a broad band at 1648 cm-1 (Fig. 2a), indicating a largely unordered structure (19). This result is generally consistent with NMR data for the full-length prion protein showing that, apart from the short β-strand 128–131, the entire 23–144 region is devoid of any α-helices or β-sheet structure (20–22). The FTIR spectrum for the oligomeric protein is quite different, displaying bands at 1,628, 1,638, 1,649, and 1,664 cm-1 (Fig. 2b). The first two bands are highly characteristic of a β-sheet structure, whereas the components at 1,649 and 1,664 cm-1 are believed to represent random conformation and turns, respectively (19). Overall, infrared spectroscopic data provide clear evidence that the formation of ThT-positive huPrP145Stop oligomers is accompanied by a transition of a substantial portion of polypeptide chain from an unordered conformation to β-sheet structure.
Fig. 2.
Infrared spectra (amide I region) for huPrP23–144 and synthetic peptide PrP106–126. (a) Monomeric huPrP23–144. (b) Fibrillar form of huPrP23–144 obtained after incubating the protein for 24 h. (c) Fibrillar form of peptide PrP106–126 after 3 days of incubation. The samples were incubated at a concentration of 400 μM (huPrP23–144) or 2.6 mM (PrP106–126) in deuterated 50 mM potassium phosphate buffer, pD 6.5, at 25°C. The spectra were deconvolved by using a Lorentzian band and a resolution enhancement factor of 2.
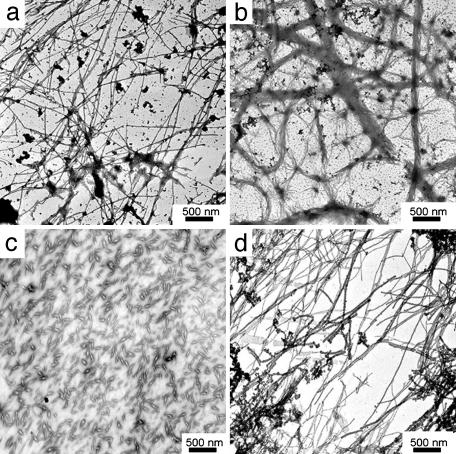
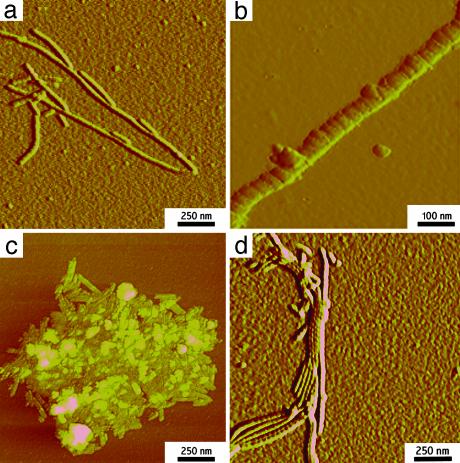
Morphology of Fibrillar Structures. The morphology of the self-assembled huPrP23–144 oligomers was studied by electron microscopy and AFM. The electron micrographs show an extensive network of fibrillar structures (Fig. 3a). The fibrils have variable length and a width of ≈20–40 nm. On longer incubation, these fibrils further assembled into thick ribbon-like structures (Fig. 3b). Examination by AFM (Fig. 4 a and b) revealed that individual fibrils actually consist of well resolved subunits (Fig. 4b). In AFM images, the fibrils display average height of ≈3 nm and axial periodicity of 20–25 nm. The fibrillar structures observed here for huPrP23–144 are morphologically similar to fibrils formed by yeast prion proteins Sup35 and Ure2p (23, 24).
Fig. 3.
Electron micrographs of oligomeric forms of huPrP N–terminal fragments. (a) Fibrils of huPrP23–144 after 4 h of incubation. (b) Bundled fibrils of huPrP23–144 after 12 h of incubation. (c) Aggregates formed by huPrP23–139 after 60 h of incubation. (d) Fibrils of huPrP23–141 after4hof incubation. All proteins were incubated at a concentration of 400 μM in 50 mM potassium phosphate buffer, pH 6.5, at 25°C.
