Abstract
In this report we describe the developmental expression and function of Sp8, a member of the Sp family of zinc finger transcription factors, and provide evidence that the legless transgene insertional mutant is a hypomorphic allele of the Sp8 gene. Sp8 is expressed during embryogenesis in the forming apical ectodermal ridge (AER), restricted regions of the central nervous system, and tail bud. Targeted deletion of the Sp8 gene gives a striking phenotype, with severe truncation of both forelimbs and hindlimbs, absent tail, as well as defects in anterior and posterior neuropore closure leading to exencephaly and spina bifida. Outgrowth of the limb depends on formation of the AER, a signaling center that forms at the limb bud apex. In Sp8 mutants, the AER precursor cells are induced and initially express multiple appropriate marker genes, but expression of these genes is not maintained and progression to a mature AER is blocked. These observations indicate that Sp8 functions downstream of Wnt3, Fgf10, and Bmpr1a in the signaling cascade that mediates AER formation.
Keywords: apical ectodermal ridge, exencephaly, spina bifida, zinc finger, legless
Outgrowth of the vertebrate limb requires the formation and maintenance of the apical ectodermal ridge (AER) at the distal limb bud margin. The initial induction of the murine AER precursor cells requires canonical Wnt signaling through ectodermal expression of Wnt3 (1), ectodermal Bmp signaling (2), and the interaction of Fgf10 expressed by the limb mesoderm with its receptor Fgfr2b expressed in the limb ectoderm (3–5). The induced AER precursor population of cells is initially localized on the ventral surface of the emerging murine limb bud and expresses several specific markers, including Fgf8 (6, 7). With subsequent growth, this population of cells becomes localized at the ventro-distal margin of the limb bud before constricting to form a mature AER.
The progression of the AER precursor into a definitive AER is poorly understood. It has been observed, however, that a disruption of this process occurs in the legless (lgl) transgene insertional mutant. In lgl mutant hindlimbs, the AER precursor cells are induced, but fail to localize at the ventro-distal limb margin and an AER subsequently fails to form (6). In contrast, in the forelimbs, an AER does form, but with irregularities in the anterior margin. These observations correlate well with the lgl homozygous mutant limb phenotype, with the hindlimbs missing all structures distal to the femur and with variable loss of distal-anterior forelimb structures (8, 9). Other aspects of the lgl mutant phenotype include randomized left–right axis formation, missing olfactory bulbs, encephalocoeles or exencephaly, and cleft lip. Previous studies showed that both the zinc finger transcription factor Sp4 and the left right dynein (Lrd) genes (10, 11) are deleted by the lgl transgene insertion, but gene targeting results indicated that neither gene is responsible for the limb truncation or craniofacial malformations observed in lgl mutants (Fig. 1A).
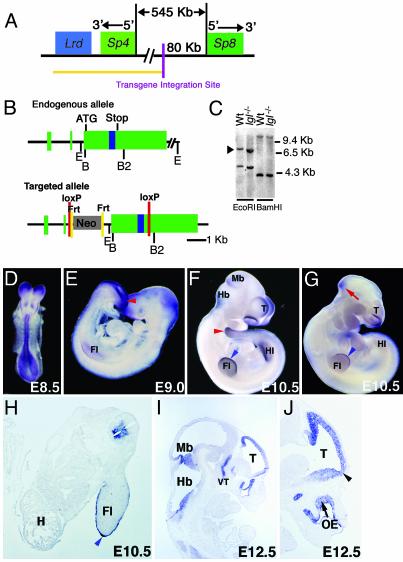
Fig. 1.
Structural characterization and expression of the Sp8 gene. (A) Relationship between the Sp8 and Sp4 genes. Yellow line indicates genomic region known to be deleted in the lgl transgene insertional mutant. (B) Structure of the Sp8 gene and the targeting vector. Green boxes (exons), blue box (zinc finger domain). Restriction sites: E, EcoRI; B, BamHI; B2, BglII. (C) Southern blot analysis of wild-type (Wt) and lgl mutant DNA. The blot was hybridized with a 435-bp Sp8 cDNA fragment containing sequences found within exon 2 and the beginning of exon 3. Arrowhead indicates the EcoRI fragment containing the coding region. (D–J) Whole mount and sectioned in situ hybridization using an Sp8 riboprobe. (D–F and H–J) Wild-type embryos. (D) Expression is detected in the neuroepithelium at E8.5. (E) On E9.0, expression is heavy around the posterior neuropore/tailbud (red arrowhead) and is first evident in the forelimb (Fl) AER precursor. (F and H) By E10.5, expression is detectable in the telencephalon (T), dorsal and ventral forelimb ectoderm, AER (blue arrowhead), hindlimb (Hl) AER precursor, tail (red arrowhead), medial and lateral nasal processes, isthmus, and ventral spinal cord. H, heart. (G) lgl–/– embryo showing reduction in the level of expression in the forelimb AER, lateral and medial nasal processes, isthmus (red arrow), and the absence of expression in the hindlimb bud AER precursor. (I and J) On E12.5, expression is detected at the midbrain (Mb)/hindbrain (Hb) border, in the olfactory epithelium (OE) and the ventral thalamus (VT). Notably, Sp8 is expressed in the olfactory bulb primordia (J, arrowhead).
