Abstract
Specification of bilateral cardiac primordia and formation of the linear heart tube are highly conserved from Drosophila to humans. However, subsequent heart morphogenesis involving nonmesodermal neural crest cells was thought to be specific for vertebrates. Here, we provide evidence that a group of nonmesodermal cells that we have named heart-anchoring cells (HANCs) contribute to heart morphogenesis in Drosophila. We show that the homeobox genes ladybird (lb) known to be involved in diversification of cardiac precursors are expressed in HANCs and required for their specification. Interestingly, the HANCs selectively contact the anterior cardiac cells, which express lb as well. Direct interaction between HANCs and cardiac cells is assisted by a pair of cardiac outflow muscles (COMs), each of which selectively attaches to both the lb-expressing cardiac cells and HANCs. COM muscles seem to ensure ventral bending of the heart tip and together with HANCs determine the spatial positioning of the cardiac outflow region. Experimentally depleted cardiac lb expression leads to the disruption of the contact between the tip of the heart and either the COM muscles or the HANC cells, indicating a pivotal morphogenetic role for the lb expression within the heart.
The understanding of coordinated patterning of different cell types that build functional organs during embryogenesis seems to be an important challenge in developmental biology. Formation of the vertebrate heart provides an example of how the cells of mesodermal (heart primordia) and ectodermal (neural crest cells) origin may contribute to creating a fully functional organ (1). Interestingly, from the molecular and embryological points of view, the early stages of cardiogenesis, including specification of the heart primordia and formation of the beating heart tube, have been preserved from Drosophila to humans (2, 3). In this context, it becomes important to know whether nonmesodermal cells in Drosophila also contribute to a functional heart.
The Drosophila heart (dorsal vessel) is a hemolymph-pumping organ that is frontally open and arranged in a repeat pattern of segmental units. At the end of cardiac morphogenesis, the posterior area of the dorsal vessel becomes enlarged and constitutes the definitive heart, whereas the anterior portion has a narrow diameter and is equivalent to the aorta. The heart is composed of two major mesodermal cell types: cardioblasts and pericardial cells (3, 4). The cardioblasts are aligned in two highly ordered rows that form the lumen of the heart. These cells are contractile and express a variety of muscle-specific proteins. The pericardial cells, whose function is unknown so far, are loosely associated with the cardioblasts and do not express any muscle-specific marker. Recent studies of genes involved in heart development (5, 6) have revealed discrete subsets of cardioblasts and pericardial cells. According to the expression domains of heart identity genes, in each abdominal segment three types of cardioblasts can be detected, the anterior tinman (tin)-expressing pair, the adjacent tin/ladybird (lb)-positive pair, and the most posterior seven up (svp)-expressing pair (5–10). As for pericardial cells, the combinatory code of tin, lb, even-skipped (eve), and odd-skipped (odd) expression makes it possible to distinguish five distinct subpopulations (6–8, 10, 11). The bilateral heart primordia arise from the most dorsally located mesodermal cells specified by the intersection of epidermal Wg and Dpp signals (3, 12). Subsequent migration of two heart primordia and the formation of the linear heart tube require a collagen IV-type protein, pericardin, that is secreted by cardiac cells and enables their direct contact with the epidermal leading edge cells during dorsal closure (13).
Several organs are closely associated with the Drosophila heart. The hematopoietic organs, called the lymph glands, are attached frontally on both sides of the aorta. As cardiac cells, the hematopoietic precursors originate from the dorsal mesoderm, but their specification is controlled by a distinct genetic pathway (14). Another heart-associated organ is situated close to the lymph glands. This is the ring gland (15), which consists of cells of mesodermal and ectodermal origin. During the larval stage, the ring gland secretes ecdysone but has no apparent role in heart function.
In this article, we have focused on the later stages of heart formation and demonstrate that the cardiac outflow region in Drosophila undergoes programmed morphogenesis, which, as in the vertebrates, involves a group of nonmesodermal cells. These cells, which we have named heart-anchoring cells (HANCs), originate from the head epidermis and express the homeodomain transcription factor Lb. Using confocal microscopy, we show the direct contact between HANC cells and lb-positive cardiac cells and provide evidence for a crucial role of restricted lb expression within the heart. We also demonstrate that the proper positioning of the tip of the heart depends on the contact with a pair of cardiac outflow muscles (COMs).
