Abstract
Germ cells undergo epigenetic modifications as they develop, which suggests that they may be ideal donors for nuclear transfer (cloning). In this study, nuclei from confirmed embryonic germ cells were used as donors to determine whether they are competent for cloning and at which stage they are most competent. Embryos cloned from migrating 10.5-days-postcoitum (dpc) primordial germ cells (PGCs) showed normal morphological development to midgestation but died shortly thereafter. In contrast, embryos cloned from later-stage germ cells were developmentally delayed at midgestation. Thus, donor germ cell age inversely correlated with the developmental stage attained by cloned embryos. The methylation status of the H19- and Snrpn-imprinting control regions in germ cell clones paralleled that of the donors, and revealed that demethylation, or erasure of imprints, was already initiated in PGCs at 10.5 dpc and was complete by 13.5 dpc. Similarly, clones derived from male 15.5-dpc germ cells showed increased methylation correlating with the initiation of de novo methylation that resets imprints at this stage, and clones from neonatal germ cells showed nearly complete methylation in the H19 imprinting control region. These results indicate that the epigenetic state of the donor nucleus is retained in cloned embryos, and that germ cells are therefore inadequate nuclear donors for cloning because they are either erasing or resetting epigenetic patterns.
Somatic cell cloning has been successfully used to produce live cloned offspring in a variety of mammals (1–3). Although the rate of obtaining healthy offspring is very low, such successes illustrate the capacity of the egg cytoplasm and the donor somatic cell nucleus to support embryonic development.
As part of the normal course of development, embryonic germ cells are dynamically reprogrammed during germ cell migration and differentiation. In this context, reprogramming is defined as a stepwise process whereby the somatic-like epigenetic pattern, hypothesized to be characteristic of the earliest detectable primordial germ cells (PGCs), undergoes erasure and is transformed into the sex-specific pattern of mature germ cells (4, 5). This process is exemplified by reprogramming of methylation associated with imprinted genes during germ-cell development; the somatic methylation imprints associated with early PGCs are erased around 11.5 days postcoitum (dpc) in both male and female PGCs and are reestablished in a sex-specific manner with paternal methylation imprints acquired early and maternal-specific methylation obtained late in the gametogenic process (6–10).
Because embryonic germ cells by their very nature are in the process of reprogramming their genome, cells with different epigenetic patterns can be obtained and, in this study, were isolated by using a germ cell marker, EMA-1. Before migration into the genital ridges (10.5 dpc), PGCs are thought to possess a somatic-like epigenetic pattern, whereas, after migration (11.5 dpc and onward), germ cells are in various stages of reprogramming. Germ cell nuclei were used as donors in cloning experiments to determine whether these cells are competent to clone and at which stage they are most competent. Midgestation cloned embryos were examined for morphological development, which was complemented by an assessment of methylation status of two imprinting control regions (ICRs). We observed an inverse correlation between the increased age of the donor germ cells and the developmental stage obtained by cloned embryos. The failure to obtain full-term fetuses indicated that reprogramming of PGC nuclei had likely been initiated before entry into the genital ridge. In addition, we observed that the ICR methylation pattern of PGC-cloned embryos resembled that inherent in the donor cell populations. These results indicate that the reprogramming capabilities of PGCs were terminated in nuclear transfer embryos, rendering PGCs unsuitable as nuclear donors.
Materials and Methods
Animals. B6D2F1 (C57BL/6 × DBA/2) mice were used as both oocyte and nuclear donors. PGCs were isolated from B6D2F1 embryos at 9.5–16.5 dpc (11). Gonocytes were collected from B6D2F1 male neonates at 0–1 days postpartum. Surrogate mothers of cloned embryos were CD-1 females. The protocol of animal handling and treatment was reviewed and approved by the Animal Care and Use Committee of the University of Hawaii.
Media. DMEM (GIBCO/BRL) supplemented with 10–20% (vol/vol) FBS (HyClone) was used for dissecting embryos and isolating PGCs. Oocytes and preimplantation embryos were cultured in bicarbonate-buffered CZB medium (12) at 37°C under 5% CO2 in air. Oocyte manipulation was carried out in Hepes-buffered CZB (Hepes-CZB) (13) at room temperature.
