Abstract
Olfactory receptor (OR) genes form the largest known multigene family in the human genome. To obtain some insight into their evolutionary history, we have identified the complete set of OR genes and their chromosomal locations from the latest human genome sequences. We detected 388 potentially functional genes that have intact ORFs and 414 apparent pseudogenes. The number and the fraction (48%) of functional genes are considerably larger than the ones previously reported. The human OR genes can clearly be divided into class I and class II genes, as was previously noted. Our phylogenetic analysis has shown that the class II OR genes can further be classified into 19 phylogenetic clades supported by high bootstrap values. We have also found that there are many tandem arrays of OR genes that are phylogenetically closely related. These genes appear to have been generated by tandem gene duplication. However, the relationships between genomic clusters and phylogenetic clades are very complicated. There are a substantial number of cases in which the genes in the same phylogenetic clade are located on different chromosomal regions. In addition, OR genes belonging to distantly related phylogenetic clades are sometimes located very closely in a chromosomal region and form a tight genomic cluster. These observations can be explained by the assumption that several chromosomal rearrangements have occurred at the regions of OR gene clusters and the OR genes contained in different genomic clusters are shuffled.
Olfaction, the sense of smell, is important for mammals to find food, identify mates and offspring, and avoid danger. Mammalian olfactory systems can discriminate between thousands of different odorant molecules in the environment. These odorant molecules are detected by olfactory receptors (ORs), which are encoded by the largest multigene family in mammals. OR genes were first characterized in rats (1), and have been identified in various vertebrates, from lampreys to humans (reviewed in refs. 2–5). ORs are G protein-coupled receptors that have seven α-helical transmembrane regions and trigger a signaling cascade. Mammalian OR genes are expressed mainly in sensory neurons of olfactory epithelium in nasal cavities. It is generally believed that each olfactory neuron expresses only one OR gene (6, 7), but this mechanism is still unknown (8). Some mammalian OR genes are expressed in spermatogenic cells, and recent study indicates that they have a function in sperm chemotaxis (9, 10).
OR genes are ≈310 codons long and contain no introns in the coding region. This property facilitates the identification of OR genes from genome sequences. A total of 906 OR genes and pseudogenes were identified from draft human genome sequences and other databases by homology search (11). They were distributed on different chromosomes and typically found in clusters, although there were some singletons. Statistical analysis suggested that at least 63% of them are pseudogenes. The number of intact (potentially functional) OR genes in the human genome was reported as 322 (11) or 347 (12). In mice, 1,296–1,393 OR functional genes and pseudogenes were detected from draft genome sequences (13, 14). The fraction of pseudogenes in the mouse genome is much lower than that in the human and has been estimated to be ≈20%. This observation is likely to reflect the difference in the importance of olfaction between humans and mice (15, 16). The catfish genome is believed to contain ≈100 OR genes (17). On the basis of sequence similarity, OR genes in mammals, birds, and amphibians are classified into two groups: class I and class II genes (3, 18). All known OR genes in teleosts belong to class I. Several experiments suggested that class I ORs are specialized for recognizing water-soluble odorants, whereas class II ORs are specialized for airborne odorants in amphibians (19, 20). For this reason, class I ORs in mammals were thought to be evolutionary relics. However, the human and mouse genomes also contain a substantial number of class I OR genes that are potentially functional (11, 13). The functional significance of these class I genes in mammals is unknown.
The classification or nomenclature of OR genes is not fully established. Glusman et al. (18) proposed a hierarchical nomenclature based on families and subfamilies, which correspond to the largest phylogenetic groups with >40% and 60% amino acid identities, respectively. According to them, class I OR genes can be classified into 17 families, and class II genes were classified into 14 families. In contrast, Zozulya et al. (12) proposed another nomenclature, in which both phylogenetic grouping and chromosomal location were taken into account. They classified human OR genes into 119 families.
The evolution of OR genes is poorly understood, mainly because the number of genes is so large. The purpose of this study is to obtain some insight into the evolutionary dynamics of a large multigene family. We conducted a phylogenetic analysis of all OR genes that are putatively functional and introduced a previously undescribed system of OR gene classification. Using this classification, we investigated evolutionary relationships of OR genes from the same and different chromosomal regions.