Materials and Methods
Drosophila Strains and Misexpression Experiments. The lb mutants referred as lb-deficient were generated by an imperfect excision of the P5545 element inserted between the C15 and S59 genes. This excision produced a deletion covering the lbe, lbl, and the adjacent C15 and bap genes. Because C15 and bap are not expressed in the HANCs (unpublished observation), this deficiency seemed to be suitable to analyzing the role of lb genes in the specification of the HANCs. The UAS-RNAiC15 transgenic flies (three independent lines) were generated by injection of a PUAST vector carrying the tail-to-tail cloned 400-bp fragment of the C15 coding region. The bap208 null mutants were kindly provided by M. Frasch (Mount Sinai School of Medicine, New York), the ring gland-specific line l(2)1857 was provided by P. J. Bryant (University of California, Irvine), and the reproducing held-out-wing (how) expression how-lacZ line was provided by M. Semeriva (Laboratoire de Génétique et Physiologie du Développement, Marseille, France). The white1118 strain was used as a WT. The effects of cardiac misexpression of lb genes was analyzed in F1 embryos derived from the cross of UAS-lbe and UAS-lbl lines (11) with 24B-Gal4 driver (16) or Tin-GAL4 driver (kindly provided by R. Bodmer, University of Michigan, Ann Arbor). Attenuation of the C15 function was performed by crossing the UAS-RNAiC15 flies with a ubiquitous Arm-Gal4 driver line obtained from the Bloomington Stock Center (Indiana University, Bloomington). The loss of cardiac lb expression was experimentally induced by overexpression of even-skipped known to be able to repress lb in the heart (5). The F1 embryos derived from the cross of 24B-GAL4 driver or Tin-GAL4 driver and the UAS-eve line (obtained from R. Bodmer) were used for the analysis.
Antibody Staining and in Situ Hybridization. Embryos were stained with the following primary antibodies: monoclonal anti-Lbe 1A2, 1:1 (14); rabbit anti-β-galactosidase, 1:2,000 (Cappel); rabbit anti-Srp, 1:1,000 (provided by R. Reuter, University of Tuebingen, Tuebingen, Germany); rabbit anti-Tin, 1:500 (provided by M. Frasch); rabbit anti-myosin heavy chain, 1:500 (provided by D. Kiehart, Duke University, Durham, NC); anti-β3-tubulin, 1:1,000 (provided by R. Renkawitz-Pohl, Philipps University, Marburg, Germany); and rabbit anti-Stripe, 1:400 (provided by G. Vorbrueggen, Max Plank Institute, Gottingen, Germany).
Fluorescent labeling was revealed with secondary antibodies conjugated with dichlorotriazinyl aminofluorescein or Cy3 (Jackson ImmunoResearch). In some cases, immunostaining was amplified using biotinylated sheep anti-mouse antibody (Vector Laboratories) and streptavidin-dichlorotriazinyl aminofluorescein (Jackson ImmunoResearch). Labeled embryos were analyzed by using the Olympus FV 300 confocal microscope. The 3D reconstruction of the heart outflow region was performed by using the imaris–surpass program (Bitplane, Zurich).
The whole-mount in situ hybridizations were performed with a digoxigenin-labeled esg antisense-RNA probe according to Tautz and Pfeifle (18). The PCR-cloned esg cDNA fragment (882 bp), corresponding to the 3′ portion of the gene, was used as a template.
Results and Discussion
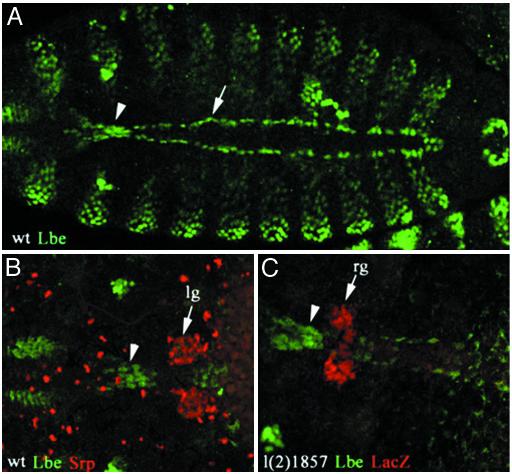
HANCs Originate from the Head Epidermis and Require lb Gene Function. Previous studies (5, 11, 17, 19) have described the role of lb genes in cell fate specification processes. In Drosophila, gain and loss of function experiments have revealed that lb genes specify the identity of a subset of cardiac and muscular cells and that this function results from the highly restricted, lineage-specific mesodermal expression of lb. In addition, precise analysis of the lbe (Fig. 1) and lbl (data not shown) expression pattern at the onset of heart fusion revealed a triangular cluster of lb-positive cells situated just anteriorly to the tip of the heart (Fig. 1 A, arrowhead). To produce a more precise description of the position of these cells with the respect to the heart-associated lymph glands and ring glands, we double-labeled embryos for lb and specific gland markers (Fig. 1 B and C). The lymph gland precursors revealed with anti-Serpent antibody flank either side of the heart tip (Fig. 1B) but do not overlap with the area labeled by lb. Similarly, the double staining of l(2)1857 enhancer trap line, which specifically expresses LacZ in the embryonic primordia of the ring gland (15), demonstrates that lb-positive cells are situated immediately in front of the ring gland (Fig. 1C).
