Abstract
The biological formation of inorganic materials with complex form (biominerals) is a widespread phenomenon in nature, yet the molecular mechanisms underlying biomineral morphogenesis are not well understood. Among the most fascinating examples of biomineral structures are the intricately patterned, silicified cell walls of diatoms, which contain tightly associated organic macromolecules. From diatom biosilica a highly polyanionic phosphoprotein, termed native silaffin-2 (natSil-2), was isolated that carries unconventional amino acid modifications. natSil-2 lacked intrinsic silica formation activity but was able to regulate the activities of the previously characterized silica-forming biomolecules natSil-1A and long-chain polyamines. Combining natSil-2 and natSil-1A (or long-chain polyamines) generated an organic matrix that mediated precipitation of porous silica within minutes after the addition of silicic acid. Remarkably, the precipitate displayed pore sizes in the range 100–1000 nm, which is characteristic for diatom biosilica nanopatterns.
The biological formation of intracellular or extracellular skeletons made of amorphous hydrated SiO2 (biosilica) is relatively frequent among the protists and also occurs in many higher plants (1, 2). To date the organic molecules involved in biosilica formation have mainly been studied in sponges and diatoms (3). From the silica spicules of the sponge Thetya aurantia, proteaselike proteins termed silicateins were characterized that catalyze silica formation from tetraethoxysilane in vitro (4, 5). Diatoms are unicellular algae that have the extraordinary capability to produce an enormous variety of biosilica structures (6). Each diatom species is characterized by a specific biosilica cell wall that contains regularly arranged slits or pores in the size range between 10 and 1,000 nm (nanopatterned biosilica). Biosilica morphogenesis takes place inside the diatom cell within a specialized membrane-bound compartment termed the silica deposition vesicle (SDV). It has been postulated that the SDV contains a matrix of organic macromolecules that not only regulate silica formation but also act as templates to mediate biosilica nanopatterning (7–9). Insight into the nature of this organic matrix has been gained through the characterization of diatom biosilica-associated peptides (silaffins) and long-chain polyamines (LCPA), both of which accelerate silica formation from a silicic acid solution in vitro (10–12). Although native silaffin-1A (natSil-1A) and LCPA, respectively, mediate the formation of unusual silica structures in vitro (networks of irregularly shaped silica bands and large spherical silica particles) (11, 12), none of these are akin to diatom biosilica nanopatterns.
Recently, it was shown that phosphorylation of natSil-1A is essential for silica formation activity (12), leading to the speculation that phosphorylated components may be generally important for biosilica morphogenesis. Therefore, a search for additional silica-associated phosphoproteins, which are able to influence silica formation, was initiated. Here we describe the isolation and chemical characterization of the polyanionic phosphoprotein natSil-2, which strongly affects the silica-forming properties of both natSil-1A and LCPA. We demonstrate that diatom-like silica nanopatterns can be generated in vitro by mixtures of natSil-2 and natSil-1A (or LCPA), which spontaneously self-assemble into an organic matrix. We suggest that the formation of organic matrices composed of polyanionic natSil-2-like phosphoproteins and polycationic silica-forming components may represent a widespread mechanism in diatom biosilica morphogenesis.
Materials and Methods
Metabolic Radiolabeling of Cylindrotheca fusiformis. Culture conditions for C. fusiformis were as described (13). Cells were pelleted by centrifugation and washed three times in phosphate-free artificial seawater culture medium (ASW-P), by repeated centrifugation and resuspension. After the last washing step cells were resuspended in ASW-P and starved of phosphate for 24 h. After adding 5 μCi (1 μCi = 37 kBq) of [33P]phosphoric acid per ml of medium and 25 μM Na2HPO4, cells were cultivated for 24 h, during which they incorporated 95% of the phosphate. For radiolabeling with [35S]sulfate, cells were washed with sulfate-free culture medium (ASW-S) and starved of sulfate in ASW-S for 24 h. Subsequently, 2.5 μCi of sodium [35S]sulfate per ml of medium and 50 μM MgSO4 were added and cells were cultivated for 24 h. During this time 54% of the sulfate was incorporated. The radiolabeled cells were pelleted by centrifugation, washed with ASW medium, and then subjected to cell wall isolation as described (14).