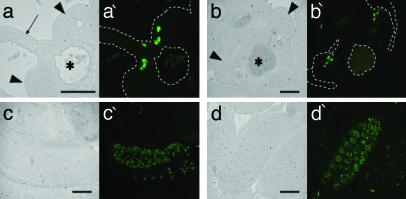
Immunohistochemistry. In situ localization of PGCs was done by using EMA-1 antibody generated by immunization of mice with Nulli SCC1 carcinoma cells. This antibody binds specifically to the surface of PGCs of 8.5- to 13.5-dpc mouse embryos (14). B6D2F1 embryos (10.5–13.5 dpc) were fixed with Bouin's fixative, embedded in paraffin, and serially sectioned at 10 μm. EMA-1 antibody was applied to the sections followed by staining with fluorescein-conjugated goat anti-mouse immunogloblin secondary antibody (Vector Laboratories). Sections were examined with a Nikon inverted microscope by using epifluorescence illumination. EMA-1-positive PGCs of 10.5-dpc embryos were located in the dorsal mesentery of the hindgut near the urogenital ridge and in the mesenchyme next to the urogenital ridge (Fig. 1 a and a′). Most PGCs were in the sexually indifferent urogenital ridges by 11.5 dpc (Fig. 1 b and b′). The size and number of PGCs in the genital ridge increased during 12.5–13.5 dpc when the gonadal sex of embryos became clearly identifiable (Fig. 1 c, c′, d, and d′).
Fig. 1.
EMA-1-positive PGCs (visualized by fluorescein staining) migrating toward the genital ridges in normal 10.5-dpc embryos. At later stages, PGCs have reached the genital ridges. Shown are a 10.5-dpc embryo (a and a′), a 11.5-dpc embryo (b and b′), a 12.5-dpc male embryo (c and c′), and a 13.5-dpc male embryo (d and d′). (a–d) Bright-field micrographs. (a′–d′) Fluorescence micrographs of the same fields. Arrow and arrowheads indicate the dorsal mesentery of hindgut and the genital ridge, respectively. *, Dorsal aorta. (Bar = 100 μm.)
Isolation and Identification of Germ Cells. Donor tissues containing PGCs (Fig. 1) were dissected from several embryos at each stage and pooled in DMEM with 20% FBS. Treatment with 0.25% trypsin and 0.04% EDTA in DMEM for 10 min at room temperature was followed by gentle pipeting to separate individual cells. After washing with DMEM containing 20% FBS, dissociated cells were treated on ice with the primary EMA-1 antibody for 30 min, followed by a 30-min treatment with FITC-labeled secondary antibody (15). After washing, cells were examined with Olympus IX70 epifluorescence microscope. Cells with strong fluorescence were selected as donor PGCs for nuclear transfer. Diameters of PGCs in 10.5-, 11.5-, 12.5-, and 13.5-dpc embryos were 13.2 ± 0.5 μm, 13.5 ± 0.5 μm, 14.6 ± 1.8 μm, and 14.5 ± 1.3 μm, respectively (n = 20–30 each). Germ cell selection in 15.5-dpc embryos was based on large size (13–15 μm), big, round nuclei, and small amounts of cytoplasm. To collect gonocytes at 0–1 days postpartum, neonatal testes were decapsulated and cell suspensions were prepared by digestion of seminiferous tubules in Hanks' balanced salt solution containing trypsin (0.17%) and DNase I (2.3 mg/ml) for 5 min at 37°C followed by pipeting. After dispersed cells were suspended in DMEM with 20% FBS, gonocytes were identified by large size and round morphology.
Preparation of Adult and Embryonic Somatic Cells. Cumulus cells were collected as described (16). Briefly, ovulated oocyte-cumulus complexes were treated with 0.1% hyaluronidase (300 units/mg, Sigma) for 5 min to disperse cumulus cells. Embryonic somatic cells were collected from normal B6D2F1 embryonic trunks between 10.5 and 13.5 dpc. After decapitation and evisceration, embryos were minced and treated with 0.25% trypsin plus 0.04% EDTA for 10 min at room temperature. Cells were dissociated by repeated pipeting. After washing, both cumulus cells and embryonic somatic cells were treated with EMA-1 and FITC-labeled antibodies and exposed under fluorescence for 15 seconds, as described for PGCs.
Nuclear Transfer and Oocyte Activation. Donor cells were placed in Hepes-CZB medium containing 12% (wt/vol) polyvinylpyrrolidone (PVP, average Mr, 360,000; ICN). Donor nuclei were individually injected into mouse oocytes from which metaphase II chromosomes had been removed (16). Reconstructed oocytes were incubated in CZB medium for 2 h (37°C) followed by a 6-h culture in Ca2+ -free CZB containing 10 mM Sr2+ and 5 μg/ml cytochalasin B. Activated oocytes were cultured in CZB medium for 1–3 days before transfer to surrogate mothers.
Intracytoplasmic Sperm Injection. Intracytoplasmic sperm injection was performed as described (17). Cauda epididymal spermatozoa were suspended in Hepes-CZB medium. A drop of sperm suspension was mixed with Hepes-CZB with 12% PVP. After injection, oocytes were cultured in CZB medium for 1 day before transfer to surrogate mothers.