Materials and Methods
Detection of OR Genes and Pseudogenes. To detect OR functional genes and pseudogenes from the complete human genome sequences, a homology search was conducted. The DNA sequences of all human chromosomes were downloaded from genome.ucsc.edu (hg15, the April 10, 2003, version; ref. 21). Human OR gene sequences were obtained from Zozulya et al. (12) and the Human Olfactory Receptor Data Exploratorium (HORDE), which is available on the web site, bioinformatics.weizmann.ac.il/HORDE (11). We merged these two databases to make a nonredundant data set, including 356 intact human OR genes. We then conducted a tblastn search (22) with the E value of 10–20 against the whole human genome sequences by using each of the intact human OR genes as a query. We regarded all of the matches detected by the homology search as OR functional genes or pseudogenes. The criterion of the E value of 10–20 is similar to that previously used for searching mouse OR pseudogenes (14), but ours is more stringent for short matches of <150 amino acids and is slightly weaker for the matches covering almost the entire sequence. The matches obtained by the homology search were classified into functional genes and pseudogenes in the following way: We first regarded matches that were shorter than 250 amino acids, and those containing interrupting stop codons or frameshifts, as pseudogenes. The other matches were used for further analysis. For each of the matches, we extended the DNA sequence to both 3′ and 5′ directions along the chromosome to extract the longest sequence that starts with the initiation codon ATG and ends with the stop codon. All these sequences were translated and aligned by using the program fft-ns-i (23), and the most appropriate start codon positions were chosen through visual inspection. We then assigned the transmembrane regions according to Zozulya et al. (12), and the sequences having long (>3 amino acids) deletions or insertions within transmembrane regions and those lacking the extracellular region completely before the first transmembrane region were regarded as pseudogenes. The remaining sequences were defined as functional genes.
Phylogenetic Analysis. Phylogenetic trees in Figs. 2 and 5 were constructed in the following way: Multiple amino acid sequence alignment was made by the program fft-ns-i (23). Poisson correction distances among amino acid sequences were calculated after gaps were completely deleted. A phylogenetic tree was constructed from these distances by using the neighbor-joining method (24). The tree construction was conducted by the program lintree, which was downloaded from www.bio.psu.edu/People/Faculty/Nei/Lab (25).
Fig. 2.
Phylogenetic tree of 388 functional OR genes. Filled circles at internal nodes represent the clades supported with >90% bootstrap values. Bootstrap values are shown only for phylogenetic clades A–S, and for three branches, a–c. Gene names are omitted for brevity. The family names defined by Glusman et al. (18) are shown in brackets. The OR genes in clade E are not found in the Human Olfactory Receptor Data Exploratorium database. The scale bar indicates the estimated number of amino acid substitutions per site. The number of amino acid sites used is 235.
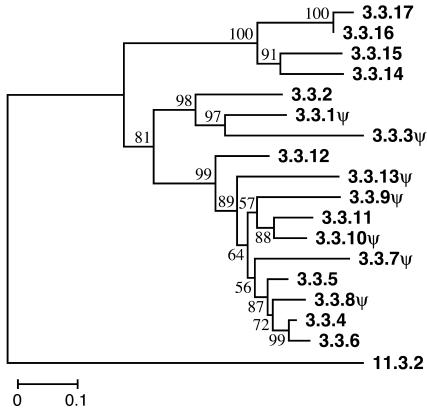
Fig. 5.
Phylogenetic tree of OR functional genes and pseudogenes located in genomic cluster 3.3. Each OR gene is named by using the cluster name defined in this study, and by the additional index number showing the gene order within the cluster. Pseudogenes are indicated by a ψ at the end of the gene name. A class I gene (11.3.2) was used as an outgroup. The number of codon sites used is 198.
Classification of Pseudogenes into Class I and Class II. We conducted a blastp search (22) for all of the 414 OR pseudogenes detected above, against all of the 388 functional OR genes. Each pseudogene is classified into class I and class II, when the best hit belongs to class I and class II, respectively.