Isolation of natSil-2. Ammonium fluoride extraction of purified cell walls and dialysis of the extract were as described (12). To remove positively charged organic components the cation-exchange resin HighS (Bio-Rad) was added to the dialysate and the suspension was mixed by gentle rotation for 1 h at 20°C. The resin was removed by filtration and washed with 50 mM ammonium acetate. The combined supernatant and wash solution were concentrated by rotary evaporation under reduced pressure and subjected to fractionation on a size-exclusion column [Superose 12 HR-10/30 (Amersham Pharmacia Biotech), 500 mM NaCl/50 mM ammonium acetate buffer, pH 6.5, 0.25 ml/min flow rate, detection wavelength 226 nm]. Fractions were analyzed by N-tris(hydroxymethyl)methylglycine (Tricine)/SDS/PAGE (15) and staining with Stains-All (Sigma). Fractions containing natSil-2 were pooled, diluted 3-fold by adding water, and loaded onto a Mono Q HR-5/5 column (Amersham Pharmacia Biotech) that was equilibrated with 50 mM Tris·HCl, pH 8.0 (buffer A). Elution was performed at a flow rate of 0.5 ml/min by increasing the concentration of buffer B (1 M NaCl/50 mM Tris·HCl, pH 8.0) to 100% within 1 h. natSil-2 fractions eluting around 500 mM NaCl were combined, exhaustively dialyzed against 10 mM ammonium acetate, and lyophilized. The dry residue was dissolved in water, adjusted to pH 5.5 with acetic acid/NaOH, and stored frozen at -20°C until use.
Chemical Analysis of natSil-2. Phosphate (16), sulfate (17), and carbohydrate (18) contents were determined according to standard procedures. For analysis of the carbohydrate composition natSil-2 was hydrolyzed in vacuo (4 M trifluoroacetic acid, 100°C, 4 h) and subsequently lyophilized. The residue was dissolved in 15 mM NaOH and subjected to high-pressure anion-exchange chromatography at a flow rate of 1 ml/min in a CarboPac PA1 column (Dionex). Separation of carbohydrates was achieved by the following program: 15 min of 15 mM NaOH (isocratic), within 20 min up to 80 mM NaOH/80 mM sodium acetate (linear gradient), and 15 min of 80 mM NaOH/80 mM sodium acetate (isocratic). The eluate was monitored by pulsed amperometric detection (ED40, Dionex).
Treatment of natSil-2 with Trifluoromethanesulfonic Acid (TFMS). A dry pellet of natSil-2 was dissolved in 300 μl of TFMS (Aldrich) and incubated for 20 min at 0°C. Subsequently, the solution was neutralized by dropwise addition of 6.2 M pyridine (in ethanol), which caused the formation of a precipitate after 300 μl of the pyridine solution was added. The precipitate was completely dissolved by adding 200 μl of water, and neutralization was quickly completed by adding 400 μl of the pyridine solution. The neutral solution was exhaustively dialyzed against 10 mM ammonium acetate, the dialysate was centrifuged, and the supernatant (containing the deglycosylated natSil-2) was lyophilized to dryness. The dried residue was either dissolved in sample buffer for SDS/PAGE analysis (in the case of radiolabeled material) or further purified for silica precipitation assays. For further purification the residue was dissolved in 500 μl of water and subjected to size-exclusion chromatography (Superose 12 HR-10/30, 250 mM ammonium acetate buffer, 0.25 ml/min flow rate, detection wavelength 226 nm]. Fractions eluting between 12.5 and 15.5 ml contained deglycosylated natSil-2 and were combined. After drying by lyophilization the residue was dissolved in water, adjusted to pH 5.5 with acetic acid/NaOH, and stored frozen at -20°C until use. When standard assays (17, 18) were used, neither carbohydrate nor sulfate was detected in 60 nmol of deglycosylated natSil-2 containing 60 nmol of chemically bound phosphate.
Preparation of the 9-Fluorenylmethoxycarbonyl (Fmoc) derivative of Phosphohydroxyproline (HypP). The HypP-containing peptide H2N-Leu-Lys-Arg-Ala-HypP-Leu-Gly-amide (Biomedical Research Facilities, University of Michigan, Ann Arbor) was subjected to gentle acid hydrolysis (3 h, 110°C, 6 M HCl) and subsequently lyophilized. The dry residue was dissolved in 100 mM formic acid and passed through a Dowex 50W-X8 (H+ form) column to enrich for acidic compounds. The flow-through was lyophilized and derivatized with Fmoc according to a standard procedure (19).
Silica Formation in Vitro. Reaction mixtures were prepared in 50 mM sodium acetate/acetic acid, pH 5.5, without silicic acid and incubated for 30 min at 18°C. A stock solution of monosilicic acid was prepared by hydrolyzing 1 M tetramethoxysilane in 1 mM HCl for 10 min at 25°C. Immediately after incubation, aliquots of the silicic acid solution were withdrawn and added to the reaction mixtures to yield a final concentration of 100 mM silicic acid. Subsequently, the reaction mixtures were incubated for a further 10 min at 18°C and then immediately processed either for quantification of silica as described (10) or for scanning electron microscopy (SEM) analysis. To process the samples for SEM analysis the reaction mixtures were centrifuged for 2 min at 16,000 × g and resuspended in 5 μl of water. The suspension was loaded on a 200-mesh copper grid (diameter 3.05 mm; Agar Scientific, Stansted, Essex, U.K.), washed, and dried as described (12).