Fig. 1.
lb-expressing HANCs. Confocal micrographs show dorsal views of WT embryos (A and B) and an enhancer trap l(2)1857 line embryo of stage 15 just before fusion of two heart primordi (C). (A) Immunostaining with anti-Lbe antibody revealing the lb-positive cardiac cells (arrow) and a cluster of lb-expressing cells (arrowhead) that lie just in front of the tip of the heart. (B and C) Double immunostainings revealing the position of Serpent-positive lymph gland primordia (lg) (B) and lacZ-positive ring glands (rg) (C) with respect to the heart-anchoring lb-expressing cells (arrowheads). Both the lymph gland and the ring gland precursors are located posteriorly to the cluster of lb-positive cells. Embryos are oriented anterior to the left. (Magnification: A, ×200; B and C, ×400.)
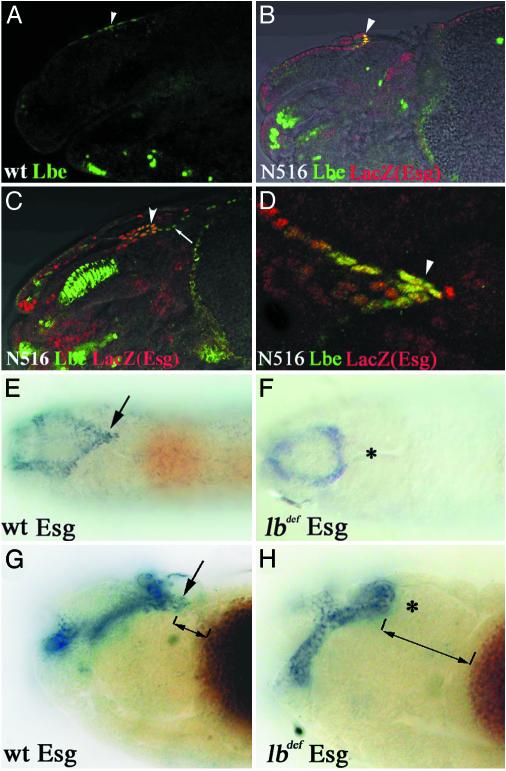
To determine the origin of heart-associated lb-positive cells, we followed the lb expression in the anterior part of the embryos from the early stage onward. At the embryonic stages that precede the heart fusion, we were unable to detect lb expression associated with the anterior part of two heart primordia. This finding suggested that the lb-positive cells are not originally specified in the heart area and may represent cells of different origin that become associated with the cardiac cells at later stages. Indeed, at late stage 11, a group of lb-positive cells in the dorsal head epidermis (Fig. 2A) can be detected. These cells start to invaginate slightly later and form the leading part of folded epidermis (Fig. 2B). The position and timing of this invagination resemble a folding of head epidermis, giving rise to the posterior part of the dorsal pouch called the frontal sac (20). To check whether lb-expressing cells correspond to the tip of frontal sac, we used escargot-LacZ line N516 that expresses lacZ in dorsal pouch-forming cells (20). Double stainings of stage 14 (Fig. 2B) and stage 16 (Fig. 2C) N516 embryos using anti-Lbe and anti-β-galactosidase antibodies clearly demonstrate that lb and esg are coexpressed in the tip of invaginating frontal sac. A dorsal view of the embryonic head region (Fig. 2D) confirms that these cells display the same triangular arrangement as the heart-associated lb-positive cells shown previously (compare Figs. 2D and 1).
Fig. 2.
HANCs originate from the head epidermis and are not specified in lb-deficient embryos. (A–D) Confocal micrographs combined with transmitted light channels showing lateral (A–C) and dorsal (D) views of the head region of WT (A) and escargot enhancer trap N516 (B–D) embryos double-labeled for Lb (green) and β-galactosidase (red). (A) In the stage 11 embryo, a few cells in the dorsal head epidermis express lb (arrowhead). At stage 14 (B) and stage 16 (C), the lb-positive cells coexpress escargot (arrowheads) and localize to the tip of the invaginating frontal sac. (D) Dorsal view of stage 16 N516 embryo, showing the coexpression of lb and LacZ in the distal part of the frontal sac (arrowhead). (E and F) Nomarski micrographs representing the dorsal views of stage 16 embryos and (G and H) dorsolateral views of stage 14 embryos stained for escargot transcripts. (E and G) esg expression in WT embryos. The arrow indicates the HANCs. (F and H) In lb-deficient embryos, esg expression in the tip of the frontal sac is absent (*). The distance between the most distal frontal sac cells and the yolk sac (displaying brown background color) is much larger in lb-deficient embryos (compare bidirectional arrows in G and H). (Magnification: A and B, ×300; C, ×350; D, ×600; E and F, ×100; G and H, ×350.)
Taken together, these data indicate that the lb-expressing cells lying immediately in front of the heart originate from the head epidermis and migrate toward the heart tip during frontal sac invagination. This observation raises an additional question regarding the role of lb genes in the specification of this group of cells. We have previously shown that lb genes play an important role in the specification of a subset of cardiac and muscular precursors (11, 19). Functional analysis of the mouse lb ortholog Lbx1 gene (21, 22) has also revealed its implication in the control of specification and directed migration of appendicular muscle precursors. Moreover, the most recent data indicate that Lbx1 in mice specifies a subpopulation of cardiac neural crest cells necessary for normal heart development (23).