Embryo Transfer and Isolation of Embryos. Embryos at the two-cell or morula/blastocyst stages were transferred into the oviducts of pseudopregnant females. The day of transfer was considered day 0.5 of pregnancy. Cloned and control embryos were recovered on day 10.5 of pregnancy. In some experiments involving 10.5-dpc PGCs (379 reconstructed oocytes, 172 two-cell embryos transferred), cloned embryos were examined on days 11.5–17.5 (33 implantation sites).
Genotyping the Sex of PGC Clones. DNA was extracted from yolk sacs by incubating at 55°C overnight in lysis buffer followed by heat inactivation at 95°C for 10 min (18). To a Ready-To-Go PCR bead (Amersham Biosciences), 1–4 μl of DNA lysate and 0.5 μM primers Zfy-5 (5′-AAGATAAGCTTACATAATCACATGGA-3′) and Zfy-3 (5′-CCTATGAAATCCTTTGCTGCACATGT-3′)or0.3 μM primers Zf1 (5′-GACAGCCTTACCGAGGTCGC-3′) and Zf2 (5′-CATGGGGGTATGCACACCTG-3′) were added. The Zfy primers (Zfy-3 and Zfy-5) assayed the sex of PGC cloned embryos (i.e., presence of Y chromosome) whereas primers for Mkrn3 (Zf1 and Zf2) served as a control for DNA extraction. Amplification was at 95°C for 2 min followed by 35 cycles at 95°C for 15 s, 58°C (Zfy) or 55°C (Mkrn3) for 10 s, and 72°C for 20 s.
DNA Preparation and Bisulfite Modification. DNA was isolated and subjected to bisulfite modification, PCR amplification, subcloning, and sequencing for the H19 and Snrpn ICRs as described (19, 20).
Results
Development of Embryos Cloned from PGCs and Somatic Cells. Before determining the competency of germ cells using nuclear transfer, it was necessary to select the appropriate donor cell type for proper interpretation of the cloning experiments (3, 21). Here, EMA-1 selection, a cell surface antigen for embryonic germ cells (14), was used to isolate live PGCs among the numerous cell types in the developing embryo (15). To test the effectiveness of this selection system, adult cumulus cells were similarly treated and used for nuclear transfer. Cloned pups were obtained at the expected rate (2.0% of two-cell cloned embryos). Thus, this antibody-selection system was reliably used to identify donor cells without effecting damage that may have been detrimental to the cloning process.
After selection, developmental competence of donor PGCs was evaluated before (10.5 dpc; somatic-like epigenotype) and after (11.5 dpc and onward; reprogramming epigenotype) migration into the genital ridges (Table 1) in nuclear transfer embryos. On average, 60–70% of the oocytes survived nuclear injection, and >30% of reconstructed oocytes cleaved. Seven live embryos derived from 10.5-dpc PGC donor nuclei were observed on day 10.5 of pregnancy. All embryos had a heartbeat and appeared normal by morphological criteria (Figs. 2 and 3). When embryos derived from 10.5-dpc PGC nuclei were allowed to develop further (days 11.5–17.5), we obtained three cloned embryos on days 11.5 (2) and 12.5 (1) of pregnancy (data not shown). Although one embryo showed normal development on day 11.5, its placenta was large and partially degenerating. Histological examination revealed two massive placental hemorrhages that would likely have been fatal by midgestation due to placental dysfunction (data not shown). The other two cloned embryos were dead and degenerated. Thus far, a full-term fetus has not been generated from a 10.5-dpc PGC donor nucleus. Of six embryos cloned from 11.5-dpc PGC nuclei, only one displayed normal morphology at day 10.5 of pregnancy; others were retarded in their development (Figs. 2 and 3). Retardation in embryonic development was more distinct when either male or female PGCs of older embryos (12.5–13.5 dpc) were used for cloning.
Table 1. Development of cloned germ cell embryos.