Electron Microscopy. SEM images were obtained on a LEO 1530 field emission scanning electron microscope (Oberkochen, Germany). Energy-dispersive x-ray analysis (EDXA) was performed by using the INCA 200 system equipped with a Si(Li)-detector (Oxford).
Dynamic Light Scattering. Determination of natSil-2 particle size was performed in aqueous solutions containing 50 mM sodium acetate/acetic acid, pH 5.5, by using the HPPS 5001 system (Malvern Instruments, Worcestershire, U.K.). Three independent preparations of natSil-2 were analyzed, and each measurement was repeated three times.
MS. LCPA were isolated as described (11) and analyzed by electrospray ionization mass spectrometry (ESI-MS) using a single quadrupole instrument (SSQ 7000, Finnigan). Samples were infused with 1 mM ammonium acetate/50% acetonitrile. The following [M + H]+ masses (Da) were detected: 830.8, 887.2, 944.2, 1,001.8, and 1,058.1. The complete acid hydrolysate of natSil-2 (24 h, 110°C, 6 M HCl) was analyzed by using the same instrument, but 0.5% acetic acid/50% acetonitrile was used for infusion. Analysis of Fmoc-derivatized phosphoamino acids was performed by using an Ion Trap Esquire LC instrument (Bruker, Billerica, MA) in the negative-ion mode. Samples were infused by a nanospray source in 0.5 mM ammonium acetate/50% acetonitrile.
Results and Discussion
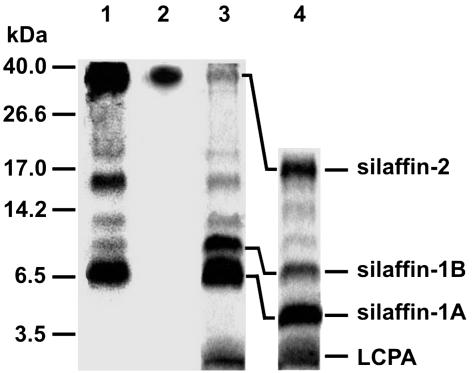
To identify biosilica-associated phosphoproteins, metabolic labeling of a cell culture of the diatom C. fusiformis by using [33P]phosphate was performed. After isolation of biosilica and extraction with ammonium fluoride, phosphoproteins were detected by SDS/PAGE and subsequent fluorography. Besides natSil-1A (6.5 kDa), strongly labeled phosphoproteins with apparent molecular masses of 40 and 16 kDa were identified as well as weakly labeled phosphoproteins of 20, 12, and 10 kDa (Fig. 1, lane 1). Staining of the SDS/polyacrylamide gel with Coomassie blue detected all these phosphoproteins and a <3.5-kDa nonphosphorylated component (Fig. 1, lane 3). Previously, extraction of the C. fusiformis cell walls by using anhydrous hydrogen fluoride (HF), a reagent that dissolves silica and cleaves phosphate esters and O-glycosidic bonds (20), led to the identification of silaffin-1A (4 kDa), silaffin-1B (8 kDa), and silaffin-2 (17 kDa) (10). In agreement with this result, HF treatment of the ammonium fluoride extract converted natSil-1A, the 10-kDa phosphoprotein, and the 40-kDa phosphoprotein into silaffin-1A, -1B, and -2, respectively, by cleaving off HF-labile modifications (Fig. 1, lane 4). Accordingly, the 10- and 40-kDa phosphoproteins were termed natSil-1B and natSil-2. Analysis by ESI-MS revealed that the <3.5-kDa nonphosphorylated component in the ammonium fluoride extract represents the LCPA previously characterized from the HF cell wall extract of C. fusiformis (11).
Fig. 1.
Tricine/SDS/PAGE analysis (15) of ammonium fluoride extracts from C. fusiformis biosilica. Preparation of biosilica and extraction with ammonium fluoride were performed as described (12). Lane 1, ammonium fluoride extract from 33P-labeled biosilica, fluorography; lane 2, ammonium fluoride extract, Stained-All stained; lane 3, ammonium fluoride extract, Coomassie bluestained; lane 4, HF-treated ammonium fluoride extract, Coomassie bluestained.
natSil-2 is the only component of the ammonium fluoride extract that is stained by the polycationic cyanine dye Stains-All (Fig. 1, lane 2), which is diagnostic of a high negative charge density (21). All other phosphoproteins exhibited basic isoelectric points as revealed by their quantitative adsorption to a cation-exchange resin at pH 7 (unpublished results). This striking charge difference prompted the assumption that natSil-2 may exhibit silica formation properties that differ from those of natSil-1A and LCPA. Surprisingly, natSil-2 proved to be incapable of precipitating silica from a silicic acid solution in vitro, yet a mixture of natSil-2 and LCPA rapidly precipitates silica under conditions at which neither of the two organic components alone is able to form silica (Table 1). The activating effect of natSil-2 on LCPA-dependent silica formation can be mimicked by multivalent inorganic anions such as phosphate (Table 1) or sulfate (unpublished results), suggesting that silica formation is elicited by the binding of the polycationic LCPA molecules to the numerous negatively charged sites in natSil-2. This interpretation is in accordance with the structural requirements for natSil-1A-dependent silica precipitation, because in this molecule the polypeptide backbone is covalently linked with both LCPA moieties and multiple phosphate groups (12). Unexpectedly, the silica precipitation activity of natSil-1A was impeded by natSil-2 (Table 1), demonstrating that it can also act as an inhibitor of silica formation.