To test whether Drosophila lb are required for the specification of heart-associated cells, we have analyzed dorsal pouch formation in embryos deficient for both lb genes (see Materials and Methods). Using the esg RNA probe as a marker, we demonstrated that in lbdef embryos the esg expression in the distal part of the frontal sac bearing the heart-associated cells is absent (compare Fig. 2 E and G with F and H). In addition, as shown by the dorsolateral views of stage 14 embryos (Fig. 2 G and H), the distance between the yolk sac and the most distal esg-positive cells is significantly enlarged in lb-deficient embryos. This finding, supported by the Nomarski optics observation revealing that the posterior part of the frontal sac has altered morphology (data not shown), indicated that the HANCs are not specified or do not migrate properly in lbdef embryos. Because the analyzed deficiency also covers adjacent bap and C15 genes, we tested frontal sac formation in bap mutant embryos and Arm-GAL4>UAS-RNAiC15 embryos. In both bap208 and RNAiC15 embryos, the esg-stained frontal sac was of normal shape (data not shown), thus excluding the influence of bap and C15 genes on differentiation or migration of heart-associated cells. These data strongly suggest that the abnormal pattern of distal esg-positive frontal sac cells in lbdef embryos results from the loss of lb function.
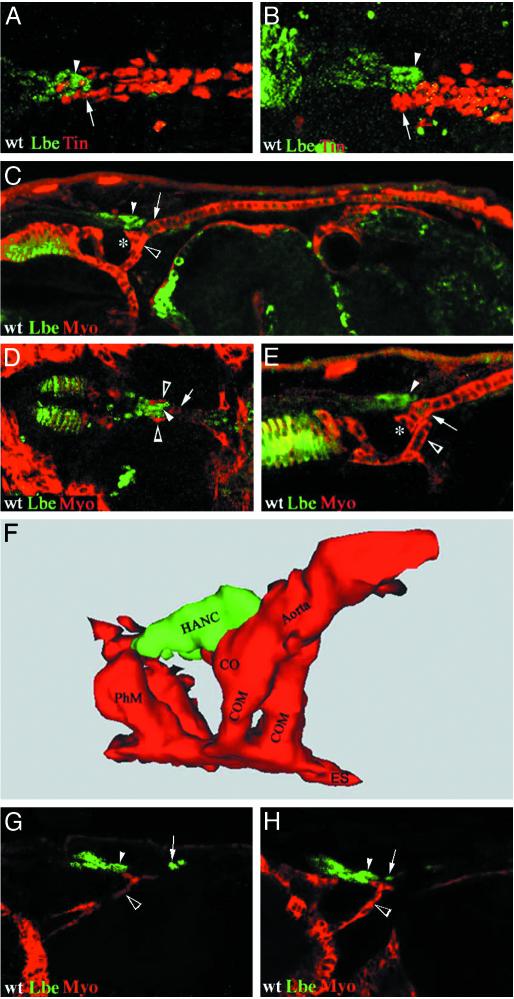
Mesodermal and Nonmesodermal Components Involved in the Patterning of the Cardiac Outflow Region. Our immunostaining experiments showed that the heart-anchoring lb-positive cells lie very close to the lb-expressing cardioblasts (Fig. 1). Because the most anterior cardioblasts express tin (5), we double-labeled embryos with anti-Tin and anti-Lbe antibodies to visualize the respective positions of the heart tip and the most distal part of the frontal sac. Surprisingly, these two structures overlap by about three cell lengths when observed from the dorsal side (Fig. 3A). Moreover, the lateral confocal view of the anterior heart region (Fig. 3B) clearly shows that the tip of the frontal sac, which consists of nonmesodermal lb-expressing cells, is directly attached to the heart. More precisely, we observed that the anterior part of the aorta (partly overlapping the heart-associated lb cells) bends ventrally, forming a morphologically distinct area corresponding to the cardiac outflow region. This morphology suggests that there might be an additional cellular component favoring ventral bending of the heart. We decided to look for the presence of a ventrally located muscle attached to the tip of the aorta. The lateral view of the embryo, double-stained for myosin heavy chain (Myo) and Lbe (Fig. 3C), clearly shows that the heart outflow is indeed attached to a head muscle, which we propose to call the COM. More precise analysis of the confocal sections revealed that the cardiac primordium is, in fact, linked by two closely lying COMs that extend from the esophagus and attach to either side of the cardiac outflow region (open arrowheads in Fig. 3D). Surprisingly, these embryonic head muscles have not been described previously to our knowledge. The lack of published documentation for COMs is probably because the morphology and the origin of somatic head muscles in the Drosophila embryo have not been systematically analyzed. Using the only available description of adult head musculature done by Miller (24), we were unable to identify adult head muscles that might correspond to the embryonic COMs presented here.
Fig. 3.