| Age of donor cells | Reconstructed oocytes | Two-cell embryos (%)* | Morulae and blastocysts (%)† | No. of embryos transferred (stage) | Total implantation sites (%)† | Embryos on day 10.5 of pregnancy (%)† | Normally developed embryos |
|---|---|---|---|---|---|---|---|
| 10.5 dpc | 415 | 171 (41.2) | 20/47 (42) | 124 (2-c), 20 (M/B) | 48 (28.1) | 7 (4.1) | 7 |
| 11.5 dpc | 226 | 104 (46.0) | — | 104 (2-c) | 46 (44.2) | 6 (5.8) | 1 |
| 12.5 dpc (M) | 191 | 70 (36.6) | 7/13 (54) | 57 (2-c), 7 (M/B) | 31 (44.3) | 4 (5.7) | 0 |
| 12.5 dpc (F) | 63 | 24 (38.1) | — | 24 (2-c) | 11 (45.8) | 3 (12.5) | 0 |
| 13.5 dpc (M) | 146 | 64 (43.8) | 15/25 (60) | 39 (2-c), 15 (M/B) | 26 (40.6) | 3 (4.7) | 0 |
| 13.5 dpc (F) | 131 | 68 (51.9) | — | 68 (2-c) | 35 (51.5) | 9 (13.2) | 0 |
| 15.5 dpc (M) | 132 | 100 (75.8) | 47/47 (100) | 53 (2-c), 47 (M/B) | 44 (44.0) | 26 (26.0) | 0 |
| 15.5 dpc (F) | 95 | 33 (34.7) | 0/9 (0) | 24 (2-c) | 5 (15.2) | 1 (3.0) | 0 |
| 0-1 dpp (M) | 230 | 207 (90.0) | 85/112 (76) | 95 (2-c), 85 (M/B) | 92 (44.4) | 33 (15.9) | 0 |
| Embryo somatic cell | 270 | 144 (53.3) | 10/49 (20) | 95 (2-c), 10 (M/B) | 24 (16.7) | 5 (3.5) | 3 |
| Adult cumulus cell | 156 | 132 (84.6) | — | 132 (2-c) | 51 (38.6) | 7 (5.3) | 3 |
2-c, two-cell embryos; M/B, morulae and blastocysts; M, male; F, female; dpp, days postpartum.
% of reconstructed oocytes.
% of two-cell embryos.
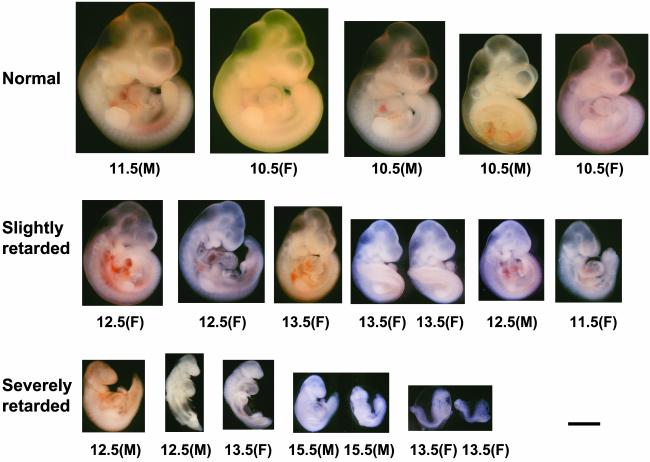
Fig. 2.
Gross morphology of embryos cloned with PGC nuclei. Embryos were examined on day 10.5 of pregnancy and classified according to ref. 33. Normal: phenotypically similar to normal embryos at 10–10.5 dpc; forelimb and hindlimb buds are formed and the telencephalon is expanded. Slightly retarded: developmental stage corresponds to that of normal embryos at ≈9.5 dpc; forelimb buds are well developed. Severely retarded: developmental stage corresponds to that of 8.0- to 9.0-dpc normal embryos. The ages of PGC donors are indicated under each embryo. M, male; F, female. (Bar = 1 mm.)
Fig. 3.
Categorization of normal and developmentally retarded PGC and somatic cell clones. Embryos on day 10.5 of pregnancy were classified into three groups with respect to their development status (normal, slightly retarded, and severely retarded; Fig. 2). Age of PGC nuclei is indicated. Each circle represents one cloned embryo. F, female; M, male.
When 15.5-dpc PGC nuclei were used as donors, almost all cloned embryos were severely delayed in their development. The rate of obtaining embryos, however, varied between male and female PGCs. Although nuclei from males supported a relatively high rate of development (26% of two-cell cloned embryos advanced to 10.5 days of pregnancy; Table 1), few 10.5-day embryos were derived from female donors (3.0% compared with 12.5% and 13.2% for female PGCs at 12.5 and 13.5 dpc, respectively; Table 1). Chromosomal analysis of female 16.5-dpc germ cells confirmed that these cells had entered meiosis (data not shown). Female germ cells begin to enter meiosis at 13.5 dpc. The entire population reaches the diplotene of first meiotic prophase by 15.5 dpc and then undergoes meiotic arrest. Thus, the decrease in developmental potential observed for female germ cell nuclei between 13.5 and 15.5 dpc is likely associated with the transition from a predominantly mitotic to a predominantly meiotic state. Conversely, male germ cells remain mitotic throughout embryonic development but cease dividing between 13.5 and 15.5 dpc. Thus, the transition of these cells into a G0 cell-cycle state may account for the increased developmental potential of 15.5-dpc PGC clones. When male gonocytes (0–1 day postpartum) were used as a source of donor nuclei, 15.9% of two-cell cloned embryos developed to day 10.5 of pregnancy (Table 1). However, almost all cloned embryos showed severe developmental delay.