Table 1. Silica precipitation activity of silaffins and LCPA.
| SiO2 formed, mM
|
|||
|---|---|---|---|
| Silaffin/polyamine | No additions | Phosphate (30 mM) | natSil-2 (2 units/μl) |
| natSil-2 (4 units/μl) | 0 | 0 | ND |
| LCPA (0.6 μg/μl) | 0 | 31.7 | 30.6 |
| natSil-1A (0.3 mM) | 40.9 | ND | 28.2 |
| Deglycosylated natSil-2 (4 units/μl) | 33.2 | ND | ND |
Silica formation assays were performed in the presence of the components indicated (the final concentration of each component is given in parentheses). Where indicated, phosphate was added as sodium phosphate, pH 5.5. Concentrations of silica, natSil-1A, and LCPA were determined as described (11, 12). Concentrations of natSil-2 and deglycosylated natSil-2 are expressed according to phosphate content (1 unit is defined as 1 nmol of silaffin-bound phosphate). ND, not determined.
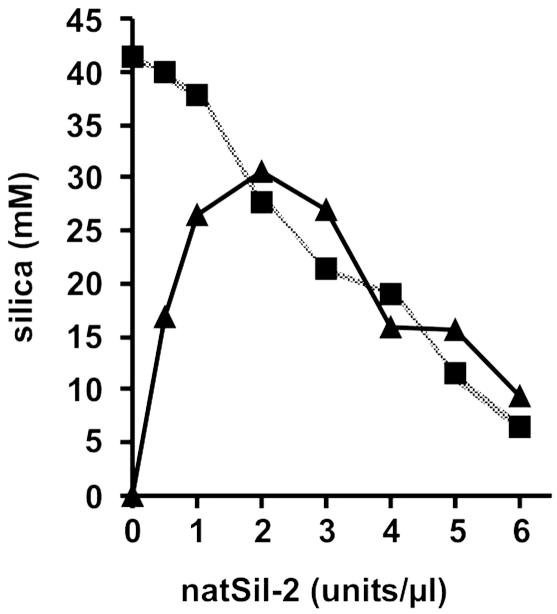
To investigate these seemingly contradictory properties of natSil-2, the silica formation activities of natSil-1A and LCPA were analyzed as a function of natSil-2 concentration. In the LCPA/natSil-2 system both components are required for silica formation and therefore the activity initially increased with increasing concentration of the limiting component natSil-2. However, at concentrations beyond 2 units/μl, natSil-2 did not function exclusively as an activator but also exerted an inhibitory effect, leading to a linear decrease (slope: -4.3 nmol/unit) of silica precipitation activity (Fig. 2). The same constant decline in activity (slope: -4.8 nmol/unit) was observed in the natSil-1A/natSil-2 system throughout the entire natSil-2 concentration range, suggesting that inhibition in both systems is caused by the same molecular mechanism. If so, the inhibitory effect of the polyanionic natSil-2 molecules is likely exerted by means of electrostatic shielding of the polycationic polyamine chains, because these represent the common structural elements of LCPA and natSil-1A, and polyamines are known to catalyze silica formation (10, 11, 22, 23). Thus, the high negative charge density of natSil-2 appears to be an essential feature for regulating the silica precipitation activities of natSil-1A and LCPA.
Fig. 2.
Influence of natSil-2 on the silica precipitation activities of natSil-1A and LCPA. Silica precipitation was performed under standard assay conditions in the presence of a constant concentration of LCPA (0.6 μg/μl, ▴) or natSil-1A (0.3 mM, ▪).
To identify the negatively charged groups in natSil-2, we analyzed its chemical structure. HF treatment converts natSil-2 into silaffin-2 (see Fig. 1), a strongly positively charged protein (unpublished results), demonstrating that the high negative charge density of natSil-2 results solely from its HF-labile post-translational modifications. These modifications include phosphate, sulfate, and carbohydrate residues in a molar ratio of 1:1:7.8. The carbohydrate composition is rather complex: galactose, rhamnose, glucuronic acid, fucose, glucosamine, and a monomethylated deoxyhexose were determined in a molecular ratio of 9:7:6:2:1:2. Because of the abundance of glucuronic acid, the carbohydrate moieties significantly add to the high negative charge density of natSil-2.