Fig. 4.
AFM images (amplitude mode) of oligomeric forms of huPrP N-terminal fragments. (a) huPrP23–144 fibrils. (b) Enlarged image of an individual huPrP23–144 fibril. (c) Aggregates formed by huPrP23–139. (d) Bundled huPrP23–141 fibrils. Incubation conditions were identical to those described in the legend to Fig. 3.
Proteinase K Digestion. The fibrillar protein was characterized by greatly increased resistance to proteinase K digestion. Monomeric huPrP23–144 was fully degraded on a 30-min incubation in the presence of as little as 1 μg/ml of proteinase K. In the fibrillar protein, a number of fragments of ≈6–10 kDa persisted at 1 μg/ml of the enzyme. At higher proteinase K concentrations, the larger fragments disappeared, leaving a species of ≈6 kDa. The latter fragment persisted up to at least 20 μg/ml of proteinase K (Fig. 5).
Fig. 5.
Proteinase K digestion of huPrP23-144 before (A) and after (B) fibrillization. Molecular weight markers are shown on the left, and the concentration of proteinase K is indicated below each line. Protein was converted to fibrillar form by incubating at a concentration of 400 μM for 12 h at 25°C in 50 mM phosphate buffer, pH 6.5. Samples of monomeric and fibrillar protein (125 μM) were digested with various amounts of proteinase K for 30 min at 37°C. The digestion was terminated by boiling for 10 min in SDS sample loading buffer. Protein fragments were separated by SDS/PAGE and visualized by Coomassie blue staining.
Specificity of Seeding. It was previously shown that a short synthetic peptide corresponding to residues 106–126 of the prion protein aggregates into oligomeric structures with amyloid-like morphology (25–27). Therefore, it was of interest to compare the latter oligomers with those formed by the much longer and physiologically more relevant (disease-associated) protein huPrP23–144. Notably, the FTIR spectrum for the oligomeric form of PrP106–126 is quite different from that for the huPrP23–144 oligomer. In particular, the spectrum for the short peptide is characterized by a low-frequency amide I band at 1,620 cm-1 (Fig. 2c) whereas the bands representing β-sheet structure in fibrillar huPrP23–144 are at 1,628 and 1,638 cm-1 (Fig. 2b). This finding clearly suggests conformational non-equivalence of the β-structures in the oligomeric forms of huPrP23–144 and the short synthetic peptide PrP106–126. Prompted by this observation, we have tested whether PrP106–126 fibrils could act as seeds for huPrP23–144 polymerization. As shown in Fig. 1, addition of preformed huPrP23–144 fibrils to the solution of huPrP23–144 monomer greatly accelerates the fibrillization of the latter protein by removing the lag phase. However, preformed PrP106–126 fibrils had no measurable effect on the polymerization rate of huPrP23–144: neither the lag phase nor the amyloid growth phase for huPrP23–144 was accelerated in the presence of as much as 5% (wt/wt) of the preformed PrP106–126 fibrils (Fig. 1b). The latter observation, together with FTIR data, indicates considerable conformational specificity in the seeding of huPrP23–144 polymerization.