Results
OR Functional Genes and Pseudogenes in the Human Genome. Conducting an extensive homology search, we detected 388 potentially functional OR genes that have intact ORFs and determined their exact positions in the human genome. This number is considerably larger than the previous report, i.e., 322 OR genes detected by Glusman et al. (11) or 347 OR genes detected by Zozulya et al. (12). We also identified 414 apparent pseudogenes and their locations in the human genome. Although Glusman et al. (11) reported >900 human OR genes and pseudogenes, they included the sequences that were detected from EST databases. According to them, the total number of the OR genes and pseudogenes of which the genomic positions were assigned was 764, which is smaller than ours (802). Our analysis suggests that the proportion of pseudogenes in the human genome is ≈52%, which is significantly smaller than the previous estimate, i.e., 72% by Rouquier et al. (26) or 63% by Glusman et al. (11). The nucleotide and amino acid sequences and the genomic locations for OR genes are available from our web site, mep.bio.psu.edu/databases.
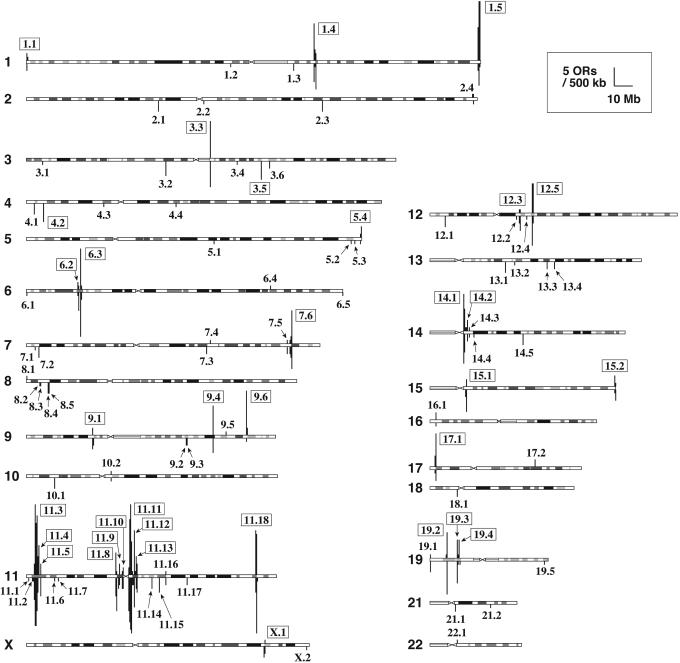
Genomic Distribution of Human OR Genes. Fig. 1 shows the distribution of OR genes on human chromosomes. The OR genes form genomic clusters, many of which are located in subtelomeric or pericentromeric regions as reported (26). In this study, we defined an OR genomic cluster by using the criterion that any distances between two neighboring OR genes (including pseudogenes) in a cluster are <500 kb, regardless of the presence of other genes within the cluster. Previous studies used the criterion that any distances between two neighboring OR genes or pseudogenes in a cluster are <1 Mb (11, 13) or 500 kb (14). Using this definition, we identified 95 OR genomic clusters (Fig. 1). In this article, even if an OR gene exists alone as a singleton, we call it a genomic cluster, for simplicity. We named these genomic clusters by using the chromosome number on which the cluster is located, and the index number of a cluster for the chromosome. Of these OR genomic clusters, 34 included at least five OR genes or pseudogenes (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), and 29 clusters contained only one gene or pseudogene. The largest cluster, 11.3, contained >100 OR genes, including pseudogenes, and occupied a 2-Mb region on chromosome 11. Several genomic clusters such as 1.5, 9.6, and 11.4 contained a significantly (P < 1%) higher fraction of functional genes than the average (Table 1). The distribution of the distances between two consecutive OR genes (including pseudogenes) on the same chromosomal arm is shown in Fig. 6, which is published as supporting information on the PNAS web site. A sharp peak was found at ≈11 kb, indicating that the interval between consecutive OR genes is nearly the same for a majority of them.
Fig. 1.