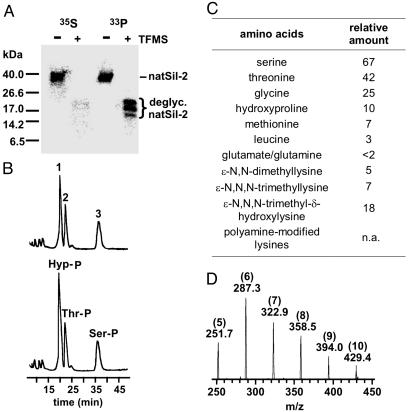
Treatment of natSil-2 with the deglycosylating agent trifluoromethanesulfonic acid completely removes the carbohydrate as well as the sulfate moieties, yet phosphorylation is fully retained (Fig. 3A), demonstrating that all phosphate groups in natSil-2 are directly linked to the polypeptide backbone. The phosphorylated amino acids of 33P-labeled natSil-2 were analyzed by anion-exchange chromatography of the respective Fmoc derivatives by following standard procedures (19, 24). According to elution time and mass spectrometric analysis, fractions 2 and 3 are identical to Fmoc-phosphothreonine (420 Da) and Fmoc-phosphoserine (406 Da), respectively (Fig. 3B). The molecular mass of fraction 1 (432 Da) corresponded to the Fmoc derivative of phosphorylated hydroxyproline, an amino acid derivative that has not yet been found in any other biogenic protein (25). Hydroxyproline is indeed a constituent of the natSil-2 polypeptide backbone (Fig. 3C) and the presence of its phosphorylated derivative in natSil-2 was confirmed by cochromatography of fraction 1 with synthetic Fmoc-phosphohydroxyproline (Fig. 3B). The remarkable occurrence of phosphorylated hydroxyproline residues adds further support to the hypothesis that a high negative charge density is important for natSil-2 function.
Fig. 3.
Chemical composition of natSil-2. (A) Analysis of natSil-2 labeled with 35S and 33P, respectively, before (-) and after (+) treatment with trifluoromethanesulfonic acid (Tricine/SDS/PAGE, fluorography). Deglycosylated (deglyc.) natSil-2 is composed of phosphorylation isoforms that appear as three discrete bands on SDS/PAGE. Residual 35S label in deglycosylated [35S]natSil-2 is due to its methionine content. (B) Upper trace, analysis of Fmoc-derivatized phosphoamino acids of natSil-2 by anion-exchange chromatography (photometric detection at 256 nm). Fraction numbers are indicated. Lower trace, synthetic Fmoc-phosphohydroxyproline (Hyp-P) was added before chromatography. Ser-P, Fmoc-phosphoserine; Thr-P, Fmoc-phosphothreonine. (C) Amino acid composition of natSil-2 after complete acid hydrolysis (24 h, 110°C, 6 M HCl). Relative amounts were determined from the peak areas in the reverse-phase HPLC chromatogram of the phenyl isothiocyanate (PITC) derivatives (PITC-hydroxyproline peak area defined as 10). Polyamine-modified lysines could not be quantified, because the corresponding PITC derivatives do not show up in the chromatogram. n.a., Not applicable. (D) ESI-MS spectrum of natSil-2 after complete acid hydrolysis (24 h, 110°C, 6 M HCl). Each peak represents a doubly charged positive ion. Note the constant difference of m/z = 35.5 between neighboring peaks, which is indicative of a repeated unit element of 71 Da. The absolute molecular masses, in Da, of the corresponding uncharged molecules (501.4, 272.6, 643.8, 715.0, 786.0, and 856.8) reveal 5–10 repeats (see numbers in parentheses) of N-methylpropylenimine (71 Da) attached to a lysine residue (10).
Because of the extremely high number of modified amino acids, peptide mapping of natSil-2 did not yield sufficient sequence information to clone the corresponding gene. Thus, the primary structure of the natSil-2 polypeptide backbone is so far unknown. To characterize its amino acid composition, exhaustive acid hydrolysis of natSil-2 was performed (24 h, 110°C, 6 M HCl), and the hydrolysate obtained was analyzed by two methods. Method A involved phenyl isothiocyanate (PITC) derivatization and subsequent separation by reverse-phase HPLC to identify standard amino acids and to determine their relative quantities. In method B ESI-MS without prior derivatization was used to identify modified amino acids that were not detected by method A. The amino acid composition of the natSil-2 polypeptide is dominated by hydroxyamino acids and glycine, which is also characteristic for natSil-1A (26), but the presence of significant amounts of methionine and leucine is a distinct feature of natSil-2 (Fig. 3C). Surprisingly, ESI-MS detected in natSil-2 the same type of LCPA-modified lysines (Fig. 3D) that confer silica precipitation activity on natSil-1A (10), yet natSil-2 itself is incapable of precipitating silica (Table 1). This apparent contradiction was resolved by the finding that deglycosylated natSil-2 has an intrinsic silica precipitation activity (Table 1), demonstrating that in natSil-2 the high number of negative charges conferred by the carbohydrate and sulfate moieties autoinhibit the silica formation activity of the polyamine side chains. This observation clearly confirms the proposed inhibitory effect of a high negative charge density on polyamine-dependent silica formation. The polyamine moieties in natSil-2 may therefore be exclusively devoted to mediating the self-assembly of natSil-2 molecules (see below) by electrostatic interactions with the numerous negatively charged moieties.