Release of the human genome sequence revealed the presence of a second zinc finger transcription factor gene, Sp8, in a region syntenic to the lgl transgene insertion on mouse chromosome 12. This gene was of particular interest because of its homology to the Drosophila D-Sp1 gene, implicated in leg development (12). In mammals, seven other Sp family members have been described (for review see ref. 13). All Sp proteins possess a highly conserved DNA-binding domain comprised of three Cys2His2 zinc fingers that interact with GC and GT boxes commonly found within the promoter regions of genes. Targeted mutations of mouse Sp1, 3, 4, 5, and 7 have previously been reported. Absence of the ubiquitously expressed Sp1gene results in embryo lethality at embryonic day 10 (E10) (14). Sp3-deficient mice die at birth of respiratory failure and have delayed bone ossification and impaired tooth development (15, 16). Targeted disruption of Sp4 results in a reduction in viability and growth, cardiac malformations, and a failure of males to mate (10, 17). No overt phenotype is observed in Sp5 null mice (18), whereas Sp7 (osterix) mutants show a striking inability to form bone (19).
In this report we describe the expression and function of the Sp8 gene. In the developing limbs Sp8 showed restricted expression in the ectoderm, including the AER precursor cells. Of further interest, in lgl mice the Sp8 coding sequences are intact, but the gene showed reduced levels of expression, particularly in the developing hindlimbs, suggesting an Sp8 hypomorphic allele. Consistent with this, mice with a targeted deletion of Sp8 gave a dramatic phenotype including the absence of tails and severely truncated forelimbs and hindlimbs. CNS defects included a failure to close both the anterior and posterior neuropores, leading to exencephaly and spina bifida. Molecular analysis showed that Sp8 is not necessary for induction of the AER precursor, but is required for its maintenance and maturation into a definitive AER.
Methods
Gene Targeting. The targeting vector was generated by using a Cre/lox targeting vector in which the neomycin resistance gene is immediately flanked by Frt sequences. 5′ to Neo is a unique Fse1 site, a loxP sequence, and a unique Pme1 site. Sequences 3′ to Neo include a second loxP sequence and a unique Srf1 site. The following Sp8 gene specific primers were used to PCR amplify genomic regions for subcloning into each of these sites. A 4.2-kb fragment was amplified by using the primers 5′-ATTGCCCGGGCCCCAGCACCAAAACTGCCT-3′ and 5′-ATTGCCCGGGCTCCCCCTCAACCCATCCTTA-3′ and subcloned into the Srf1 site. A 2.1-kb fragment was amplified by using the primers 5′-TTAAGTTTAAACGGGGAGAAGCAACTAAGGA-3′ and 5′-TTAAGTTAAACCAAGGATTCAGCCACCGATG-3′ and subcloned into the Pme1 site. A 3.6-kb fragment was amplified by using the primers 5′-TAAGGCCGGCCGACAGTAAGTAGCACACAT-3′ and 5′-TAAGGCCGGCGATAAAGAAGCCAGGAGAAA-3′ and subcloned into the Fse1 site. Mice were generated from R1ES cells carrying the modified allele and were bred to either black Swiss mice or germ line Cre mice (20). PCR genotyping was performed on genomic DNA isolated from either embryonic yolk sacs or fetal visceral tissue. The endogenous allele was detected by using the primer pair 5′-TCCTCCCACGAGTGTAATGCTCAG-3′ and 5′-GCGTTCTTTCCCCCAACTTC-3′ under cycling conditions of 95°C for 30 s, 63°C for 30 s, and 72°C for 1 min. The recombined null allele was detected by using the primer pair 5′-CCACAGCCTCTTCAAAGTTCCG-3′ and 5′-GCGAGTAAGTTTTTCCCTCCTGG-3′ under cycling conditions of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Mice from two distinct targeted embryonic stem cell clones were studied and found to give identical phenotypes.
In Situ Hybridization. A NotI/ApaI fragment of the Sp8 cDNA was used for whole mount and sectioned in situ assays. Antisense riboprobes were kindly provided by Gail Martin (University of California, San Francisco) (Fgf8), Alexandra Joyner (New York University School of Medicine, New York) (En1), Robert Maxson (University of Southern California, Los Angeles) (Msx2), and Allan Bradley (Sanger Center, Cambridge, U.K.) (p63). Whole mount and sectioned in situ hybridization assays were performed as described (6, 21).
Skeletal Preparation. Alizarin red and alcian blue skeletal staining was performed as described (22).