Mapping the Amyloidogenic Determinant. To identify the specific region of huPrP23–144 that is essential for self-assembly into amyloid fibrils, we expressed and purified a number of C-terminally truncated fragments of the protein and studied them by the ThT assay, FTIR spectroscopy, and electron and AFM. In view of the previous data for short synthetic peptides (25–27), our expectation was that the critical amyloidogenic determinant for huPrP144Stop is largely within the region encompassing residues 106–126. However, to our surprise, the truncated proteins huPrP23–124, huPrP23–126, huPrP23–134, and huPrP23–137 did not show any propensity to undergo a spontaneous conversion to ThT-positive structures (Fig. 6a). In addition to ThT measurements, the inability of these truncated proteins to self-assemble into oligomeric structure was further confirmed by electron microscopy. Importantly, no fibrillization of these proteins could be detected even in the presence of fibrillar seeds of huPrP23–144 (Fig. 6a). The protein truncated at residue 139 (huPrP23–139) formed, after very long incubation periods, oligomers that precipitated from solution. These aggregates were not totally amorphous. However, their morphology (Figs. 3c and 4c) was clearly different from long amyloid fibrils. Furthermore, the huPrP23–139 oligomers did not show any appreciable content of β-sheet structure and could not seed polymerization of the monomeric huPrP23–139 or huPrP23–144 (data not shown for brevity). The shortest C-truncated variant of huPrP145Stop that was able to undergo self-propagating, nucleation-dependent conversion to β-sheet-rich fibrillar structures was huPrP23–141 (Fig. 6b). The properties of the latter protein were indistinguishable from those of huPrP23–144, both with respect to kinetics of conformational conversion, the requirement for oligomeric seeds (Figs. 1b and 6b), as well as the morphology of fibrillar aggregates (Figs. 3 and 4). Furthermore, as indicated by FTIR spectra, the secondary structure of fibrillar huPrP23–141 was indistinguishable from that of huPrP23–144 fibrils (FTIR spectrum for huPrP23–141 not shown for brevity). Consistent with these morphological and conformational similarities, preformed fibrils of either of the two proteins could effectively seed polymerization of the other protein (Fig. 6b). Overall, these data indicate that residues within a relatively short 138–141 region are essential for fibrillization of huPrP23–144.
Fig. 6.
Time course of fibrillization of huPrP N-terminal fragments monitored by ThT fluorescence. (a) huPrP23–124 (squares) and huPrP23–137 (circles) incubated in the absence (open symbols) or presence (filled symbols) of huPrP23–144 seeds. (b) huPrP23–141 in the absence of preformed seeds (○) and in the presence of huPrP23–141 seeds (•) or huPrP23–144 seeds (▵). All samples were incubated at a concentration of 400 μM under identical conditions to those described in legend to Fig. 1. In case of seeded reactions, the amount of preformed fibrillar seeds was 1% wt/wt.
Discussion
The pathogenesis of prion diseases seems to depend on a conversion of the cellular prion protein into abnormally folded oligomeric form(s). However, despite many years of research, the mechanism of this conformational transition remains poorly understood. One approach to gain mechanistic insight into the conversion reaction is to take advantage of the link between inherited prion diseases and specific mutations in the prion protein. Because familial Creutzfeldt–Jakob disease or GSS pathology develops spontaneously (i.e., without introduction of exogenous prion agent), understanding how mutations affect biophysical properties of prion protein could provide insight into the molecular basis of the disease in general, and the mechanism of prion protein conversion in particular.
In the present study, we have focused on the prion protein variant with 145Stop mutation. Although this mutation is unique in that it eliminates the entire α-helical region of PrP, the disease phenotype associated with this variant is similar to other subtypes of GSS disease (12). Our data clearly demonstrate that the purified recombinant protein corresponding to huPrP145Stop spontaneously polymerizes into highly ordered fibrils which bind ThT and are rich in β-sheet structure. The self-propagating conversion of the huPrP145Stop monomer to a fibrillar state occurs after a lag phase, whose duration strongly depends on the concentration of the protein. Importantly, addition of a small quantity of preformed fibrils abolishes the lag phase, indicating that conformational conversion of huPrP145Stop is a nucleation-dependent reaction.
Previous studies have shown that short synthetic peptides corresponding to various segments within the N-terminal part of PrP can also form amyloid fibrils (25–29). The major focus of most of these studies was on amino acid residues 106–126. The short synthetic peptide PrP106–126 is indeed amyloidogenic (25–27) and seems to be neurotoxic (26, 30). However, our data show that, when present in the context of a larger, disease-associated prion protein variant huPrP145Stop, the sequence 106–126 alone is insufficient to induce protein fibrillization. The present results demonstrate that the critical amyloidogenic determinant in huPrP145Stop extends well beyond the 106–126 region and includes residues within the 138–141 segment. Consistent with their propensity to form β-structure (31), the latter residues seem to be an integral part of a specific recognition site that is essential for nucleating intermolecular interactions leading to the assembly of amyloid fibrils.