Spatial positioning of the cardiac outflow region. Confocal micrographs showing dorsal (A and D) and lateral (B, C, E, G, and H) views of WT embryos double-stained for Lbe (green), Tin (A and B), or Myo (C–E, G, and H) (red). (A and B) lb-positive HANCs (arrowhead) overlap with the tin-positive cardiac cells (arrow). Note that at stage 16 (A and B) lb is no longer expressed within the heart. (C) General lateral view of a stage 15 embryo showing that the tip of the heart (asterisk) bends ventrally when attached to COMs (open arrowhead) and HANCs (arrowhead). (D) Dorsal view showing that COMs (open arrowhead) overlap at both sides of the heart tip (arrow) and the lb-positive HANCs (arrowhead). (E) The HANCs (arrowhead) attach selectively to the second pair of cardioblasts (arrow) expressing lb. Note that the cardiac outflow (*) is encompassed by HANCs from the dorsal side and COMs from the posterior-dorsal side. (F) Computer-assisted 3D reconstruction of the heart outflow region of a stage 16 embryo showing spatial positioning of the heart tip, COMs, and HANCs. Note that the HANCs that initially attach to the second pair of cardiac cells expressing lb at later stages also establish contact with the most anterior lb-negative cardiac cells. (G) At stage 14, COMs (open arrowhead) extend their fillopodia toward both the invaginating lb-positive HANCs (arrowhead) and the lb-expressing cardioblasts (arrow), which are not yet associated with the HANCs. The COMs at this stage are thinner and much longer than after heart fusion. (H) At the beginning of stage 15, COMs (open arrowhead) are definitively attached to cardiac cells (arrow) and HANCs (arrowhead), and this contact seems to precede the attachment of HANCs to lb-expressing cardiac cells. CO, cardiac outflow; ES, esophagus; PhM, pharyngeal muscle. (Magnification: A and B, ×400; C, ×250; D and E, ×300; G and H, ×350.)
Interestingly, Myo/Lbe double staining also revealed that the nonmesodermal HANCs are attached to both the cardiac cells (Fig. 3E) and the COMs (Fig. 3D). Because the HANCs express lb and associate selectively with the most anterior lb-positive pair of cardiac cells (Figs. 3E and 4 D–F), we speculate that establishment of this contact may involve homophilic-type cell interactions. Testing the cell adhesion molecules involved in intercellular signaling between homotypic cells (25, 26) will help in defining mechanisms underlying the establishment of connectivity between the lb-expressing HANCs and cardiac cells.
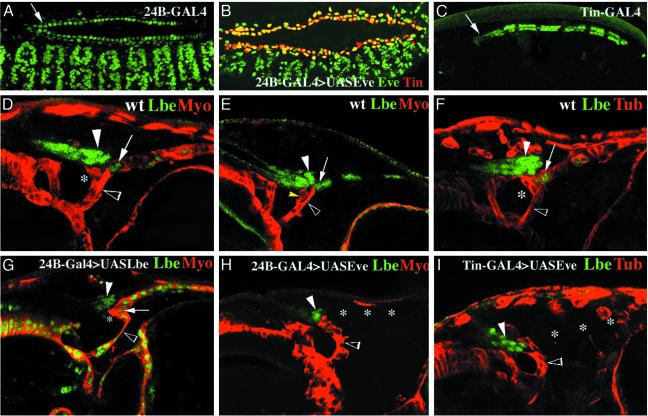
Fig. 4.
Role of cardiac lb expression in the patterning of the cardiac outflow region. (A and C) Dorsolateral views of 24B-GAL4>UAS-GFP (A) and Tin-GAL4>UAS-GFP (C) embryos showing the GFP-revealed profile of expression driven by these effector lines. Arrows point to the cardiac outflow region. Note the relatively low GFP expression driven in this region by Tin-GAL4. (B) Dorsolateral view of a late stage 14 24B-GAL4>UAS-eve embryo double-stained for Eve and Tin to show that the heart primordium is normally formed in the context of mesodermal Eve misexpression. (D–I) Confocal micrographs showing lateral views of the dorsal part of the embryonic head region. (D, F, G, and I) Stage 15 embryos. (E and H) Stage 14 embryos. (D–I)WT(D–F), (G) heterozygous 24B-Gal4>UASLbe, (H) 24B-GAL4>UAS-Eve, and (I) Tin-GAL4>UASEve embryos stained for myosin heavy chain and Lbe (D, E, G, and H)or β3-tubulin and Lbe (F and I). (D–F)InWT, the main COM branch (open arrowheads) attaches to the second most anterior pair of cardiac cells expressing lb. A separated COM branch (yellow arrowhead in E) displaying weaker level of myosin expression contacts HANCs (filled arrowheads). (G) In embryos with ubiquitous cardiac expression of lbe, COM (open arrowhead) attaches to the tip of the heart and not to the second pair of cardioblasts. COM displays an abnormal shape, and its two branches are difficult to detect. The abnormal contact between COM and the tip of the heart leads to the increased ventral bending of the cardiac outflow region. (H and I) In embryos in which cardiac lb expression was depleted (*), the COM (open arrowheads) extended anteriorly and attached to HANCs (filled arrowheads). (Magnification: A–C, ×200; D–I, ×350.)