Nile Blue Sulfate Staining. Dissected embryos were placed in Nile blue sulfate (1:20,000) in PBS for 15 min, rinsed in PBS for 30 min, and immediately photographed.
Results and Discussion
Release of the human 7p15–21 genomic sequence revealed the linkage of Sp4 with Sp8 in a region syntenic to the lgl transgene insertion. In the mouse, Sp4 and Sp8 are separated by ≈545 kb and arranged with their promoters facing each other (Fig. 1A), similar to the Sp2–Sp6 and Sp1–Sp7 gene pairs. Although the entire Sp4 gene is deleted from the lgl genome, Southern blot analysis indicated that the Sp8 coding region is intact (Fig. 1C). Additional PCR and Southern analysis positioned the transgene integration site ≈80 kb upstream of the Sp8 coding region (data not shown).
The Sp8 gene consists of two 5′ UTR exons and a third exon containing all of the coding and 3′ UTR sequences (Fig. 1B). This structure was determined by comparison of the mouse genomic sequence with an RT-PCR-derived Sp8 cDNA from E10 mouse embryos as well as RIKEN and EST clones, EST BB663595 and the Fantom-DB clone 5730507L14 (NCBI accession no. AK030745) (23). This genomic structure closely resembles that observed for the Sp5, 6, and 7 genes (18, 19, 24).
The murine and human (NCBI accession no. NM_182700) Sp8 cDNAs encode highly conserved proteins of 486 and 466 aa, respectively, with 97% identity. The human protein includes an additional 18 amino acids at the N terminus; the last four of these amino acids comprise the beginning of the conserved “Sp box” sequence TPLAMLAATCNKIGSP present in other family members (18). The mouse protein contains an additional 42-aa insertion after amino acid 103. Like other members of the Sp family, the buttonhead and the highly conserved zinc finger domains are at the C terminus (13, 18). In Sp8, these domains are most homologous to the Drosophila protein D-Sp1 and murine Sp7 (data not shown) (12, 19). Variability exists at the N termini, where Sp1–4 all possess large activation domains, whereas Sp8, like Sp5, 6, and 7, have significantly smaller domains of unknown function (13).
Sp8 expression during embryogenesis is distinct from other Sp family members (13, 18, 19, 24, 25). In E8 embryos, Sp8 was expressed in the forming neural tube. On E9–10, localized expression was detected at the telencephalic midline, mid-hindbrain junction, ventral spinal cord, posterior neuropore, genital tubercle, tail bud, and lateral and medial nasal processes (Fig. 1 D–H). In the fore- and hindlimb buds, expression was observed in the AER precursor cells/AER and in the adjacent dorsal and ventral limb ectoderm (Fig. 1 E, F, and H). Notably, a dramatic reduction in Sp8 expression in the lateral and medial nasal processes, forebrain, limbs, and tail bud was observed in E10–11 lgl–/– embryos (Fig. 1G and data not shown). At E12.5, Sp8 expression remained in the mid-hindbrain boundary, the olfactory epithelia, and telencephalic midline and included the primordia of the olfactory bulbs (Fig. 1 I and J), which are missing in lgl mutants (9). Expression in the olfactory bulbs at later stages (e.g., E16.5, data not shown) was also evident. The reduced expression of Sp8 in multiple sites of malformation in the lgl mutant suggested that integration of the lgl transgene may have inactivated a distant regulatory element or that the hypermethylation associated with this transgene might have spread to flanking sequences repressing Sp8 expression (26).
The interesting expression pattern of Sp8 in the limb and craniofacial regions suggested that the observed severe reduction of Sp8 expression in lgl mutants was responsible for their limb and craniofacial abnormalities. To determine Sp8 function in these tissues, we targeted loxP sequences into the second intron and the 3′ UTR (Fig. 1B). Sp8loxP/+ mice were mated to mice having germ-line Cre expression resulting in deletion of all Sp8 coding sequences as well as the Neo selectable marker. The resulting Sp8+/– mice were normal.
We performed a complementation test by interbreeding lgl+/– and Sp8+/– mice to make lgl+/–/Sp8+/– double heterozygotes. The resulting mice showed hindlimb malformations similar to those observed in lgl–/– mutants, with only the femur present. In addition, the forelimb and craniofacial malformations of double heterozygotes were more severe than for lgl–/– mutants (Table 1 and Fig. 2), thereby confirming a role for Sp8 in these aspects of the lgl–/– phenotype.
