Abstract
We have mapped the chromosomal binding site distribution of a transcription factor in human cells. The NF-κB family of transcription factors plays an essential role in regulating the induction of genes involved in several physiological processes, including apoptosis, immunity, and inflammation. The binding sites of the NF-κB family member p65 were determined by using chromatin immunoprecipitation and a genomic microarray of human chromosome 22 DNA. Sites of binding were observed along the entire chromosome in both coding and noncoding regions, with an enrichment at the 5′ end of genes. Strikingly, a significant proportion of binding was seen in intronic regions, demonstrating that transcription factor binding is not restricted to promoter regions. NF-κB binding was also found at genes whose expression was regulated by tumor necrosis factor α, a known inducer of NF-κB-dependent gene expression, as well as adjacent to genes whose expression is not affected by tumor necrosis factor α. Many of these latter genes are either known to be activated by NF-κB under other conditions or are consistent with NF-κB's role in the immune and apoptotic responses. Our results suggest that binding is not restricted to promoter regions and that NF-κB binding occurs at a significant number of genes whose expression is not altered, thereby suggesting that binding alone is not sufficient for gene activation.
Understanding the targets regulated by transcription factors and where they bind relative to these targets in an unbiased fashion in mammalian cells is highly desirable. We and others have developed a procedure for mapping in vivo targets of transcription factors by chromatin immunoprecipitation (ChIP) with antibodies to a transcription factor of interest to isolate protein-bound DNA, followed by probing a microarray containing genomic DNA sequences with the immunoprecipitated DNA (ChIP chip) (1–3). This approach was first used successfully in yeast and has more recently been used in a limited fashion to identify transcription factor binding sites in mammalian cells (4–6). However, a large-scale, unbiased global analysis of the distribution of mammalian transcription factor binding sites along large genomic regions has not been previously explored.
In this study we employ a microarray containing the entire nonrepetitive sequence of chromosome 22 to determine the chromosome-wide binding profile for the transcription factor NF-κB. The NF-κB/Rel family of transcription factors plays an essential role in regulating the induction of genes involved in several physiological processes, including immune and inflammatory responses (7, 8), and the activation pathway has been studied extensively over the last two decades (9, 10). Numerous NF-κB target genes have also been identified; however, it remains unclear how many of these are direct targets of the transcription factor (11).
There are five mammalian NF-κB family members (p50, p52, RelA/p65, RelB, and c-rel), all of which function as homo- or heterodimers. The different dimers exhibit varying binding affinities for κB sites (GGGRNNYYCC; R is purine, Y is pyrimidine, and N is any base). They also differ in their ability to activate transcription; only p65 and c-Rel have been shown to be potent transcriptional activators, where complexes containing p50 homodimers are thought to repress transcription (12).
In the present study, we examine the binding distribution of p65 along human chromosome 22 in response to tumor necrosis factor (TNF) α. We find that p65 has many binding sites on chromosome 22, corresponding to a number of interesting gene loci. Binding occurs at several locations relative to these targets, but primarily at 5′ ends and introns; consensus- and nonconsensus-sequence binding sites are used at equal frequency. Finally, we detect p65 binding in previously unannotated regions of the chromosome, thereby providing insight into the potential function of these regions.
Materials and Methods
Protein Extracts and Immunoblots. HeLa suspension cells (American Type Culture Collection clone S3) cultured in S-MEM (GIBCO) were either treated with 20 ng/ml TNF-α (Sigma) for 90 min or left untreated. The cells were harvested by centrifugation, resuspended in hypotonic buffer (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EGTA/0.1 mM EDTA/1mMDTT/0.5 mM PMSF) and incubated on ice for 15 min. Nonidet P-40 (0.5%) was added, and cells were vortexed vigorously and pelleted at 3,000 × g for 15 min. Nuclei were resuspended in RIPA lysis buffer [10 mM Tris·Cl, pH 8/140 mM NaCl/1% Triton X-100/0.1% SDS/1% Na-deoxycholate/1 mM PMSF with protease inhibitors (Roche Molecular Biochemicals)], incubated on ice for 15 min, passed through a 20-gauge needle five times, and incubated an additional 30 min on ice with fresh PMSF. Extracts were clearified by centrifugation at 14,000 × g at 4°C for 15 min.