The results of the present experiments in vitro correlate with previous observations regarding amyloid deposits in GSS diseases (12). Regardless of the specific PRNP mutation, GSS disorders are invariably associated with the presence of PrP amyloid plaques although the abundance of these plaques and their distribution depend on the disease genotype. The deposits seem to contain a variety of prion protein fragments ranging in size from ≈7 to ≈15 kDa. Analysis of these fragments revealed that many of them are C-terminally truncated at residues 146–153, but not before residue 141 (29, 32). The above profile of amyloid plaques in GSS patients is consistent with, and may be rationalized by, the present finding regarding the location of amyloidogenic determinant within the N-terminal part of the prion protein.
The protein-only hypothesis of prion diseases centers on the conversion of prion protein into the PrPSc conformer. Attempts to propagate the infectious PrPSc state in vitro have not yet been successful. Nevertheless, previous observations suggest the importance of a broadly defined N-terminal region of PrP in conformational conversion, especially between residues 90 and ≈140 (29, 33–37). Our findings regarding self-perpetuating conversion of C-terminally truncated PrP variants provide direct experimental validation and more precise structural definition of this model. Given the present data, we propose that, like for the huPrP145Stop variant, a determinant for self-propagating conformational conversion of the full-length PrP includes the region around residues 138–141. In the native PrPC, this region constitutes part of the loop between β-strand (residues 128–131) and first α-helix (residues 144–154) (22). The importance of the 138–141 segment is in line with the reports that residues within or around this region influence abnormal PrP formation in cell culture and in animals (35, 38). Furthermore, recent data have identified the region around residues ≈119 to 138 as a potential recognition site for the initial binding of PrPC to PrPSc (39), and a synthetic peptide comprised of residues 119–141 has been shown to be a potent inhibitor of the prion protein conversion reaction in cell-free systems (40).
Studies aimed at dissecting molecular mechanisms of prion replication have been hampered by difficulties encountered in propagating the PrPC → PrPSc conversion in vitro. Recent data indicate that, in the presence of low concentrations of chemical denaturants, the recombinant prion protein can be transformed to an oligomeric β-sheet-rich structure with physicochemical properties resembling those of the PrPSc isoform (41–44). However, attempts to use these recombinant PrPSc-like oligomers as seeds that could catalyze the conversion of PrPC under native conditions have not yet been successful. The most advanced and best documented system for PrPSc-templated prion protein conversion outside the living cell is the one developed by Caughey and coworkers (39, 45, 46). However, even in this case, the yields of the conversion are perplexingly low, limited to substoichiometric quantities of a newly converted protein in relation to input PrPSc. Such yields are insufficient to sustain continuous self-propagation of the prion state. This apparent conversion resistance of the full-length prion protein sharply contrasts with the behavior of the huPrP145Stop variant explored in this study. In this case, a very small amount of preformed fibrils is sufficient to initiate the conversion reaction. Furthermore, once triggered, this conversion propagates until essentially complete depletion of the monomeric protein is observed.
The striking difference in the conversion propensity of the full-length PrP vs. the C-terminally truncated variant huPrP145Stop suggests that, in the full-length protein, the site(s) critical for nucleating the conversion may be “protected” by intramolecular contacts with the C-terminal domain and, thus, unavailable for intermolecular interactions. This model is consistent with the present finding regarding the importance in the conversion of the region around residues 138–141. Although unrestricted in huPrP145Stop, in the full-length protein, this region maps to the loop that constitutes an integral part of the folded domain (20, 21). Residues in this loop are anchored to the rest of the folded domain by nearby α-helix 1 and β-strand 1. Furthermore, inspection of NMR data indicates close proximity and likely interactions between the side chains of Tyr-150 and Phe-141. Consequently, we hypothesize that the full-length PrP could acquire a conversion-competent state only on removal or destabilization of intramolecular interactions that restrict the loop region. Such a conversion-competent state could correspond to the fully unfolded protein or to a partially structured folding intermediate. The latter possibility seems to be more likely, especially in view of recent experimental evidence for the population of intermediate states in prion protein folding (47, 48). Partially structured folding intermediates are also believed to play a central role in amyloid formation by proteins such as transthyretin or lysozyme variants (49, 50).