Table 1. Phenotype comparison of lgl-/-, lgl+/-/Sp8+/-, and Sp8-/- fetuses.
| Forelimb malformations
|
Cranial malformations
|
Caudal malformations
|
||||
|---|---|---|---|---|---|---|
| Genotype | Missing/dysplastic radius | Adactyly or monodactyly | Encephalocoeles | Exencephaly | Curled/bifurcated tail or vertebral fusions | Spina bifida/absent tail |
| lgl-/- | 4/36 (11%) | 5/36 (14%) | 10/18 (56%) | 1/18 (6%) | 7/18 (39%) | 0/18 (0%) |
| lgl+/-/Sp8+/- | 27/54 (50%) | 38/54 (70%) | 7/27 (26%) | 16/27 (59%) | 23/27 (85%) | 0/27 (0%) |
| Sp8-/- | 22/22 (100%) | 22/22 (100%) | 0/11 (0%) | 10/11 (91%) | 0/11 (0%) | 11/11 (100%) |
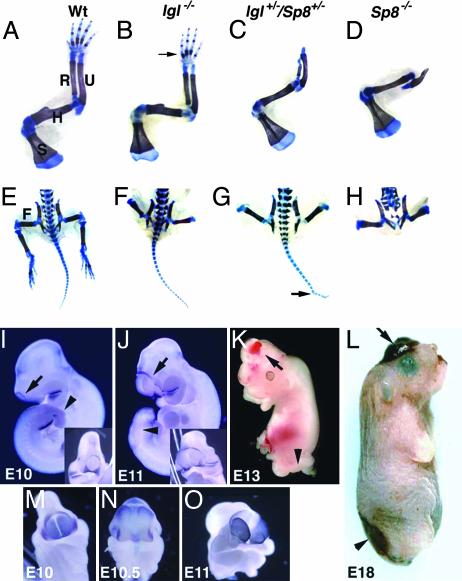
Fig. 2.
Malformations in Sp8 mutants. (A–H) Alizarin red and alcian blue limb skeletal analysis of E18 fetuses. (A–D) Forelimb skeletons. (A) S, scapula; H, humerus; R, radius; U, ulna. Arrow in B indicates absence of digit 1. (C and D) The humerus forms normally; however, the ulna is distally truncated. The radius is dysplastic (C) or absent (D) and the autopod is absent. (E–H) Caudal portion of fetuses. (E) F, femur. (F–H) Structures distal to the femur are absent. (G) Tail vertebral fusions (arrow). (H) In the Sp8–/– fetus, sacral and caudal vertebrae are absent and the lumbar vertebrae are disorganized and abnormal. (I–M and O) Sp8 null embryos at the ages indicated. (N) Wild type. (I–L) Arrows indicate open anterior neuropore/exencephaly. Arrowheads indicate open posterior neuropore/spina bifida. (I and J) Whole mount in situ with Fgf8 riboprobe arrows indicate ring of cells around open anterior neuropore. (Inset) Frontal view. (M–O) Whole mount in situ with Bf1 riboprobe emphasizing progression of telencephalon tissue through open anterior neuropore.
To further define the developmental functions of Sp8, we interbred Sp8+/– mice to make Sp8–/– progeny. Like lgl–/– and lgl+/–/Sp8+/–, Sp8–/– individuals lacked hindlimb structures distal to the femur (Fig. 2 E–H). However, the complete absence of Sp8 led to more severely affected forelimbs in which the radius and entire autopod were absent and the ulna was severely truncated (Table 1 and Fig. 2 A–D and L).
An effect of Sp8 gene dosage was observed not only in limb morphogenesis but also in closure of the neural tube. The neural tube defects, exencephaly and spina bifida, were both observed in nearly all Sp8–/– fetuses (Fig. 2L, Table 1). In contrast, only 85% of the lgl+/–/Sp8+/– fetuses had neural tube closure defects, with 59% exhibiting exencephaly and 26% possessing encephalocoeles. These defects were least evident in lgl–/– fetuses, with 56% exhibiting encephalocoeles and 6% exencephaly, with or without facial clefting (Table 1). Skeletal examinations indicated that there was a gene dosage related retardation or loss of cranial skeletal bones when comparing Sp8–/– to lgl+/–/Sp8+/– perinates that included the frontal, parietal, nasal, premaxilla, and maxilla anteriorly as well as the supraoccipital posteriorly.
In mice, the neural tube defects spina bifida, split face, exencephaly, and rachischisis are caused by failure of neural tube closure along one of four zones of the neural tube. Although a number of mouse models exhibit exencephaly with or without spina bifida, in only a few of these is the exencephaly the result of a failure in closure at position 3, the anterior neuropore (for review see ref. 27). The exencephaly resulting from lowered or absent Sp8 expression is attributable to a defect in anterior neuropore closure. In murine embryos, the anterior neuropore is normally closed by E9 of gestation (28) but remains open in E9.5 and older Sp8–/– embryos as emphasized by continued expression of Fgf8 in a ring of cells surrounding the open anterior neuropore (Fig. 2 I and J). Whole mount in situ hybridizations on E10–11 embryos with the telencephalon-specific probe Bf1 indicated that the forebrain protrudes progressively through the open anterior neuropore resulting in the exencephalic phenotype at E13 and later (Fig. 2 I–L).