p65 was immunoprecipitated overnight at 4° with anti-p65 polyclonal antibodies (Santa Cruz Biotechnology) at a final concentration of 1:500 and then incubated with protein A/G bead for 45 min. The beads were washed twice with RIPA, once with LiCl detergent solution (10 mM Tris·Cl, pH 8/500 mM NaCl/0.025% sodium azide/1% Triton X-100/0.1% SDS/1% Na-deoxycholate), and twice with 1× TBS (20 mM Tris·Cl, pH 7.6/150 mM NaCl). Protein extracts were separated on a 10% denaturing polyacrylamide gel and analyzed by immunoblot analysis with a 1:1,000 dilution of p65 primary monoclonal antibody (F-6, Santa Cruz Biotechnology) and a HRP-conjugated secondary antibody. Immunocomplexes were visualized by using the ECL system (Amersham Biosciences).
ChIP. HeLa S3 cells (5 × 108) were either treated with 20 ng/ml TNF-α (human, recombinant, Sigma) for 90 min or left untreated. Cells were treated with formaldehyde, and p65–DNA complexes were immunoprecipitated and purified as described by Horak et al. (6), using the p65 antibody as described above.
Expression Analysis. Poly(A) RNA was isolated from untreated cells and cells treated with 20 ng/ml TNF-α for 1.5 and 4 h at 37°C by using the MicroPoly(A)Pure kit (Ambion, Austin, TX). cDNA was synthesized by using the Amino Allyl cDNA labeling kit (Ambion), using both random decamers and oligo(dT) primers in the reverse transcription reaction.
Probe Labeling and Microarray Hybridization. Chromatin-immunoprecipitated DNA was labeled as described by Boyd and Farnham (13). Labeled DNA was further purified from free dye by using the QIAquick PCR purification kit (Qiagen, Valencia, CA), ethanol-precipitated, and resuspended in hybridization solution (5× SSC/0.1% SDS/25% formamide; 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7).
Chromosome 22 microarray slides were prehybridized in 5× SSC, 0.1% SDS, 25% formamide, and 1 mg/ml BSA in HybChambers (GeneMachines, San Carlos, CA) at 42°C for 45 min. After prehybridization, slides were rinsed briefly in water and dried by centrifugation. Suspended probe was applied directly to the microarray and hybridized for 12–16 h at 42°C. Slides were then washed in 2× SSC, two times for 5 min in 2× SSC/0.1% SDS, two times for 5 min in 0.1× SSC/0.1% SDS, and four times for 1 min in 0.1× SSC.
Microarray signals were normalized by using the expressyourself web site (http://bioinfo.mbb.yale.edu/ExpressYourself) with default parameters to correct for spatial, intensity-based, and dye-specific artifacts (14). Positively hybridizing fragments were identified as those with logged Cy5/Cy3 ratios more than 2.5 standard deviations above the mean for fragments with similar total intensities (15).
PCR Analysis. Primers were designed to amplify a 400-bp region upstream from the TATA box in the IL-8, intercellular adhesion molecule (ICAM), and p65 promoters. For p65 chromosome 22 targets, primers were designed to tile the microarray fragments, which range in size from 300 to 1,400 bp, and generate a product of average size 300 bp. Immunoprecipitated DNA was amplified, and the products were visualized by using a 1.5% agarose gel.
Chromatin-immunoprecipitated DNA was also analyzed by quantitative PCR. Each PCR reaction was carried out in duplicate in a 20-μl reaction volume by using 0.02% of the total immunoprecipitated DNA and SYBR Green Master Mix (MJ Research, Cambridge, MA), using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Dissociation curves were analyzed as a means to ensure quality of amplicon and to monitor primer dimers. Enrichment was determined based on critical threshold (Ct) measurements (changes in fluorescence per PCR cycle number at a given threshold).
Results
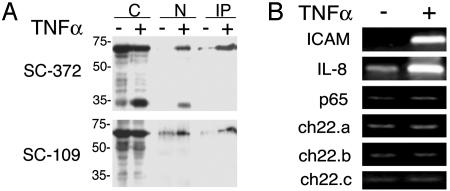
Specific ChIP with Anti-p65 Antibodies. To identify regions on chromosome 22 bound by p65 in response to TNF-α in vivo, we first tested p65 antibodies for their ability to selectively immunoprecipitate the p65 protein. Nuclear protein extracts were prepared from cells stimulated with TNF-α for 90 min and untreated cells, and immunoprecipitated with two anti-65 antibodies, SC-372 and SC-109, that recognize two distinct regions of p65. As shown in Fig. 1A, a band at 65 kDa is selectively immunoprecipitated from the nuclei of TNF-α-stimulated cells, but not from those of unstimulated cells, by the anti-p65 antibodies. Control sera did not immunoprecipitate this band. A presumed degradation product at 35 kDa is also recognized in the cytoplasmic and nuclear extracts (but not the IP) with the SC-372 antibody but not with the SC-109 antibody.