A number of studies have noted overall similarities in structural organization of mammalian PrP and prion-like proteins from yeast and fungi (see ref. 9 and references therein). All these proteins are characterized by a folded globular domain and a highly flexible, largely unstructured N-terminal (PrP, Sup35, Ure2p) or C-terminal (HET-s) region. However, the present data suggest that this similarity does not fully extend to the functional role of individual structural domains in prion propagation. In yeast and fungal prion proteins, the determinants responsible for prion state formation map entirely to the unstructured domains although the efficiency of prion appearance may be modulated by elements outside these regions (9). Furthermore, these unstructured prionogenic domains are fully capable of self-propagating conversion, even in the absence of the folded domains (9, 23, 51, 52). In contrast to these proteins, the polypeptide encompassing the entire flexible N-terminal domain of mammalian PrP (residues 23–124) is not amyloidogenic, and the region critical for PrP conversion seems to extend into the folded C-terminal domain. It is tempting to speculate that such a “shielding” of the amyloidogenic region in PrP by the folded domain may reflect evolutionary selection toward preventing protein conversion into the extremely toxic PrPSc state.
Acknowledgments
We thank Dr. Mark Bell for help with preliminary FTIR experiments and Dr. Frank Sonnichsen for helpful discussions. This work was supported by National Institutes of Health Grant NS 44158.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GSS disease, Gerstmann–Straussler–Scheinker disease; PrP, prion protein; PrPC, cellular PrP isoform; PrPSc, scrapie PrP isoform; huPrP, human prion protein; ThT, thioflavine T; FTIR, Fourier-transform infrared; AFM, atomic force microscopy
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissman, C. (1996) FEBS Lett. 398, 3–11. [Google Scholar]
- 3.Caughey B. & Chesebro, B. (2001) Adv. Virus Res. 56, 277–311. [DOI] [PubMed] [Google Scholar]
- 4.Collinge, J. (2001) Annu. Rev. Neurosci. 24, 519–550. [DOI] [PubMed] [Google Scholar]
- 5.Meyer, R. K., McKinley, M. P., Bowman, K. A., Braunfeld, M. B., Barry, R. A. & Prusiner, S. B. (1986) Proc. Natl. Acad. Sci. USA 83, 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caughey, B. W., Dong, A., Bhat, K. S., Ernst, D., Hayes, S. F. & Caughey, W. S. (1991) Biochemistry 30, 7672–7680. [DOI] [PubMed] [Google Scholar]
- 7.Gasset, M., Baldwin, M. A., Fletterick, R. J. & Prusiner, S. B. (1993) Proc. Natl. Acad. Sci. USA 90, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prusiner, S. B. (1982) Science 216, 136–144. [DOI] [PubMed] [Google Scholar]
- 9.Wickner, R. B., Taylor, K. L., Edskes, H. K., Maddelein, M. L., Moriyama, H. & Roberts, T. J. (2000) J. Struct. Biol. 130, 310–322. [DOI] [PubMed] [Google Scholar]
- 10.Lindquist, S. (1997) Cell 89, 495–498. [DOI] [PubMed] [Google Scholar]
- 11.Maddelein, M.-L., Dos Reis, S., Duvezin-Caubet, S., Coulary-Salin, B. & Saupe, S. J. (2002) Proc. Natl. Acad. Sci. USA 99, 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambetti, P., Petersen, R. B., Parchi, P., Chen, S. G., Capellari, S., Goldfarb, L., Gabizon, R., Montagna, P., Lugaresi, E., Picardo, P. & Ghetti, B. (1999) in Prion Biology and Diseases, ed. Prusiner, S. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 509–583.