To visualize the spatial arrangement of the tip of aorta, the COMs, and the lb-expressing HANCs, we used computer-assisted 3D analysis and reconstruction of confocal scans. This approach fully confirmed our observations revealing that, in addition to HANCs, the heart outflow region is tightly attached to a pair of COMs (Fig. 3F). These muscles overlap the tip of the heart from both sides and contribute to its ventral bending.
To understand how the contact between heart, COMs, and HANCs is established during development, we monitored these structures in early and late stage 14 embryos (Fig. 3 G and H). Our data indicate that both the lb-positive cardiac cells and lb-expressing HANCs attract extending COMs.
The contact between COMs and HANCs seems to slightly precede COM's attachment to the tip of aorta and is seen at the beginning of stage 14 (Fig. 3G). At that time, COM's fillopodia extend in the direction of lb-positive cardiac cells (Fig. 3G) and become definitively attached to these cells at late stage 14 at the onset of heart fusion (Fig. 3H). Because the COMs come into contact with cardiac cells and HANCs before the HANCs become attached to the heart, we speculate that COMs, in addition to bending the tip of the heart, facilitate its contact with HANCs. The establishment of contact between somatic muscles and their epidermal attachment sites (tendon cells) has been extensively studied (27), revealing the key role of the zinc finger transcription factor Stripe (28) and an RNA-binding protein How (29). We have used both these markers to test whether the HANCs and cardiac cells to which the COMs attach display properties of tendon-like cells. Double labeling performed on WT embryos with anti-Lbe/anti-Stripe and on how-lacZ embryos with anti-Lbe/anti-LacZ antibodies revealed that the HANCs and the COM-contacting cardiac cells do not express tendon cell markers (data not shown). This finding indicates that lb-positive cells attract COMs by using a mechanism different from that used by tendon cells. The most interesting candidates for guiding the COMs are the secreted protein Slit possessing the multiple-protein binding motifs and its receptors Robo and Robo2 (30). Slit and Robo emerged recently as pivotal components controlling attraction and repulsion processes during morphogenesis of somatic muscles (31), suggesting that they might also be implicated in COM attraction.
The Most Anterior lb-Expressing Cardiac Cells Play a Central Role in the Patterning of the Cardiac Outflow Region. The observation that both COMs and HANCs chose to attach to the most anterior lb-positive cardiac cells prompted us to test whether the deregulation of lb expression within the heart might influence patterning of the cardiac outflow region. To perform this test, we used the Gal4/UAS-targeted expression system (16). Two different GAL4 effector lines, 24B-GAL4 and Tin-GAL4, were used (Fig. 4 A and C). The 24B-GAL4 line allows targeted expression in all cardiac and muscular cells (Fig. 4A) with a uniform and high level of expression in the most anterior heart cells (arrow in Fig. 4A). The Tin-GAL4 line (kindly provided by R. Bodmer) induces the UAS transgene expression selectively in four cardioblasts in each hemisegment (these include the lb-positive cells), but the level of induction in the most anterior cardiac cells is lower (arrow in Fig. 4C) than that of 24B-GAL4 line. To expand cardiac lb expression, both GAL4 effector lines have been crossed with the UAS-lbe line. The ectopic lb cardiac expression driven by the Tin-GAL4 line was weaker than the endogenous lb expression within the heart, and no obvious gain-of-function induced alterations in the patterning of the cardiac outflow region have been observed (data not shown). In contrast, the 24B-GAL4>UAS-lbe embryos showed marked expansion of lb expression within the heart (Fig. 4G), leading to the abnormal contact of the COMs with the tip of the heart. Most precisely, we found that COMs attach directly to the most anterior cardiac cells that form the cardiac outflow (asterisk in Fig. 4G) and not to the second pair of cardiac cells as in the WT (Fig. 4D). The altered connection of COMs most probably results from the ectopic attractive signals generated by the enlarged cardiac expression of lb. As a consequence, the ventral bending of the most anterior part of the heart was more pronounced (compare Fig. 4 D and G). We have also noticed an altered morphology of COMs (open arrowhead in Fig. 4 D and G), which might result from the 24B-GAL4-driven ectopic muscular expression of lb. The contact between HANCs and cardiac cells was not affected by the experimental misexpression conditions. To test whether cardiac lb expression is required for the attraction of COMs, we took advantage from the previously described negative regulatory influence of even-skipped (eve) that is able to specifically repress lb in cardiac cells (5). Again, the 24B-GAL4 and the Tin-GAL4 effector lines were used to drive eve expression within the heart. We found that both of these drivers crossed with UAS-eve line (Tin-GAL4 with lower penetrance) lead to the repression of lb activity within the heart (Fig. 4 H and I). Despite the misspecification defects, cardiac primordia are formed normally in UAS-eve gain of function embryos (ref. 5 and Fig. 4B), allowing