Throughout these experiments, embryonic somatic cells and cumulus cells were used to control for the nuclear transfer procedure. About half of somatic cell-cloned embryos were normal in appearance on day 10.5 of pregnancy; others were considerably delayed in their development (Fig. 3), consistent with previous experiments (22). In contrast to 10.5-dpc PGCs, cloned pups were obtained at the expected rate (2.0%) when cumulus cells were used as nuclear donors.
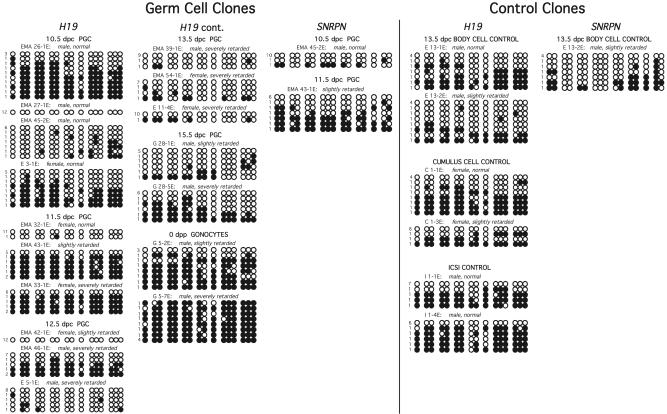
Methylation Analysis of ICRs in Germ Cell-Cloned Embryos. To assess the epigenetic state of the PGC-cloned embryos, we analyzed the methylation status of the ICRs of the H19 and Snrpn genes. In somatic cells of normal embryos, the differentially methylated domain (DMD) of the H19 gene is paternally hypermethylated (23), whereas the Snrpn promoter-exon 1 region is maternally hypermethylated (24). Cloned embryos isolated on day 10.5 of pregnancy were assayed at the 5′ portion of the H19 DMD by using the bisulfite mutagenesis assay (Fig. 4). Two of four and two of three cloned embryos derived from 10.5- and 11.5-dpc PGC donor nuclei, respectively, yielded a biparental, somatic-type pattern of methylation with approximately half the strands hypermethylated (>50% of cytosine residues methylated) and the other half hypomethylated. These embryos were likely derived from PGCs that had not initiated erasure at the H19 locus. The other embryos showed hypomethylated patterns, suggesting that the donors had initiated the characteristic genomewide erasure that occurs in PGCs that have entered the genital ridge. No difference was observed in the methylation state of clones derived from male or female PGCs. Two cloned embryos, one each from the 10.5- and 11.5-dpc PGC donor populations, were also assayed by bisulfite mutagenesis at the Snrpn ICR (Fig. 4). These embryos showed the same pattern noted for the H19 DMD. Embryo 45-2E was hypomethylated both for H19 and Snrpn, and embryo 43-1E exhibited the biparental, somatic-type methylation pattern for both regions.
Fig. 4.
Methylation status of cloned PGC embryos at the H19 and Snrpn ICRs. Unmethylated CpGs are represented as open circles, whereas methylated CpGs are depicted as filled circles. Each line denotes an individual strand of DNA in the H19 upstream DMD and Snrpn promoter-exon 1 region with the number of strands showing a given pattern indicated to the left. One to four clones were analyzed for each donor type. The identity, sex, and morphology of the cloned embryos are indicated above each sample. Placentas were also assayed and gave similar results to the corresponding embryo (data not shown).
The analysis of H19 DMD methylation in cloned embryos derived from 12.5- and 13.5-dpc PGC nuclei revealed that 2/3 and 3/3, respectively, exhibited a hypomethylation pattern (Fig. 4). As suggested by the methylation analysis of germ cells, genomewide erasure is likely complete by 13.5 dpc (6–8, 25, 26). Although one of the two clones generated from male 15.5-dpc PGCs was hypomethylated at the H19 DMD, the other clone exhibited a biparental, somatic pattern of methylation. It was shown that male 15.5-dpc germ cells had initiated remethylation on most but not all of the paternal alleles, whereas the maternal allele remained unmethylated (7). It is likely that the former clone represented one of those germ cells that had neither allele methylated and the latter clone was derived from one of the cells that had initiated methylation of the H19 paternal allele. Finally, as reported for the germ cells of newborn male mice (19, 25, 27), both clones derived from newborn spermatogenic cells were hypermethylated at the H19 DMD. Similar results were obtained for the corresponding placentas of these PGC clones (data not shown).