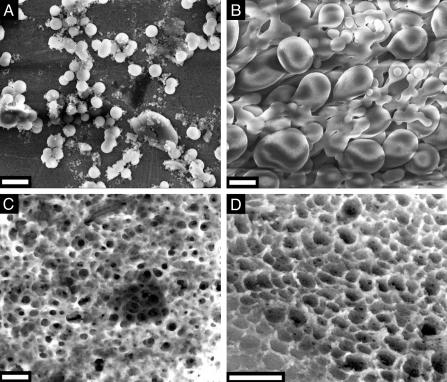
Despite its lack of silica formation activity, natSil-2 may be able to influence silica morphogenesis by means of the interaction with silica-forming molecules. To investigate this possibility, the structures of silica precipitates formed by mixtures of natSil-1A and natSil-2 were analyzed by SEM. At low and high natSil-2/natSil-1 concentration ratios, polydisperse silica spheres (Fig. 4A; diameters 100–1,000 nm) and large interconnected spherical or pear-shaped silica particles (Fig. 4B), respectively, were obtained. These structures are larger than but structurally similar to the silica spheres formed by pure natSil-1A (diameter 400–700 nm) (12). Thus, under these conditions natSil-2 does not have a marked effect on silica morphogenesis. However, at intermediate natSil-1A/natSil-2 concentration ratios totally different structures were formed, representing silica blocks that contain numerous irregularly arranged pores (Fig. 4 C and D). The pore diameters are confined to the range of 100–1,000 nm, matching the characteristic pore size range of diatom biosilica nanopatterns (8). Similar porous silica structures were also obtained with appropriate mixtures of natSil-2 and LCPA (unpublished results).
Fig. 4.
SEM analysis of silica precipitates formed by mixtures of natSil-1A and natSil-2. (A) natSil-2 at 0.5 units/μl, natSil-1A at 0.3 mM. (B) natSil-2 at 5.0 units/μl, natSil-1A at 0.3 mM. (C) natSil-2 at 2.0 units/μl, natSil-1A at 0.3 mM. (D) natSil-2 at 1.6 units/μl, natSil-1A at 0.2 mM. (Bars = 2 μm.)
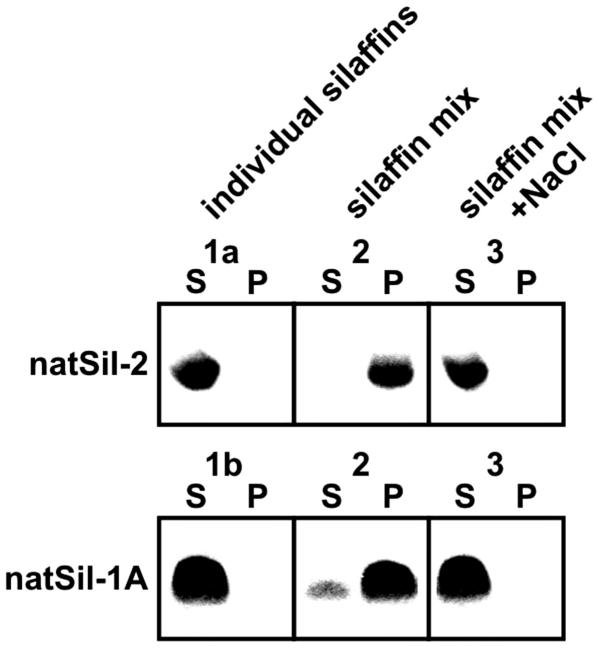
We propose that morphogenesis of the porous silica structures by mixtures of natSil-2 and natSil-1A (or LCPA) is mechanistically similar to the synthesis of mesoporous silica by surfactants and block copolymers. Mesoporous silica synthesis relies on the ability of organic molecules to build up 3D-nanostructured phases by self-assembly or cooperative aggregation with polysilicic acid molecules, thus acting as templates for the forming silica. In this way, nanopatterns with pore diameters in the range 2–50 nm can be produced, which correspond to the building block sizes of the nanostructured organic templates (27–30). When natSil-2 and natSil-1A are mixed in the absence of silicic acid a silaffin phase distinct from the bulk aqueous phase is formed, as is evident from the fact that both natSil-1A and natSil-2 become almost quantitatively pelleted by centrifugation (Fig. 5, sample 2). No silaffin pellet is formed by the individual components (Fig. 5, samples 1a and 1b) or in a mixture of both silaffins at elevated ionic strength (Fig. 5, sample 3), indicating that formation of the silaffin phase depends on ionic interactions between the oppositely charged constituents. If the silaffin phase does indeed act as an organic matrix in silica morphogenesis, the silica pore sizes of 100–1,000 nm should reflect the building block size of the silaffin matrix. Dynamic light scattering revealed that natSil-2 molecules form extremely large assemblies of 90 nm diameter, and the natSil-1A molecules have been shown to form 16-nm particles (12). These assemblies and particles cannot be pelleted by centrifugation. The lower silica pore size limit of 100 nm may be directly correlated to the size of the natSil-2 assemblies, and larger building blocks could be generated by fusion of a varying number of natSil-2 and natSil-1A particles attributable to the electrostatic interactions between their constituents (see above). At appropriate stoichiometric ratios of silaffins, unfused natSil-1A particles may remain, which catalyze silica formation around the natSil-2-containing building blocks, thus generating the porous silica structure. However, at extremes of natSil-2/natSil-1A concentration ratio, the predominance of natSil-2 or natSil-1A particles, respectively, may result in differently structured organic matrices that are unable to direct the formation of porous silica.