us to monitor COMs and HANCs in a context in which cardiac expression of lb is depleted. Our data show that in such lb-depleted embryos (see the absence of cardiac lb expression in Fig. 4 H and I) the COMs do not attach to the tip of the heart. We observed that COMs extended anteriorly and attached to invaginating HANCs (Fig. 4 H and I), which are no longer able to anchor properly the tip of the heart. The main COM branch that normally attaches to the cardiac outflow region (compare Fig. 4 E and F with H and I) was absent or fused with the HANC-attaching branch. Thus, we believe that the abnormal shape of COM and a complete loss of its contact with the tip of the heart are caused by cardiac lb depletion. This assumption is supported by the fact that the same phenotypes were observed in embryos with ectopic cardiac (Tin-GAL4-driven) and cardiac plus muscular (24B-GAL4-driven) expression of eve (compare Fig. 4 H and I). The altered contact and positioning of COMs appeared in late stage 14 embryos when myosin heavy chain is not yet detectable in cardioblasts (Fig. 4 E and H). The analysis of stage 15 embryos revealed in addition that the loss of cardiac lb expression is accompanied by the down-regulation of β3-tubulin (compare Fig. 4 F and I) in the cardiac outflow region, indicating that lb expression within the heart has an important impact on final cardiac morphogenesis. These data complement recent reports describing morphogenetic roles of a COUP-TF receptor family member, Seven-up, expressed in the posterior pair of cardioblasts in each segment and required for the formation of ostia in the abdominal segments A5–A7 (32, 33). In this context, the demonstration of the role of lb in the patterning of the cardiac outflow region rises a possibility that in abdominal segments lb plays a similar role in establishing contact between cardiac cells and the heart-anchoring alary muscles. This hypothesis remains to be investigated.
Moreover, the presented phenotypes (Fig. 4) suggest that the lb expression experimentally depleted within the heart influences the emission or reception of attractive signals by cardiac cells and in consequence leads to the disruption of the contact with COMs. The same or similar muscle attractant is likely to be produced or received by lb-positive HANCs, thus suggesting that lb regulates expression of genes involved in cell–cell contact. Interestingly, in mouse lb homologue, the Lbx1 gene was found to be required for directed migration of a subset of muscle precursors (21, 22), indicating that the conserved cell-movement mechanisms might exist that are controlled by lb genes.
Taken together, through analysis of embryos in which cardiac lb expression was expanded or experimentally depleted, we have demonstrated that the lb is required for the correct attachment of HANCs and COMs and in consequence for the proper patterning of the cardiac outflow region. This finding provides insights into functional meaning of anterior–posterior diversification of cardiac precursors within each segment of the Drosophila heart.
Can HANCs Represent a Drosophila Prototype of Neural Crest Cells? Vertebrate heart morphogenesis involves two different cell types, the mesodermal heart primordia and a subpopulation of neural crest cells that migrate from the head region. In this article, we have shown that in Drosophila a group of nonmesodermal cells originating from the head epidermis (which we have named the HANCs) contributes to the final morphogenesis of the heart. As in vertebrate neural crest cells, the HANCs undergo directed movements, enter in contact with cardiac cells, and participate in the patterning of the cardiac outflow region. We have demonstrated that the homeobox genes lb known to be involved in diversification of cardiac precursors (5, 11) are expressed in HANCs and are required for their specification. Similarly, the ortholog of lb, the Lbx1 gene, was recently found to be required for the specification of a subset of cardiac neural crest cells in mouse (23), indicating that lb/Lbx1 genes play a conserved role in the specification of nonmesodermal components of the heart. In this view, the HANCs might indeed be compared with vertebrate neural crest cells. However, our data indicate also that the HANCs display at least two features that are specific for Drosophila only: (i) they move as a part of folded head ectoderm and not as delaminated neural crest cells; and (ii) they form a functional complex with COMs, which have no homologous structures in vertebrates. Thus, further analyses are required to elucidate the exact role of HANCs and their potential neural crest cell-like functions.
Acknowledgments
We thank C. Soler for assistance in 3D reconstruction and F. De Graeve for critical reading of the manuscript. M.Z. was supported by the Fondation pour la Recherche Medicale and the Association Française contre les Myopathies.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HANC, heart-anchoring cell; COM, cardiac outflow muscle; lb, ladybird.
References
- 1.Kelly, R. G. & Buckingham, M. E. (2002) Trends Genet. 18, 210–216. [DOI] [PubMed] [Google Scholar]
- 2.Jagla, K. & Bellard, M. (1998) Med. Sci. 14, 1067–1071. [Google Scholar]
- 3.Bodmer, R. & Frasch, M. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, San Diego), pp. 65–90.