Distribution of OR genes on human chromosomes. Vertical bars above and below the chromosomes indicate the locations of OR functional genes and pseudogenes. The height of each bar represents the number of OR genes in a nonoverlapping 500-kb window at the position. Genomic clusters containing five or more OR genes (including pseudogenes) are represented by boxes. The genomic clusters are presented in relation to the Giemsa-stained bands at an 800-band resolution, which was obtained from the web site genome.ucsc.edu (36). Crosses show centromeres, and horizontal lines show the regions of noncentromeric heterochromatin. Chromosomes 20 and Y are not shown because OR genes were not found on these chromosomes.
Phylogenetic Analysis. To classify human OR genes into related groups of sequences, we conducted a phylogenetic analysis. Here, we confined our analysis to functional genes only, because most pseudogenes contained deletions and were much shorter than functional genes. Fig. 2 shows the phylogenetic tree for the 388 functional OR genes. It is seen that functional OR genes are clearly separated into class I and class II (18) with 100% bootstrap support. The numbers of class I and class II genes are 57 and 331, respectively. Class II genes can further be subdivided into phylogenetic clades, each of which is supported by a bootstrap value of >90% and contains at least five members. When a clade was nested in another larger clade, we used the latter clade, ignoring smaller clades. In this way, we identified 19 phylogenetic clades and named them clades A–S, from the largest to the smallest clade (Fig. 2). (Our classification did not change significantly when currently known mouse OR genes were added to the phylogenetic tree.) However, the bootstrap values for the interior branches relating these clades are generally quite low. There are only two interior branches (branches a and b in Fig. 2) that represent interclade relationships with a bootstrap value of >50%. Interestingly, the bootstrap value for branch c in Fig. 2 is quite high (73%), suggesting that one gene diverged earlier than the other class II OR genes. The average amino acid identity of OR genes within a phylogenetic clade was from 47.4% (clade L) to 70.3% (clade K).
Fig. 2 also shows the families (numbered in brackets) proposed by Glusman et al. (18). There are some differences between their classification and ours. For example, the members of family 2 by Glusman et al. (18) are divided into clades B, C, F, and K in our classification, but clade F also contains the members of family 13. Similarly, their family 5, 8, or 10 does not correspond to a monophyletic clade in our classification. These differences between the two classifications apparently occurred, because Glusman et al. (18) defined a family as the largest subtree for which the average amino acid identity is >40%, regardless of the bootstrap value of the subtree.
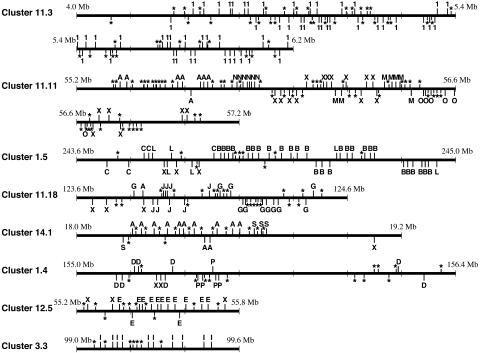
Relationships Between Genomic Clusters and Phylogenetic Clades. Fig. 3 shows the physical maps of OR functional genes and pseudogenes in several genomic clusters. It is clear that functional OR genes belonging to the same phylogenetic clade tend to be located close to one another on a chromosome, forming a tandem array. For example, all of the functional OR genes in genomic cluster 3.3 belong to clade I, and all genes, including pseudogenes, are located on the same strand. However, the transcriptional directions of OR genes belonging to the same phylogenetic clade are often different, as in the case of some genes from clade E in genomic cluster 12.5.
Fig. 3.
Physical maps of OR functional genes and pseudogenes in several genomic clusters. The position of each OR gene is represented by a vertical bar above or below a horizontal line, the latter indicating the opposite transcriptional direction to the former. *, pseudogenes. Phylogenetic clades are shown for functional genes. 1, a class I functional gene; X, an unclassified functional gene in class II.
Fig. 3 also indicates that one genomic cluster often includes OR genes from two or more phylogenetic clades. For example, genomic cluster 11.11 contains OR genes from phylogenetic clades A, M, N, and O and many unclassified genes. The genes belonging to each of the clades A, M, N, and O form tandem arrays. The phylogenetic tree (Fig. 2) shows that the genes belonging to clade A and clades M, N, and O are distantly related. Similarly, genomic cluster 1.5 contains functional OR genes from clades B, C, and L, but the genes from these clades are distantly related (Fig. 2). Furthermore, clades G and J in cluster 11.18, clades A and S in cluster 14.1, and clades D and P in cluster 1.4 are distantly related to each other. In each genomic cluster, neighboring OR genes are generally closely located, even if they belong to different phylogenetic clades.