Fig. 5.
Formation of a silaffin phase by ionic interactions between natSil-1A and natSil-2. Samples 1a, 1b, 2, and 3 were incubated for 2 h at 18°C and subsequently centrifuged (15 min, 20,000 × g, 18°C). Aliquots of the pellets (P) and supernatants (S) were analyzed by Tricine/SDS/PAGE for natSil-1A (Coomassie blue staining) and natSil-2 (Stains-All staining). Composition of the samples: 1a, 2.0 units/μl natSil-2; 1b, 0.3 mM natSil-1A; 2, 2.0 units/μl natSil-2 plus 0.3 mM natSil-1A; 3, same as sample 2 plus 500 mM NaCl. Each sample contained 50 mM sodium acetate/acetic acid, pH 5.5.
The data presented in this study demonstrate that silica structures containing pores of a biologically relevant size range can be produced by a simple in vitro system that requires only two types of organic molecules: a polycationic component that is able to accelerate silica formation (natSil-1A or LCPA) and a polyanionic regulator of silica formation (natSil-2). We propose that the formation of a self-assembled organic matrix by means of electrostatic interactions of these molecules represents a widespread mechanism of diatoms to generate porous biosilica patterns. LCPA have been found in all diatom species investigated, and silaffins also are widespread (11) although not ubiquitously present in diatoms (31). In diatom species that lack silaffins, arrays of LCPA-containing microdroplets have been postulated as templates for the formation of hexagonal porous biosilica patterns (31). However, in a large number of diatom genera less symmetrical biosilica nanopatterns are observed (6), a fact that demands additional components that break up the inherently hexagonal arrangement of the proposed LCPA microdroplets. Presumably, this breakup is achieved by the formation of an organic matrix composed of LCPA and polyanionic silaffins.
Apart from their biological significance, the properties of silaffins may also be of interest within the area of materials engineering. Previously, a synthetic silaffin peptide has been used to generate ordered arrays of silica nanospheres within a polymer hologram, which may be of practical use for the fabrication of photonic devices (32). The silaffin phase discovered in the present study represents, to our knowledge, the only self-assembled matrix of organic macromolecules that is able to form macroporous silica (pore diameters >50 nm) within only a few minutes under extremely mild reaction conditions (pH 5.5, ambient pressure and temperature). Therefore, exploiting the properties of silaffins by using biomimetic approaches may in the future lead to the development of novel synthetic routes to silica-based macroporous materials.
Acknowledgments
We thank R. Deutzmann and E. Hochmuth for MS analysis and G. Lehmann for technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 521-A2) and the Fonds der Chemischen Industrie.
Abbreviations: LCPA, long-chain polyamines; Tricine, N-tris(hydroxymethyl)methylglycine; ESI-MS, electrospray ionization MS; Fmoc, 9-fluorenylmethoxycarbonyl; SEM, scanning electron microscopy.
References
- 1.Lowenstam, H. A. & Weiner, S. (1989) On Biomineralization (Oxford Univ. Press, Oxford).
- 2.Perry, C. C. & Keeling-Tucker, T. (2000) J. Biol. Inorg. Chem. 5, 537–550. [DOI] [PubMed] [Google Scholar]
- 3.Bäuerlein, E. (2000) Biomineralization (Wiley–VCH, Weinheim, Germany).