- 4.Rugendorff, A., Younossi-Hartenstein, A. & Hartenstein, V. (1994) Roux's Arch. Dev. Biol. 203, 266–280. [DOI] [PubMed] [Google Scholar]
- 5.Jagla, T., Bidet, Y., Da Ponte, J. P., Dastugue, B. & Jagla, K. (2002) Development (Cambridge, U.K.) 129, 1037–1047. [DOI] [PubMed] [Google Scholar]
- 6.Ward, E. J. & Skeath, J. B. (2000) Development (Cambridge, U.K.) 127, 4959–4969. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez, A. D., Shi, W., Wilson, B. A. & Skeath, J. B. (2003) Development (Cambridge, U.K.) 130, 3015–3026. [DOI] [PubMed] [Google Scholar]
- 8.Han, Z. & Bodmer, R. (2003) Development (Cambridge, U.K.) 130, 3039–3051. [DOI] [PubMed] [Google Scholar]
- 9.Klinedinst, S. L. & Bodmer, R. (2003) Development (Cambridge, U.K.) 130, 3027–3038. [DOI] [PubMed] [Google Scholar]
- 10.Gajewski, K., Choi, C. Y., Kim, Y. & Schulz, R. A. (2000) Genesis 28, 36–43. [DOI] [PubMed] [Google Scholar]
- 11.Jagla, K., Frasch, M., Jagla, T., Bellard, F., Dretzen, G. & Bellard, M. (1997) Development (Cambridge, U.K.) 124, 3471–3479. [DOI] [PubMed] [Google Scholar]
- 12.Lockwood, W. K. & Bodmer, R. (2002) Mech. Dev. 114, 13–26. [DOI] [PubMed] [Google Scholar]
- 13.Chartier, A., Zaffran, S., Astier, M., Semeriva, M. & Gratecos, D. (2002) Development (Cambridge, U.K.) 129, 3241–3253. [DOI] [PubMed] [Google Scholar]
- 14.Lebetsky, T., Chang, T., Hartenstein, V. & Banerjee, U. (2000) Science 288, 146–149. [DOI] [PubMed] [Google Scholar]
- 15.Harvie, P. D., Filippova, M. & Bryant, P. J. (1998) Genetics 149, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 17.Jagla, K., Jagla, T., Heitzler, P., Dretzen, G., Bellard, F. & Bellard, M. (1997) Development (Cambridge, U.K.) 124, 91–100. [DOI] [PubMed] [Google Scholar]
- 18.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 19.Jagla, T., Bellard, F., Lutz, Y., Dretzen, G., Bellard, M. & Jagla, K. (1998) Development (Cambridge, U.K.) 125, 3699–3708. [DOI] [PubMed] [Google Scholar]
- 20.Nassif, C., Daniel, A., Lengyel, J. A. & Hartenstein, V. (1998) Dev. Biol. 197, 170–186. [DOI] [PubMed] [Google Scholar]
- 21.Brohmann, H., Jagla, K. & Birchmeier, C. (2000) Development (Cambridge, U.K.) 127, 435–445. [DOI] [PubMed] [Google Scholar]
- 22.Gross, M. K., Moran-Rivard, L., Velasquez, T., Nakatsu, M. N., Jagla, K. & M. Goulding, M. (2000) Development (Cambridge, U.K.) 127, 413–424. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer, K., Neuhaus, P., Kruse, J. & Braun, T. (2003) Circ. Res. 92, 73–80. [DOI] [PubMed] [Google Scholar]
- 24.Miller, A. (1950) in Biology of Drosophila, ed. Demerec, M. (Wiley, New York), pp. 420–531.
- 25.Nose, A., Umeda, T. & Takeichi, M. (1997) Development (Cambridge, U.K.) 124, 1433–1441. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa, S. & Takeichi, M. (1995) Development (Cambridge, U.K.) 121, 1321–1332. [DOI] [PubMed] [Google Scholar]
- 27.Volk, T. (1999) Trends Genet. 15, 448–453. [DOI] [PubMed] [Google Scholar]
- 28.Frommer, G., Vorbruggen, G., Pasca, G., Jackle, H. & Volk, T. (1996) EMBO J. 15, 1642–1649. [PMC free article] [PubMed] [Google Scholar]
- 29.Nabel-Rosen, H., Volohonsky, G., Reuveny, A., Zaidel-Bar, R. & Volk, T. (2002) Dev. Cell 2, 183–193. [DOI] [PubMed] [Google Scholar]
- 30.Piper, M. & Little, M. (2002) BioEssays 25, 32–38. [DOI] [PubMed] [Google Scholar]
- 31.Kramer, S. G., Kidd, T., Simpson, J. H. & Goodman, C. S. (2001) Science 292, 737–740. [DOI] [PubMed] [Google Scholar]
- 32.Ponzielli, R., Astier, M., Chartier, A., Gallet, A., Therond, P. & Semeriva, M. (2002) Development (Cambridge, U.K.) 129, 4509–4521. [DOI] [PubMed] [Google Scholar]
- 33.gLovato, T. L., Nguyen, T. P., Molina, M. R. & Cripps, R. M. (2002) Development (Cambridge, U.K.) 129, 5019–5027. [DOI] [PubMed] [Google Scholar]