Control embryos cloned by using a 13.5-dpc embryo somatic cell and adult cumulus cell nuclei as donors were examined to determine whether the nuclear transfer procedure affected methylation patterns. Additionally, intracytoplasmic sperm injection-generated embryos were assayed. In general, although control embryos exhibited a biparental, somatic pattern of methylation as expected for the donor cell populations, a slightly lower-than-expected frequency of hypermethylated strands was observed in clones that survived to 10.5 days of pregnancy (Fig. 4). It is likely that embryos experiencing major epigenetic disregulation because of the cloning procedure failed to survive to this point of development.
Discussion
Previous cloning success was reported by using germ cells that had reached the genital ridge (11.5–16.5 dpc) and were selected solely by morphological criteria (9, 28). These cloned embryos were typically growth retarded on days 9.5–10.5 of pregnancy (9, 28). In the current study, after successfully applying the above selection system, the developmental potential of donor germ cell nuclei was assessed in cloned embryos on day 10.5 of pregnancy by morphology analysis. We observed the same growth retardation in 11.5- to 15.5-dpc PGC clones with an inverse correlation between increased age of the donor PGCs population and developmental stage obtained by PGC cloned embryos. We also extended these findings by generating cloned embryos derived from 10.5-dpc migrating PGCs. All embryos appeared morphologically normal on day 10.5 of pregnancy. However, these PGC nuclei failed to support full-term clonal development, indicating that reprogramming of PGC nuclei had likely been initiated before entry into the genital ridge and that these cells were not truly somatic in their epigenotype. The substantial difference in morphological development in clones derived from 10.5 dpc and older PGC nuclei indicates that migrating and postmigratory germ cells possess different developmental potential. Once PGCs reach the embryonic gonad, they rapidly lose the developmental potential they possess before that stage. We suggest that cessation of development likely reflects the degree to which nuclei have departed from a somatic epigenetic pattern, with clones derived from early PGCs having the greatest developmental potential (≈10.5 of pregnancy) and the least reprogramming, and the most developmentally delayed embryos having complete erasure of the somatic epigenotype.
To assess the epigenetic state of embryos cloned with PGCs in pre- and postmigratory phases, we analyzed the methylation status of the H19 and Snrpn ICRs. Specifically, half of the clones exhibited extensive hypomethylation of the H19 DMD in embryonic and placental tissues, indicating that individual migrating PGCs at 10.5 dpc had initiated demethylation of the H19 and Snrpn imprinting control regions. Genome demethylation continued in a subset of cloned embryos derived from 11.5- and 12.5-dpc PGCs. Finally, all clones derived from 13.5-dpc PGCs were essentially devoid of methylation at the H19 DMD, in agreement with previous studies showing that this germ-cell population has completed genomewide demethylation (6–9, 25, 26, 29). Similar to demethylation events, methylation of the H19 DMD in male PGC clones paralleled that of donor PGCs. Specifically, a proportion of clones derived from 15.5-dpc PGCs displayed a biparental methylation pattern whereas all clones generated from newborn male germ cells were hypermethylated, consistent with the paternal allele becoming methylated at ≈15.5 dpc and the maternal allele becoming methylated thereafter (7).
Although all studies indicate that methylation erasure is completed by 13.5 dpc, discordance exists for the timing of demethylation initiation. Methylation analysis of PGCs (9, 30) and PGC clones (this study) indicated that some 10.5-dpc PGCs had initiated demethylation, whereas others reported that demethylation had not commenced until 11.5 dpc (8). These contrasting results may be due to differences in experimental strategies, to differences in the mouse strains used, or to the analysis of a population of PGCs compared with this study where clones derived from single PGCs were assayed. With respect to the latter, however, early demethylation was observed in a population of PGCs by others (9, 30). Interestingly, few genes were actually analyzed in 10.5-dpc PGCs in the study by Hajkova et al. (8). Two assayed genes, Igf2 and Snrpn, did exhibit some demethylation in 10.5-dpc PGCs. In addition, several genes, such as H19 and Peg3, had lower levels of methylation than expected in 11.5-dpc PGCs, leaving open the possibility for earlier commencement of demethylation.
Demethylation at imprinted loci in early-stage PGCs is supported by imprinted gene expression studies. Four imprinted genes, including H19 and Snrpn, exhibited imprinted expression in 9.5-dpc PGCs (31). By 10.5 days, PGCs begin to switch to a biallelic mode of expression, with contributions from the normally silent alleles of H19 and Snprn increasing thereafter. This report supports the conclusion that methylation erasure begins before PGCs reached the genital ridge in a proportion of PGCs (9, 30, 31).
Based on the coincidence of demethylation of imprinted and nonimprinted loci after entry of the PGCs into the genital ridge, Hajkova et al. (8) proposed two hypotheses for demethylation initiation. In the first, a signal(s) from somatic cells in the genital ridge influences the onset of reprogramming induced demethylation, and, in the second, onset of reprogramming may be regulated by a developmental clock that is intrinsic to germ cells. The initiation of demethylation in some PGCs at 10.5 dpc supports an intrinsic clock mechanism, although both mechanisms may be involved.