- 4.Shimizu, K., Cha, J., Stucky, G. D. & Morse, D. E. (1998) Proc. Natl. Acad. Sci. USA 95, 6234–6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha, J., Shimizu, K., Zhou, Y., Christiansen, S., Chmelka, B. F., Stucky, G. D. & Morse, D. E. (1999) Proc. Natl. Acad. Sci. USA 96, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Round, F., Crawford, R. & Mann, D. (1990) The Diatoms (Cambridge Univ. Press, Cambridge, U.K.).
- 7.Robinson, D. & Sullivan, C. (1987) Trends Biochem. Sci. 12, 151–154. [Google Scholar]
- 8.Pickett-Heaps, J., Schmid, A. M. M. & Edgar, L. A. (1990) in Progress in Phycological Research, eds. Round, F. E. & Chapman, D. J. (Biopress, Bristol, U.K.), Vol. 7, pp. 1–168. [Google Scholar]
- 9.Swift, D. M. & Wheeler, A. P. (1992) J. Phycol. 35, 202–209. [Google Scholar]
- 10.Kröger, N., Deutzmann, R. & Sumper, M. (1999) Science 286, 1129–1132. [DOI] [PubMed] [Google Scholar]
- 11.Kröger, N., Deutzmann, R., Bergsdorf, C. & Sumper, M. (2000) Proc. Natl. Acad. Sci. USA 97, 14133–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kröger, N., Lorenz, S., Brunner, E. & Sumper, M. (2002) Science 298, 584–586. [DOI] [PubMed] [Google Scholar]
- 13.Kröger, N., Bergsdorf, C. & Sumper, M. (1994) EMBO J. 13, 4676–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kröger, N., Lehmann, G., Rachel, R. & Sumper, M. (1997) Eur. J. Biochem. 250, 99–105. [DOI] [PubMed] [Google Scholar]
- 15.Schägger, H. & von Jagow, G. (1987) Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- 16.Buss, J. E. & Stull, J. T. (1983) Methods Enzymol. 99, 7–14. [DOI] [PubMed] [Google Scholar]
- 17.Terho, T. T. & Hartiala, K. (1971) Anal. Biochem. 41, 471–476. [DOI] [PubMed] [Google Scholar]
- 18.Dubios, M., Gilles, K. A., Hamilton, J. K. & Reber, P. A. (1956) Anal. Chem. 28, 350–356. [Google Scholar]
- 19.Ringer, D. P. (1991) Methods Enzymol. 201, 3–27. [DOI] [PubMed] [Google Scholar]
- 20.Mort, A. J. & Lamport, D. T. A. (1977) Anal. Biochem. 82, 289–309. [DOI] [PubMed] [Google Scholar]
- 21.Myers, J. M., Veis, A., Sabsay, B. & Wheeler, A. P. (1996) Anal. Biochem. 240, 300–302. [DOI] [PubMed] [Google Scholar]
- 22.Patwardhan, S. V. & Clarson, S. J. (2002) Silicon Chem. 1, 207–214. [Google Scholar]
- 23.Mitzutani, T., Nagase, H., Fujiwara, N. & Ogoshi, H. (1998) Bull. Chem. Soc. Jpn. 71, 2017–2022. [Google Scholar]
- 24.Niedbalski, J. S. & Ringer, D. P. (1986) Anal. Biochem. 158, 138–145. [DOI] [PubMed] [Google Scholar]
- 25.Sickmann, A. & Meyer, H. E. (2001) Proteomics 1, 200–206. [DOI] [PubMed] [Google Scholar]
- 26.Kröger, N., Deutzmann, R. & Sumper, M. (2001) J. Biol. Chem. 276, 26066–26070. [DOI] [PubMed] [Google Scholar]
- 27.Monnier, A. F., Schüth, F., Huo, Q., Kumar, D., Margolese, D., Maxwell, R. S., Stucky, G. D., Krishnamurty, M., Petroff, P., Firouzi, A., et al. (1993) Science 261, 1299–1303. [DOI] [PubMed] [Google Scholar]
- 28.Polarz, S. & Antonietti, M. (2002) Chem. Commun. 22, 2593–2604. [DOI] [PubMed] [Google Scholar]
- 29.Förster, S. & Plantenberg, T. (2002) Angew. Chem. Int. Ed. 41, 688–714. [PubMed] [Google Scholar]
- 30.Cölfen, H. & Mann, S. (2003) Angew. Chem. Int. Ed. 42, 2350–2365. [DOI] [PubMed] [Google Scholar]
- 31.Sumper, M. (2002) Science 295, 2430–2433. [DOI] [PubMed] [Google Scholar]
- 32.Brott, L. L., Naik, R. R., Pikas, D. J., Kirkpatrick, S. M., Tomlin, D. W., Whitlock, P. W., Clarson, S. J. & Stone, M. O. (2001) Nature 413, 291–293. [DOI] [PubMed] [Google Scholar]