Interestingly, a relationship between exencephaly and methylation has previously been postulated based on the observations that targeted mutation of the de novo methylase Dnmt3b results in exencephaly, methionine supplementation is necessary in whole embryo culture to prohibit exencephaly, and that exencephaly is often more frequently observed in females than males (for review see ref. 27). One hypothesis is that methylation of the inactivated X-chromosome in females depletes the available methylation pool reducing the normal amount of DNA, protein, and lipid methylation events. Notably, GC-rich regions are the targets of DNA methylases and also contain DNA-binding sites for members of the Sp family of transcription factors. Furthermore, human supplementation with folic acid has been credited with reducing the incidence of neural tube defects and two genes in the folate pathway require the presence of Sp binding sites within their promoters for their expression (29, 30).
In caudal regions of the embryo, Sp8–/– progeny all exhibit spina bifida accompanied by complete absence of the tail (Table 1 and Fig. 2 H and L). Lgl+/–/Sp8+/– and lgl–/– fetuses never exhibit spina bifida but do have an Sp8 gene dosage-dependent incidence of tail malformations, 85% and 39%, respectively. Fusion of the caudal vertebrae is the most commonly observed malformation (Fig. 2G), although one bifurcated tail tip was noted.
Like the curly tail mouse mutant, the lumbosacral spina bifida observed in Sp8–/– fetuses is caused by a defect in primary neurulation (31). Posterior neuropore closure is normally completed by E10 at a position between somites 32 and 34 in the mouse, whereas secondary neurulation doesn't begin until somite 32 (32). In the Sp8–/– embryos, posterior neuropore defects are first evident on E9.5 as a failure of the posterior neuropore to close and eversion of the posterior neural folds. By E10, a lack of closure paralleling somites 25–31 is present, the tail bud is not evident, and somite formation posterior to the emerging hindlimbs has ceased (Figs. 2I and 3 F, H, and L). By E11, the patterning of somites adjacent to the hindlimbs is disrupted as evidenced by the lack of En1 expression (Fig. 3P).
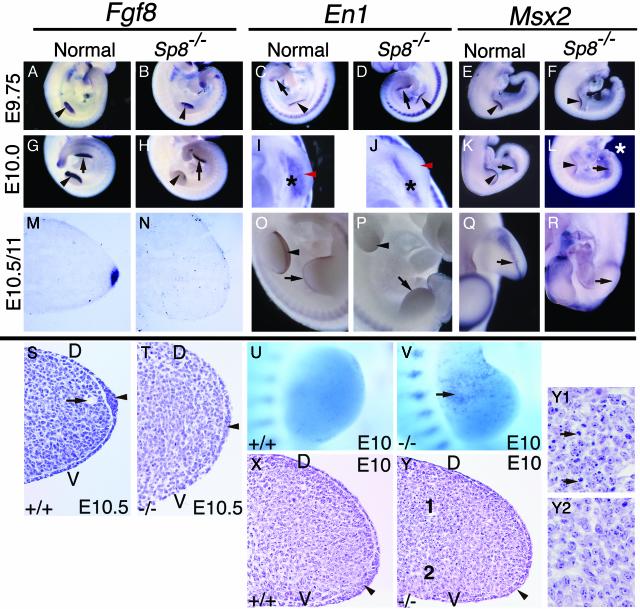
Fig. 3.
Disruption in AER morphogenesis in Sp8 null embryos. Embryo ages, genotypes, and probes used in the whole mount and section in situ hybridizations are indicated. Black arrowheads in A–Y indicate forelimb AER precursor/AER. Black arrows in A–R indicate hindlimb AER precursor. Note induction and subsequent loss of Fgf8, Msx2, and En1 expression in the fore and hindlimb buds of Sp8–/– embryos. (I and J) Black asterisks indicate ventral ectoderm expression of En1, red arrowheads indicate normal position of En1 expressing AER precursor cells and absence of expression in Sp8–/– forelimb bud. (L) White asterisk indicates truncated tail bud and open everted neural tissue adjacent to hindlimb buds. (P) Note the absence of En1 expression at distal tips of limb buds and in somites adjacent to the hindlimbs. (Q and R) Hindlimb buds, apical view. (S, T, X, Y, Y1, and Y2). Hematoxylin and eosin-stained 6-μm paraffin sections cut across the anterior/posterior axis of the forelimb. Arrowheads in S, T, X, and Y indicate AER precursor/AER. Arrow in S indicates marginal venous sinus. (U and V) Nile blue sulfate cell death staining, note punctate blue staining in anterior proximal region of Sp8–/– limb indicated by arrow in V. (Y) Note presence of pyknotic nuclei in dorsal proximal region of limb seen at higher magnification in Y1 compared with the proximal ventral mesoderm presented in Y2. D, dorsal; V, ventral.