Regardless of the actual time of initiation of demethylation, the reprogramming of PGCs appears to occur over a relative short window of time. This raises the question of whether demethylation occurs synchronously or in a gene-independent manner. We observed that, within a given PGC clone, the response of the H19 and Snrpn ICRs was closely matched. Similar results were reported for H19 and Peg10 in the majority of 11.5-dpc-PGC-cloned embryos (9). Together these results suggest that, once a cell starts, demethylation at imprinted loci may occur uniformly.
An interesting finding from this study, as well as in previous studies (9, 28), was the high cloning success rate by clones derived from male 15.5-dpc PGCs: 26% of two-cell embryos developed to 10.5 days of pregnancy. It has been suggested that the cells in G0/G1 of the cell cycle are the most suitable for cloning (16, 22, 28, 32). Although male germ cells at 15.5 dpc are in G0 mitotic arrest, the success of achieving postimplantation development was much higher than would be predicted when compared with somatic donor cells at a similar cell cycle stage; for example, only 1.7% of Sertoli cell clones (16), 5.3% of cumulus cell clones (this study), and 13.2% of fetal neural cell clones (22) develop to a similar stage. Thus, factor(s) in addition to cell cycle may influence the successful development of embryos cloned from 15.5-dpc PGCs. One additional possibility for the higher success rate may be that germ cell nuclei are beginning to establish male-specific imprinting marks and somatic genomewide methylation at nonimprinted genes (7, 26). For example, for the H19 DMD, the paternal H19 allele becomes methylated at ≈15.5 dpc, with the maternal allele becoming methylated thereafter (7). Thus, the somatic-like differential methylation pattern is transiently reestablished in male PGCs. Ultimately, these clones are severely compromised in their morphology, indicating that the potential establishment of some imprints is insufficient for normal development.
In conclusion, although germ cells by their very nature are often thought to be totipotent and therefore hypothetically ideal nuclear donors for cloning experiments, they are in fact not competent to act as nuclear donors to produce viable cloned offspring that can complete development. PGCs have typically erased or are in the process of establishing epigenetic marks. Thus germ cells should perhaps be viewed as “hemipotent” donors for nuclear transfer.
Acknowledgments
We thank Drs. Eihachiro Kawase, Hirokazu Kusakabe, and Satoshi Tanaka for histological suggestions and analyses, and Richard Chaillet, Jacquetta Trasler, Carmen Sapienza, and Howard Cedar for helpful comments on the manuscript. The EMA-1 antibody developed by Drs. Mitch Eddy and Ann Hahnel was obtained from the Developmental Studies Hybridoma Bank (Iowa City, IA). This work was supported by National Institutes of Health Grant HD042772 (to Y.Y., J.R.M., and R.Y.) and the Howard Hughes Medical Institute (M.S.B.). M.R.W.M. was supported by a grant from the Lalor Foundation.
Abbreviations: PGC, primordial germ cell; ICR, imprinting control region; dpc, days postcoitum; DMD, differentially methylated domain.
References
- 1.Solter, D. (2000) Nat. Rev. Genet. 1, 199–207. [DOI] [PubMed] [Google Scholar]
- 2.Wakayama, T. & Yanagimachi, R. (1999) Semin. Cell Dev. Biol. 10, 253–258. [DOI] [PubMed] [Google Scholar]
- 3.Hochedlinger, K. & Jaenisch, R. (2002) Curr. Opin. Cell. Biol. 14, 741–748. [DOI] [PubMed] [Google Scholar]
- 4.Reik, W., Dean, W. & Walter, J. (2001) Science 293, 1089–1093. [DOI] [PubMed] [Google Scholar]
- 5.Surani, M. A. (2001) Nature 414, 122–128. [DOI] [PubMed] [Google Scholar]
- 6.Chaillet, J. R., Vogt, T. F., Beier, D. R. & Leder, P. (1991) Cell 66, 77–83. [DOI] [PubMed] [Google Scholar]
- 7.Davis, T. L., Yang, G. J., McCarrey, J. R. & Bartolomei, M. S. (2000) Hum. Mol. Genet. 9, 2885–2894. [DOI] [PubMed] [Google Scholar]
- 8.Hajkova, P., Erhardt, S., Lane, N., Haaf, T., El-Maarri, O., Reik, W., Walter, J. & Surani, M. A. (2002) Mech. Dev. 117, 15–23. [DOI] [PubMed] [Google Scholar]
- 9.Lee, J., Inoue, K., Ono, R., Ogonuki, N., Kohda, T., Kaneko-Ishino, T., Ogura, A. & Ishino, F. (2002) Development (Cambridge, U.K.) 129, 1807–1817. [DOI] [PubMed] [Google Scholar]
- 10.Lucifero, D., Mertineit, C., Clarke, H. J., Bestor, T. H. & Trasler, J. M. (2002) Genomics 79, 530–538. [DOI] [PubMed] [Google Scholar]
- 11.McCarrey, J. R. (1993) in Cell and Molecular Biology of the Testis, eds. Desjardins, C. & Ewing, L. L. (Oxford Univ. Press, Oxford), pp. 58–89.