- 13.Tateishi, J., Kitamoto, T., Hoque, M. Z. & Furukawa, H. (1996) Neurology 46, 532–537. [DOI] [PubMed] [Google Scholar]
- 14.Kitamoto, T., Iizuka, R. & Tateishi, J. (1993) Biochem. Biophys. Res. Commun. 192, 525–531. [DOI] [PubMed] [Google Scholar]
- 15.Ghetti, B., Piccardo, P., Spillantini, M. G., Ichimiya, Y., Porro, M., Perini, F., Kitamoto, T., Tateishi, J., Seiler, C., Frangione, B., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morillas, M., Swietnicki, W., Gambetti, P. & Surewicz, W. K. (1999) J. Biol. Chem. 274, 36859–36865. [DOI] [PubMed] [Google Scholar]
- 17.Zahn, R., von Schroetter, C. & Wuthrich, K. (1997) FEBS Lett. 417, 400–404. [DOI] [PubMed] [Google Scholar]
- 18.Naiki, H., Higuchi, K., Hosokawa, M. & Takeda T. (1989) Anal. Biochem. 177, 244–249. [DOI] [PubMed] [Google Scholar]
- 19.Surewicz, W. K. & Mantsch, H. H. (1988) Biochim. Biophys. Acta 952, 115–130. [DOI] [PubMed] [Google Scholar]
- 20.Riek, R., Hornemann, S., Wider, G., Glockshuber, R. & Wuthrich, K. (1997) FEBS Lett. 413, 282–288. [DOI] [PubMed] [Google Scholar]
- 21.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahn, R., Liu, A., Luhrs, T., Riek, R., von Schroetter, C., Lopez, G. F., Billeter, M., Calzolai, L., Wider, G. & Wuthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serio, T. R., Cashikar, A. G., Kowal, A. S., Sawicki, G. J., Moslehi, J. J., Serpell, L., Arnsdorf, M. F. & Lindquist, S. L. (2000) Science 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, K. L., Cheng, N., Williams, R. W., Steven, A. C. & Wickner, R. B. (1999) Science 283, 1339–13343. [DOI] [PubMed] [Google Scholar]
- 25.Tagliavini, F., Prelli, F., Verga, L., Giaccone, G., Sarma, R., Gorevic, P., Ghetti, B., Passerini, F., Ghibaudi, E., Forloni, G., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 9678–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forloni, G., Angeretti, N., Chiesa, R., Monzani, E., Salmona, M., Bugiani, O. & Tagliavini, F. (1993) Nature 362, 543–546. [DOI] [PubMed] [Google Scholar]
- 27.Selvaggini, C., De Gioia, L., Cantu, L., Ghibaudi, E., Diomede, L., Passerini, F., Forloni, G., Bugiani, O., Tagliavini, F. & Salmona, M. (1993) Biochem. Biophys. Res. Commun. 194, 1380–1386. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, H., Kaneko, K., Nguyen, J. T., Livshits, T. L., Baldwin, M. A., Cohen, F. E., James, T. L. & Prusiner, S. B. (1995) J. Mol. Biol. 250, 514–526. [DOI] [PubMed] [Google Scholar]
- 29.Tagliavini, F., Lievens, P. M.-J., Tranchant, C., Warter, J.-M., Mohr, M., Giaconne, G., Perini, F., Rossi, G., Salmona, M., Piccardo, P., et al. (2001) J. Biol. Chem. 276, 6009–6015. [DOI] [PubMed] [Google Scholar]
- 30.Brown, D. R., Schmidt, B. & Kretzschmar, H. A. (1996) Nature 380, 345–347. [DOI] [PubMed] [Google Scholar]
- 31.Alonso, D. O. V. & Daggett, V. (2001) Adv. Protein Chem. 57, 107–137. [DOI] [PubMed] [Google Scholar]
- 32.Tagliavini, F., Prelli, F., Porro, M., Rossi, G., Giaccone, G., Farlow, M. R., Dlouhy, S. R., Ghetti, B., Bugiani, O. & Frangione, B. (1994) Cell 79, 695–703. [DOI] [PubMed] [Google Scholar]