In the emerging limb bud, Sp8 is coexpressed throughout the limb ectoderm with Wnt3, p63, and Fgfr2b. However, in the absence of Sp8, the humerus and femur do form, whereas all hindlimb structures are absent upon removal of Wnt3 (1), β-catenin (1), p63 (33, 34), Bmpr1a (2), or Fgfr2b (4, 5) expression from the limb. Furthermore, the expression of Fgf8 and other markers of the AER precursor population was not detectable in the ventro-distal ectoderm of emerging limb buds of Fgfr2b, Bmpr1a, or β-catenin-deficient embryos. We have examined this cell population in the fore- and hindlimb buds of lgl+/–/Sp8+/– and Sp8–/– embryos on E9–11 of gestation histologically and by whole mount and section in situ hybridization. In stage 1 fore- and hindlimb buds, Fgf8, En1, Msx2, and p63 expression was initiated, but the expression of these genes was rapidly lost and an AER failed to form (Fig. 3 A–T, data not shown). Although En1 expression in the disto-ventral AER precursor was undetectable in stage 2 and older Sp8–/– limb buds, expression was detectable in the proximo-ventral ectoderm (Fig. 3 I and J), confirming initial establishment of the dorsal/ventral axis in the Sp8–/– limb bud. In addition to being expressed in the AER precursor, p63 is also normally expressed by the dorsal and ventral limb ectoderm (33, 34). This expression in non-AER ectoderm was unaffected in Sp8–/– embryos (data not shown). By E10.5/11 Msx2 expression in the fore- and hindlimb buds, respectively, was undetectable in the AER precursor cells. Small expression domains were, however, transiently detectable in the anterior and posterior mesoderm (Fig. 3 Q and R).
Because an AER can still form in the absence of both Fgf8 and Fgf4 (35), we also evaluated AER formation histologically in the forelimb buds of E10 and E10.5 Sp8–/– embryos. AER precursor cells were present as a thickened population of cells at E10 (Fig. 3 X and Y). At E10.5, a distinct AER has formed across the entire anterior posterior axis of nonmutant limbs (Fig. 3 M and S). In contrast, most of the apical ectoderm in Sp8–/– embryos is indistinguishable from adjacent dorsal and ventral ectoderm. A few regions of thickened ectoderm were observed in the very middle of some limb buds (Fig. 3T); however, neither Fgf8 nor Msx2 expression was detectable by section in situ hybridization within this population of cells at E10.5 (Fig. 3N and data not shown). Consistent with our conclusion that a functional AER fails to form, in all mutant limb buds examined there was no evidence of the marginal venous sinus in the distal limb mesoderm (Fig. 3S), a structure that forms in response to AER signaling.
Surgical removal of the ridge or a ridge lacking FGF signaling results in mesenchymal cell death (35–37). Cell death in Sp8–/– forelimb buds was assayed by staining with the vital dye Nile blue sulfate. Abnormal cell death was first evident in stage 1 forelimbs of E9.5 Sp8–/– embryos (data not shown). A prominent anterior, proximal zone of cell death was evident in stage 2–3 Sp8–/– limb buds on E10, whereas no cell death was observed in this region of wild-type littermates (Fig. 3 U and V). Histological sections confirmed the presence of pyknotic nuclei in an anterior, dorsal, proximal region of the Sp8–/– forelimb (Fig. 3 Y and Y1). This zone of cell death in Sp8–/– embryos is very similar to that observed in Fgf 4/8 double knockout limbs (35), but its location contrasts with that found in the distal mesenchyme after surgical removal of the AER (36, 37).
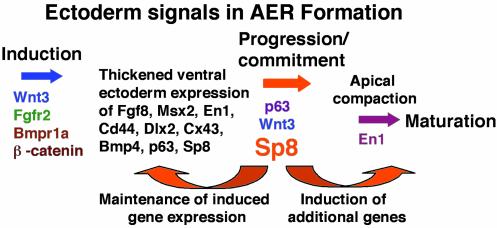
The observed early transient expression of Fgf8 in Sp8–/– embryos is consistent with the ability of the limb buds to specify the most proximal limb skeletal elements (35). The initial induction of the AER precursor population expressing Fgf8, Msx2, and En1 in Sp8–/– embryos is in agreement with our previous studies of lgl–/– hindlimb buds, which indicated that the AER precursor population was induced and expressed a variety of markers including Fgf8, En1, Dlx2, Cd44, Bmp4, Cx43, and Msx2 (6). However, by stage 2, the expression of all of these markers was lost, the AER precursor cells failed to localize at the disto-ventral margin, and limb outgrowth subsequently ceased. As proposed in the AER induction model in Fig. 4, these observations show that Sp8 functions downstream of molecules like Wnt3, β-catenin, and Bmpr1a in the signaling cascade leading to AER precursor induction. Our data indicate that Sp8 is required to maintain early AER genetic marker expression and also that developmental progression of the AER requires Sp8 expression. Sp8 may be required to maintain expression of a key signaling molecule like Wnt3 or a component of the Wnt signaling pathway. Notably, like the limb, tail bud formation also depends on Wnt signaling (38, 39), and the tail bud is also a site of Sp8 expression and dysmorphogenesis in Sp8–/– embryos.