Fig. 1.
Immunoprecipitation of p65. (A) Protein extracts from cytoplasmic (C) and nuclear (N) and immunoprecipitated (IP) samples demonstrate p65 translocation to the nucleus upon TNF-α treatment. In Upper it is shown that the αp65 polyclonal antibody (SC-372) specific for the carboxyl terminus of the protein recognizes a 65-kDa protein in TNF-α-stimulated nuclei and a presumed degradation product migrating at 35 kDa. In Lower it is shown that the SC-109 αp65 polyclonal antibody that recognizes the amino terminus of p65 also reacts with a 65-kDa protein in the nuclei of TNF-α-stimulated cells. (B) PCR analysis of chromatin-immunoprecipitated DNA. p65-bound DNA was purified by ChIP from HeLa S3 cells treated or untreated with TNF-α for 90 min. Enrichment for p65 targets in the treated samples was observed by PCR analysis of the promoter regions of ICAM and IL-8. Binding is not observed at the p65 promoter region or at three distinct chromosome 22 regions that were not identified as p65 targets in the ChIP chip experiments.
To determine whether the antibody can specifically immunoprecipitate p65 bound to DNA at its target sites in HeLa S3 cells, TNF-α-treated and untreated cells were treated with formaldehyde to crosslink protein–DNA complexes. Chromatin purified from nuclei was sheared to an average size of 500 bp and immunoprecipitated with the SC-372 anti-p65 antibody. The crosslinks were reversed, and the DNA was purified and analyzed for the enrichment of two known NF-κB targets, IL-8 and ICAM, by using PCR with primer pairs targeting their promoter regions (16–18). These promoter fragments are selectively enriched in the TNF-α-treated cells relative to untreated cells. Enrichment was not observed at the promoter region of p65, which is not known to be activated by NF-κB, or at three individual chromosome 22 regions not identified as p65 targets in this study (Fig. 1B). These data demonstrate that we are able to selectively immunoprecipitate p65-bound DNA in vivo.
Many Binding Sites of p65 Reside on Human Chromosome 22. We next examined the chromosomal distribution of p65 by probing the chromosome 22 microarray with chromatin-immunoprecipitated DNA isolated from TNF-α-treated and untreated HeLa S3 cells. The DNA from the treated and untreated samples was differentially labeled with Cy5 and Cy3 dye, respectively, pooled, and hybridized to the chromosome 22 genomic array, consisting of 21,024 PCR products with a mean size of 700 bp (range 300–1,400 bp) and covering 93% of the nonrepetitive sequences of the chromosome (19). The microarray also contains the genomic sequences of the IκB and Cox-2 (cyclooxygenase 2) genes included as positive control fragments for p65 binding as well as many negative control fragments. Four independent immunoprecipitation experiments were performed and hybridized. Raw microarray data were analyzed by using expressyourself, and binding sites were identified following standard approaches (14, 15). The positive control fragments were observed to be consistently enriched in these experiments, whereas the negative control fragments were not enriched.