- 12.Chatot, C. L., Lewis, J. L., Torres, I. & Ziomek, C. A. (1990) Biol. Reprod. 42, 432–440. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, Y. & Yanagimachi, R. (1995) Biol. Reprod. 52, 709–720. [DOI] [PubMed] [Google Scholar]
- 14.Hahnel, A. C. & Eddy, E. M. (1986) Gamete Res. 15, 25–34. [Google Scholar]
- 15.McCarrey, J. R., Hsu, K. C., Eddy, E. M., Klevecz, R. R. & Bolen, J. L. (1987) J. Exp. Zool. 242, 107–111. [DOI] [PubMed] [Google Scholar]
- 16.Wakayama, T., Perry, A. C. F., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. (1998) Nature 394, 369–373. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, Y. & Yanagimachi, R. (1995) Development (Cambridge, U.K.) 121, 2397–2405. [DOI] [PubMed] [Google Scholar]
- 18.Percec, I., Plenge, R. M., Nadeau, J. H., Bartolomei, M. S. & Willard, H. F. (2002) Science 296, 1136–1139. [DOI] [PubMed] [Google Scholar]
- 19.Davis, T. L., Trasler, J. M., Moss, S. B., Yang, G. J. & Bartolomei, M. S. (1999) Genomics 58, 18–28. [DOI] [PubMed] [Google Scholar]
- 20.Mann, M. R. W., Chung, Y. G., Nolen, L. D., Verona, R. I., Latham, K. E. & Bartolomei, M. S. (2003) Biol. Reprod. 69, 902–914. [DOI] [PubMed] [Google Scholar]
- 21.Wakayama, T. & Yanagimachi, R. (2001) Mol. Reprod. Dev. 58, 376–383. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki, Y., Makino, H., Hamaguchi-Hamada, K., Hamada, S., Sugino, H., Kawase, E., Miyata, T., Ogawa, M., Yanagimachi, R. & Yagi, T. (2001) Proc. Natl. Acad. Sci. USA 98, 14022–14026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremblay, K. D., Duran, K. L. & Bartolomei, M. S. (1997) Mol. Cell. Biol. 17, 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shemer, R., Birger, Y., Riggs, A. D. & Razin, A. (1997) Proc. Natl. Acad. Sci. USA 94, 10267–10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandeis, M., Kafri, T., Ariel, M., Chaillet, J. R., McCarrey, J., Razin, A. & Cedar, H. (1993) EMBO J. 12, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kafri, T., Ariel, M., Brandeis, M., Shemer, R., Urven, L., McCarrey, J., Cedar, H. & Razin, A. (1992) Genes Dev. 6, 705–714. [DOI] [PubMed] [Google Scholar]
- 27.Ueda, T., Abe, K., Miura, A., Yuzuriha, M., Zubair, M., Noguchi, M., Niwa, K., Kawase, Y., Kona, T., Matsuda, Y., et al. (2000) Genes Cells 5, 649–659. [DOI] [PubMed] [Google Scholar]
- 28.Kato, Y., Rideout, W. M., III, Hilton, K., Barton, S. C., Tsunoda, Y. & Surani, M. A. (1999) Development (Cambridge, U.K.) 126, 1823–1832. [DOI] [PubMed] [Google Scholar]
- 29.Monk, M., Boubelik, M. & Lehnert, S. (1987) Development (Cambridge, U.K.) 99, 371–382. [DOI] [PubMed] [Google Scholar]
- 30.Sato, S., Yoshimizu, T., Sato, E. & Matsui, Y. (2003) Mol. Reprod. Dev. 65, 41–50. [DOI] [PubMed] [Google Scholar]
- 31.Szabo, P. E., Hubner, K., Scholer, H. & Mann, J. R. (2002) Mech. Dev. 115, 157–160. [DOI] [PubMed] [Google Scholar]
- 32.Ogura, A., Inoue, K., Ogonuki, N., Noguchi, A., Takano, K., Nagano, R., Suzuki, O., Lee, J., Ishino, F. & Matsuda, J. (2000) Biol. Reprod. 62, 1579–1584. [DOI] [PubMed] [Google Scholar]
- 33.Kaufman, M. H. (1992) The Atlas of Mouse Development (Academic, London), pp. 40–144.