Fig. 4.
Proposed model of genes involved in the process of AER formation. Only the genes in color have been implicated as being required for normal AER formation.
In summary, we have presented evidence indicating that Sp8 is a key player in the genetic pathways that mediate limb outgrowth and neural tube closure. Unlike other genes involved in early AER formation, Sp8 is critical for maintenance/progression of the induced AER precursor population to the disto-ventral margin of the limb. The observed anterior and posterior neural tube defects indicate that Sp8 is also required in the process of neuropore closure. Because neural tube defects are one of the most common human malformations observed (40) and high levels of amino acid identity are observed between the murine and human Sp8 orthologues, Sp8 represents a new candidate gene for these devastating birth defects. There are numerous case reports of chromosomal excess and deficiency that include 7p21 and are associated with craniofacial, limb, CNS, urogenital, cardiovascular, and gastrointestinal malformations (41, 42). Future studies will be needed to distinguish any involvement of Sp8 in these human syndromes and to define the genes regulated by Sp8 that are responsible for AER maturation and neuropore closure in the mouse.
Acknowledgments
This work would not have been possible without the extensive studies of the transgene integration site performed by Dr. Dorothy Supp and the technical assistance of Brad Huntsman. All animal protocols were approved by the Institutional Animal Care and Use Committee in accordance with National Institutes of Health guidelines. This work was funded by National Institutes of Health Grant HD24517.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AER, apical ectodermal ridge; En, embryonic day n.
References
- 1.Barrow, J. R., Thomas, K. R., Boussadia-Zahui, O., Moore, R., Kemler, R., Capecchi, M. R. & McMahon, A. P. (2003) Genes Dev. 17, 394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, K., Mishina, Y., Hanks, M. C., Behringer, R. R. & Crenshaw, E. B. I. (2001) Development (Cambridge, U.K.) 128, 4449–4461. [DOI] [PubMed] [Google Scholar]
- 3.Min, H., Danilenko, D. M., Scully, S. A., Bolon, B., Ring, B. D., Tarpley, J. E., DeRose, M. & Simonet, W. S. (1998) Genes Dev. 12, 3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu, X., Weinstein, M., Li, Cuiling, Naski, M., Cohen, R. I., Ornitz, D. M., Leder, P. & Deng, C. (1998) Development (Cambridge, U.K.) 125, 753–765. [DOI] [PubMed] [Google Scholar]
- 5.Moerlooze, D. L., Spencer-Dene, B., Revest, J.-M., Hajihosseini, M., Rosewell, I. & Dickson, C. (2000) Development (Cambridge, U.K.) 127, 483–492. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. M., Schreiner, C. M. & Scott, W. J. (1998) Mech. Dev. 74, 41–50. [DOI] [PubMed] [Google Scholar]
- 7.Loomis, C. A., Kimmel, R., Tong, C.-X., Michaud, J. & Joyner, A. L. (1998) Development (Cambridge, U.K.) 125, 1137–1148. [DOI] [PubMed] [Google Scholar]
- 8.McNeish, J., Scott, W. & Potter, S. (1988) Science 241, 837–839. [DOI] [PubMed] [Google Scholar]
- 9.McNeish, J, Thayer, J., Walling, K., Sulik, K., Potter, S. & Scott, W. (1990) J. Exp. Zool. 253, 151–162. [DOI] [PubMed] [Google Scholar]
- 10.Supp, D. M., Witte, D. P., Branford, W. W., Smith, E. P. & Potter, S. S. (1996) Dev. Biol. 176, 284–299. [DOI] [PubMed] [Google Scholar]
- 11.Supp, D. M., Brueckner, M., Kuehn, M. R., Witte, D. P., Lowe, L. A., McGrath, J., Corrales, J. & Potter, S. S. (1999) Development (Cambridge, U.K.) 126, 5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schock, F., Purnell, B. A., Wimmer, E. A. & Jackle, H. (1999) Mech. Dev. 89, 125–132. [DOI] [PubMed] [Google Scholar]
- 13.Bouwman, P. & Philipsen, S. (2002) Mol. Cell. Endocrinol. 195, 27–38. [DOI] [PubMed] [Google Scholar]
- 14.Marin, M., Karis, A., Visser, P., Grosveld, F. & Philipsen, S. (1997) Cell 89, 619–628. [DOI] [PubMed] [Google Scholar]