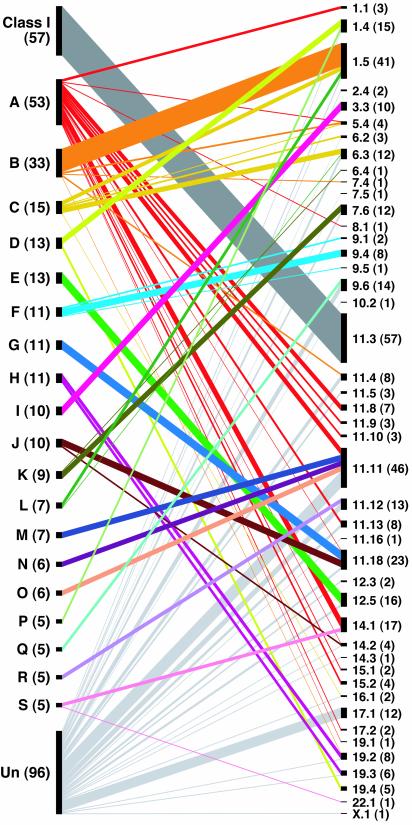
Fig. 4 shows the relationships between phylogenetic clades and genomic clusters. OR genes in one phylogenetic clade are often found in several different genomic clusters. For example, 53 functional OR genes belonging to clade A are dispersed in 15 different genomic clusters. At the same time, one genomic cluster sometimes contains genes from distantly related phylogenetic clades, as was shown in Fig. 3.
Fig. 4.
Relationships between phylogenetic clades and genomic clusters for functional OR genes. Phylogenetic clades and genomic clusters are listed at the left and right sides, respectively, with the number of OR genes contained in each clade or cluster in parentheses. When one or more OR genes belong to a particular clade and are located in a particular cluster, the clade and the cluster are joined by a line. The thickness of the line is proportional to the number of such OR genes. For example, 8 of 13 OR genes in clade D are located in cluster 1.4, and 5 OR genes are in cluster 19.4. Therefore, clade D and clusters 1.4 and 19.4 are joined by lines with different thickness. Un, unclassified.
Class I OR genes are exceptional in that all of functional class I genes are located in one cluster, 11.3, and the cluster does not contain any functional genes from class II. To see the genomic distribution of class I pseudogenes, we classified all OR pseudogenes into class I and class II on the basis of homology search against functional genes (see Materials and Methods). Of the 414 pseudogenes, 45 were classified into class I and 369 were classified into class II. The fraction of pseudogenes was 44% in class I and 53% in class II. We also found that all of the class I pseudogenes are located in genomic cluster 11.3, and genomic cluster 11.3 does not contain any class II pseudogenes. Therefore, the correspondence between the cluster 11.3 and class I genes holds true for both functional genes and pseudogenes.
Discussion
Our results can be summarized as follows: (i) A substantial fraction of human OR genes are pseudogenes. (ii) Functional OR genes that belong to one phylogenetic clade are generally located close to one another on a chromosome, and, in many cases, have the same transcriptional direction. However, (iii) functional OR genes belonging to one phylogenetic clade are often found in several different genomic clusters. (iv) One genomic cluster often contains OR genes belonging to different phylogenetic clades that are distantly related. Observation i suggests that the OR gene family is subject to the birth-and-death model of evolution, in which new genes are formed by gene duplication and some of the duplicate genes differentiate in function, whereas others are inactivated or deleted from the genome (27, 28). In fact, our joint phylogenetic analysis of human and mouse OR genes, which will be published elsewhere, has confirmed this assertion (see also Fig. 5). Observation ii can be explained by tandem duplication. In the case of genomic cluster 3.3, we can trace the evolutionary history of OR functional genes and pseudogenes. The phylogenetic tree in Fig. 5 shows that all of these genes are closely related, and that recently diverged genes are generally located closely to one another. This result suggests that these genes were generated approximately one by one by repeated tandem duplication. However, the genes diverged most recently are not necessarily located adjacently, as in the case of genes 3.3.4 and 3.3.6. The transcriptional direction of neighboring OR genes belonging to the same phylogenetic clade are often different (Fig. 3). These observations indicate that the genes in a genomic cluster were often inverted.