- 33.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R. B., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, E. E., et al. (1997) J. Mol. Biol. 273, 614–622. [DOI] [PubMed] [Google Scholar]
- 34.Holscher, C., Delius, H. & Burkle, A. (1998) J. Virol. 72, 1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priola, S. A. & Chesebro, B. (1995) J. Virol. 69, 7754–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramoto, T., Scott, M., Cohen, F. E. & Prusiner, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 15457–15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko, K., Ball, H. L., Wille, H., Zhang, H., Groth, D., Torchia, M., Tremblay, P., Safar, J., Prusiner, S. B., DeArmond, S. J., et al. (2000) J. Mol. Biol. 295, 997–1007. [DOI] [PubMed] [Google Scholar]
- 38.Goldmann, W., Martin, T., Foster, J., Hughes, S., Smith, G., Hughes, K., Dawson, M. & Hunter, N. (1996) J. Gen. Virol. 77, 2885–2891. [DOI] [PubMed] [Google Scholar]
- 39.Horiuchi, M. & Caughey B. (1999) EMBO J. 18, 3193–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chabry, J., Caughey, B. & Chesebro, B. (1998) J. Biol. Chem. 273, 13203–13207. [DOI] [PubMed] [Google Scholar]
- 41.Jackson, G. S., Hosszu, L. L. P., Power, A., Hill, A. F., Kenney, J., Saibil, H., Craven, C. J., Waltho, J. P., Clarke, A. R. & Collinge, J. (1999) Science 283, 1935–1937. [DOI] [PubMed] [Google Scholar]
- 42.Swietnicki, W., Morillas, M., Chen, S. G., Gambetti, P. & Surewicz, W. K. (2000) Biochemistry 39, 424–431. [DOI] [PubMed] [Google Scholar]
- 43.Morillas, M., Vanik, D. L. & Surewicz, W. K. (2001) Biochemistry 40, 6982–6987. [DOI] [PubMed] [Google Scholar]
- 44.Baskakov, I. V., Legname, G., Prusiner, S. B. & Cohen, F. E. (2001) J. Biol. Chem. 276, 19687–19690. [DOI] [PubMed] [Google Scholar]
- 45.Kocisko, D. A., Come, J. H., Priola, S. A., Chesebro, B., Raymond, G. J., Lansbury, P. T. & Caughey, B. (1994) Nature 370, 471–474. [DOI] [PubMed] [Google Scholar]
- 46.Caughey, B. (2001) Trends Biochem. Sci. 26, 235–242. [DOI] [PubMed] [Google Scholar]
- 47.Apetri, A. C. & Surewicz, W. K. (2002) J. Biol. Chem. 47, 44589–44592. [DOI] [PubMed] [Google Scholar]
- 48.Kuwata, K., Li, H., Yamada, H., Legname, G., Prusiner, S. B., Akasaka, K. & James, T. L. (2002) Biochemistry 41, 12277–12283. [DOI] [PubMed] [Google Scholar]
- 49.Kelly, J. W. (1997) Structure 5, 595–600. [DOI] [PubMed] [Google Scholar]
- 50.Canet, D., Last, A. M., Tito, P., Sunde, M., Spencer, A., Archer, D. B., Redfield, C., Robinson, C. V. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 308–315. [DOI] [PubMed] [Google Scholar]
- 51.Glover, J. R., Kowal, A. S., Schirmer, E. C., Patino, M. M., Liu, J.-J. & Lindquist, S. (1997) Cell 89, 811–819. [DOI] [PubMed] [Google Scholar]
- 52.Balguerie, A., Dos Reis, S., Ritter, C., Chaignepain, S., Coulary-Salin, B., Forge, V., Bathany, K., Lascu, I., Schmitter, J.-M., Riek, R., et al. (2003) EMBO J. 22, 2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]