- 15.Bouwman, P., Gollner, H., Elsasser, H. P., Eckhoff, G., Karis, A., Grosveld, F., Philipsen, S. & Suske, G. (2000) EMBO J. 19, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gollner, H., Dani, C., Phillips, B., Philipsen, S. & Suske, G. (2001) Mech. Dev. 106, 77–83. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen-Tran, V. T., Kubalak, S. W., Minamisawa, S., Fiset, C., Wollert, K. C., Brown, A. B., Ruiz-Lozano, P., Barrere-Lemaire, S., Kondo, R., Norman, L. W., et al. (2000) Cell 102, 671–682. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, S. M., Houzelstein, D., Dunwoodie, S. L. & Beddington, R. S. (2000) Dev. Biol. 227, 358–372. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R. & Crombrugghe, B. (2002) Cell 108, 17–29. [DOI] [PubMed] [Google Scholar]
- 20.Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., Alt, F. W. & Westphal, H. (1996) Proc. Natl. Acad. Sci. USA 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toresson, H., Mata de Urquiza, A., Fagerstrom, C., Perlmann, T. & Campbell, K. (1999) Development (Cambridge, U.K.) 126, 1317–1326. [DOI] [PubMed] [Google Scholar]
- 22.Kuczuk, M. H. & Scott, W. J. (1984) Teratology 29, 427–435. [DOI] [PubMed] [Google Scholar]
- 23.The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team (2002) Nature 420, 563–573.12466851 [Google Scholar]
- 24.Scohy, S. Gabant, P., Reeth, T. V., Hertveldt, V., Dreze, P.-L., Vooren, P. V., Riviere, M., Szpirer, J. & Szpirer, C. (2000) Genomics 70, 93–101. [DOI] [PubMed] [Google Scholar]
- 25.Treichel, D., Becker, M.-B. & Gruss, P. (2001) Mech. Dev. 101, 175–179. [DOI] [PubMed] [Google Scholar]
- 26.Singh, G., Supp, D. M., Schreiner, C., McNeish, J., Merker, H. J., Copeland, N. G., Jenkins, N. A., Potter, S. S. & Scott, W. (1991) Genes Dev. 5, 2245–2255. [DOI] [PubMed] [Google Scholar]
- 27.Juriloff, D. M. & Harris, M. J. (2000) Hum. Mol. Genet. 9, 993–1000. [DOI] [PubMed] [Google Scholar]
- 28.Sakai, Y. (1989) Anat. Rec. 223, 194–203. [DOI] [PubMed] [Google Scholar]
- 29.Park, K. K., Rue, S.-W., Lee, I.-S., Kim, H.-C., Lee, I.-K., Ahn, J. D., Kim, H.-S., Yu, T.-S., Kwak, J.-Y., Henitz, N. H., et al. (2003) Biochem. Biophys. Res. Commun. 306, 239–243. [DOI] [PubMed] [Google Scholar]
- 30.Hao, H., Qi, H. & Ratnam, M. (2003) Blood 101, 4551–4560. [DOI] [PubMed] [Google Scholar]
- 31.Schoenwolf, G. C. (1984) Am. J. Anat. 169, 361–374. [DOI] [PubMed] [Google Scholar]
- 32.Copp, A. J. & Brook, F. A. (1989) J. Med. Genet. 26, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills, A. A., Zheng, B., Wang, X.-J., Vogel, H., Roop, D. R. & Bradley, A. (1999) Nature 398, 708–713. [DOI] [PubMed] [Google Scholar]
- 34.Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R. T., Tabin, C., Sharpe, A., Caput, D., Crum, C. & McKeon, F. (1999) Nature 398, 714–718. [DOI] [PubMed] [Google Scholar]
- 35.Sun, X., Mariani, F. V. & Martin, G. R. (2002) Nature 418, 501–508. [DOI] [PubMed] [Google Scholar]
- 36.Rowe, D. A., Cairns, J. M. & Fallon, J. F. (1982) Dev. Biol. 93, 83–91. [DOI] [PubMed] [Google Scholar]
- 37.Dudley, A. T., Ros, M. A. & gTabin, C. J. (2002) Nature 418, 539–544. [DOI] [PubMed] [Google Scholar]
- 38.Takada, S., Stark, K. L., Shea, M. J., Vassileva, G., McMahon, J. A. & McMahon, A. P. (1994) Genes Dev. 8, 174–189. [DOI] [PubMed] [Google Scholar]
- 39.Pinson, K. I., Brennan, J., Monkley, S., Avery, B. J. & Skarnes, W. C. (2000) Nature 407, 535–538. [DOI] [PubMed] [Google Scholar]
- 40.Mulinaire, J. & Erickson, J. D. (1997) Teratology 56, 17–18. [DOI] [PubMed] [Google Scholar]
- 41.Ahrens, Y. H. J. M., Toutain, A., Engelen, J. J. M., Offermans, J. P. M., Hamers, A. J. H., Schrander, J. J. P., Pulles-Heintzberger, C. F. M. & Schrander-Stumpel, C. T. R. M. (2000) Genet. Couns. 11, 347–354. [PubMed] [Google Scholar]
- 42.Hoover-Fong, J. E. Cai, J., Cargile, C. B., Thomas, G. H., Patel, A., Griffin, C. A. & Jabs, E. W. (2003) Am. J. Med. Genet. 117A, 47–56. [DOI] [PubMed] [Google Scholar]