Observation iii implies that a single genomic cluster was fragmented by chromosomal translocation into smaller clusters that were eventually dispersed on different chromosomal regions. To explain observation iv, a mechanism that brings two clusters from different chromosomal regions into one cluster should be considered. A possible explanation is that recombination takes place between two OR gene clusters located in different chromosomal regions, and genes included in the two clusters are shuffled (see Fig. 7, which is published as supporting information on the PNAS web site). This event can occur by an inversion (when the two clusters are located on the same chromosome), or by a reciprocal translocation (when they are located on different chromosomes). It is thought that reciprocal translocations cannot easily be fixed in the population, because an organism heterozygous for a reciprocal translocation and the original chromosome usually produces only half as many offspring as the homozygotes, and thus they are deleterious (29). In the present case, however, OR genomic clusters are often located at the terminal regions of chromosomes (see Fig. 1). Therefore, a reciprocal translocation occurring between two OR gene clusters located on different chromosomes would not affect the fitness of heterozygotes seriously, because the number of genes affected by the translocation is small.
This model appears to be acceptable for the following reasons: First, mammalian species have undergone extensive chromosomal rearrangements. It has been estimated that at least ≈300 chromosomal rearrangements have occurred after the divergence of humans and mice (30). The divergence of phylogenetic clades A–S is much more ancient than the human-mouse divergence (data not shown). Therefore, chromosomal rearrangements appear to have occurred many times after the formation of OR gene clusters. Second, several studies have shown that the recombination between nonallelic low-copy repeats are responsible for chromosomal rearrangements such as deletions, duplications, inversions, and, possibly, translocations (reviewed in refs. 31 and 32). It has also been suggested that, in humans, several chromosomal rearrangements, including reciprocal translocation t(4;8)(p16;p23), have occurred by the mediation of OR gene clusters on chromosomal regions 4p16 and 8p23 (33, 34), although these clusters seem to contain only pseudogenes.
As mentioned above, OR genes belonging to one phylogenetic clade tend to form a tandem array in a genomic cluster. However, the genes from different phylogenetic clades often intermingle in a genomic cluster. For example, OR genes from clades B, C, and L are mixed to one another in a genomic cluster 1.5 (Fig. 3). This finding indicates that local chromosomal rearrangements have taken place within a genomic cluster after large-scale chromosomal rearrangements occurred to form the cluster. The occurrence of singleton OR genes can also be explained by chromosomal rearrangement, but it is also possible that these genes were generated by reverse transcription of mRNA transcripts and integration of the resultant cDNA into genomic DNA (35). Interestingly, our data showed that 24 of 29 singletons are pseudogenes.
Glusman et al. (11) proposed the “out of chromosome 11” theory of evolution for human OR genes. According to this theory, the duplication of class I OR genomic cluster on chromosome 11 resulted in the formation of the first class II genomic cluster on the same chromosome. This class II cluster was again duplicated, and one of the clusters generated by the duplication was transferred to chromosome 1. This cluster on chromosome 1 was then duplicated many times, and the resultant duplicate clusters were transferred to other chromosomes. However, this theory seems to be unreasonable, because it assumes that each genomic cluster of OR genes is an evolutionary unit. The real process of evolution of OR genes is much more complicated, as we have seen.
Supplementary Material
Acknowledgments
We thank Alex Rooney, Shozo Yokoyama, and Jianzhi Zhang for valuable comments. This work was supported by National Institutes of Health Grant GM20293 (to M.N.). Y.N. was partially supported by the Japan Society for the Promotion of Science.
Abbreviation: OR, olfactory receptor.
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- 2.Buck, L. B. (2000) Cell 100, 611–618. [DOI] [PubMed] [Google Scholar]
- 3.Dryer, L. (2000) BioEssays 22, 803–810. [DOI] [PubMed] [Google Scholar]
- 4.Firestein, S. (2001) Nature 413, 211–218. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts, P. (2001) Nat. Neurosci. 4, Suppl., 1192–1198. [DOI] [PubMed] [Google Scholar]
- 6.Chess, A., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- 7.Serizawa, S., Ishii, T., Nakatani, H., Tsuboi, A., Nagawa, F., Asano, M., Sudo, K., Sakagami, J., Sakano, H., Ijiri, T., et al. (2000) Nat. Neurosci. 3, 687–693. [DOI] [PubMed] [Google Scholar]
- 8.Kratz, E., Dugas, J. C. & Ngai, J. (2002) Trends Genet. 18, 29–34. [DOI] [PubMed] [Google Scholar]
- 9.Parmentier, M., Libert, F., Schurmans, S., Schiffmann, S., Lefort, A., Eggerickx, D., Ledent, C., Mollereau, C., Gérard, C., Perret, J., et al. (1992) Nature 355, 453–455. [DOI] [PubMed] [Google Scholar]
- 10.Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K. & Hatt, H. (2003) Science 299, 2054–2058. [DOI] [PubMed] [Google Scholar]
- 11.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685–702. [DOI] [PubMed] [Google Scholar]
- 12.Zozulya, S., Echeverri, F. & Nguyen, T. (2001) Genome Biol. 2, research0018.1–0018.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124–133. [DOI] [PubMed] [Google Scholar]
- 14.Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 535–546. [DOI] [PubMed] [Google Scholar]
- 15.Rouquier, S., Blancher, A. & Giorgi, D. (2000) Proc. Natl. Acad. Sci. USA 97, 2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilad, Y., Man, O., Pääbo, S. & Lancet, D. (2003) Proc. Natl. Acad. Sci. USA 100, 3324–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngai, J., Dowling, M. M., Buck, L., Axel, R. & Chess, A. (1993) Cell 72, 657–666. [DOI] [PubMed] [Google Scholar]
- 18.Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J. & Lancet, D. (2000) Mamm. Genome 11, 1016–1023. [DOI] [PubMed] [Google Scholar]
- 19.Freitag, J., Ludwig, G., Andreini, I., Rossler, P. & Breer, H. (1998) J. Comp. Physiol. A 183, 635–650. [DOI] [PubMed] [Google Scholar]
- 20.Mezler, M., Fleischer, J. & Breer, H. (2001) J. Exp. Biol. 204, 2987–2997. [DOI] [PubMed] [Google Scholar]
- 21.International Human Genome Sequencing Consortium (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 22.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 25.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823–833. [DOI] [PubMed] [Google Scholar]
- 26.Rouquier, S., Taviaux, S., Trask, B. J., Brand-Arpon, V., van den Engh, G., Demaille, J. & Giorgi, D. (1998) Nat. Genet. 18, 243–250. [DOI] [PubMed] [Google Scholar]
- 27.Nei, M. (1969) Nature 221, 40–42. [DOI] [PubMed] [Google Scholar]
- 28.Nei, M., Gu, X. & Sitnikova, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright, S. (1941) Am. Nat. 75, 513–522. [Google Scholar]
- 30.Mouse Genome Sequencing Consortium (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 31.Samonte, R. V. & Eichler, E. E. (2002) Nat. Rev. Genet. 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 32.Stankiewicz, P. & Lupski, J. R. (2002) Trends Genet. 18, 74–82. [DOI] [PubMed] [Google Scholar]
- 33.Giglio, S., Broman, K. W., Matsumoto, N., Calvari, V., Gimelli, G., Neumann, T., Ohashi, H., Voullaire, L., Larizza, D., Giorda, R., et al. (2001) Am. J. Hum. Genet. 68, 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giglio, S., Calvari, V., Gregato, G., Gimelli, G., Camanini, S., Giorda, R., Ragusa, A., Guerneri, S., Selicorni, A., Stumm, M., et al. (2002) Am. J. Hum. Genet. 71, 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mighell, A. J., Smith, N. R., Robinson, P. A. & Markham, A. F. (2000) FEBS Lett. 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 36.Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., Roskin, K. M., Schwartz, M., Sugnet, C. W., Thomas, D. J., et al. (2003) Nucleic Acids Res. 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.