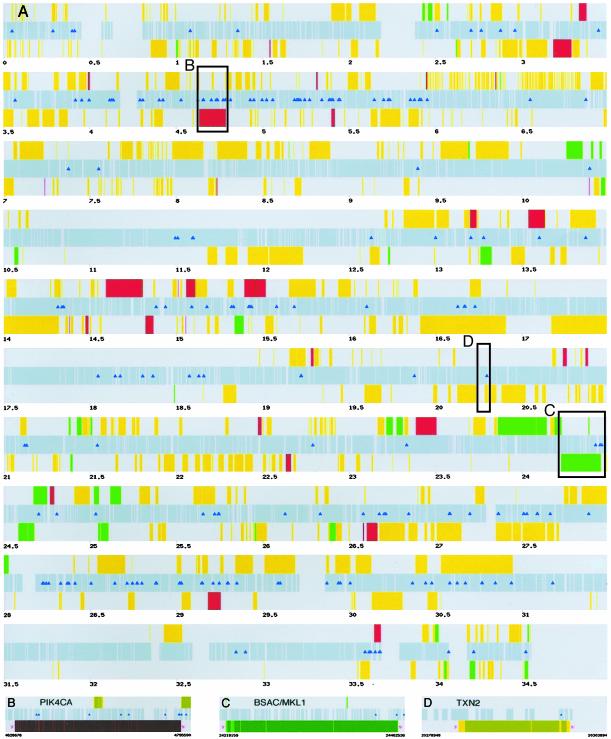
Our analysis identified 209 unique binding sites for p65 on human chromosome 22 in response to TNF-α stimulation (Fig. 2A). Chromosome 22 contains 917 distinct loci, consisting of 678 known or predicted genes and 239 pseudogenes (20, 21). Of these annotated regions, p65 binding is located within or proximal to (within 10 kb) 143 distinct loci or ≈15.5%; however, this may be an overestimate of actual target genes because it includes pseudogenes, EST clusters that are not well defined, and cases of redundancy where one fragment overlaps two genes (e.g., PI4KCA and HCF, as illustrated in Fig. 2B).
Fig. 2.
Chromosome 22q binding profile for p65. (A) The center gray bar represents the nonrepetitive sequence on chromosome 22, ordered from centromere to telomere. Genes oriented 5′ → 3′ and 3′ → 5′ are depicted above and below the center bar, respectively. Genes up-regulated (red), down-regulated (green), and nondifferentially regulated (yellow) in response to TNF-α are shown. Triangles indicate the locations of p65-bound fragments. (B–D) Higher-resolution examples of genes with p65-bound fragments. For all three examples, the lighter shade represents an exon and the darker shade represents an intron. (B) PIK4CA is an up-regulated gene that contains 12 p65-binding sites in its vicinity, including one near HCF (the yellow gene on the + strand). (C) BSAC is a down-regulated gene that contains three p65-binding sites. (D) TXN2 has one p65 site located in the first intron.
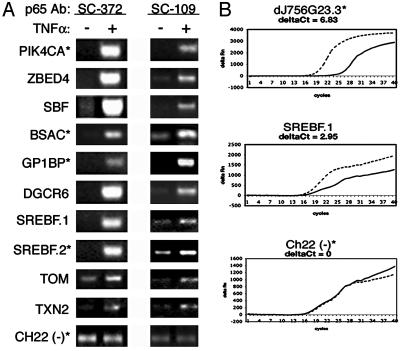
Of the 209 binding regions, 75 representative fragments were individually examined for enrichment in the TNF-α-treated samples by using a standard PCR assay. Fifty-nine of the 75 fragments show clear enrichment (Fig. 3A), irrespective of whether they contained a consensus sequence or lay in the proximal promoter region. Thirty of these same fragments were examined for enrichment by using real-time quantitative PCR analysis, and 29 exhibited enrichment (Fig. 3B). To further verify that the targets we identified are bound by p65 rather than a nonspecific antibody interaction, we tested 10 by ChIP analysis using the SC-109 antibody. Nine of the 10 targets were found to be enriched after TNF-α treatment, including five without NF-κB consensus sites (Fig. 3A). Thus, we conclude that most of the p65 targets identified are bona fide binding sites.
Fig. 3.
PCR analysis of p65 target regions. (A) Chromatin immunoprecipitates from TNF-α-treated and untreated cells were analyzed by PCR to confirm microarray results at many target loci; a subset, indicated by gene names, is shown. The two anti-p65 antibodies (SC-372 and SC-109) yield similar results. Ch22 (–) represents a region of chromosome 22 not identified as a p65 target by ChIP chip. (B) Real-time PCR analysis of chromatin immunoprecipitates. Real-time PCR was used to independently confirm a subset of the standard PCR reactions, three of which are shown. Enrichment in the TNF-α-treated samples is evident by the difference in amplification curves and is measured by the difference in fluorescence at a given cycle threshold, or deltaCt. One deltaCt unit corresponds to ≈2-fold enrichment. (Top) A down-regulated protein similar to a Drosophila transcriptional repressor protein. (Middle) The 5′-proximal SREBF p65 site (A). (Bottom) A chromosome 22 fragment not enriched for p65 binding in the ChIP chip analysis. Fragments with consensus sites are denoted by *. ———, Unstimulated; – – –, TNF-α.
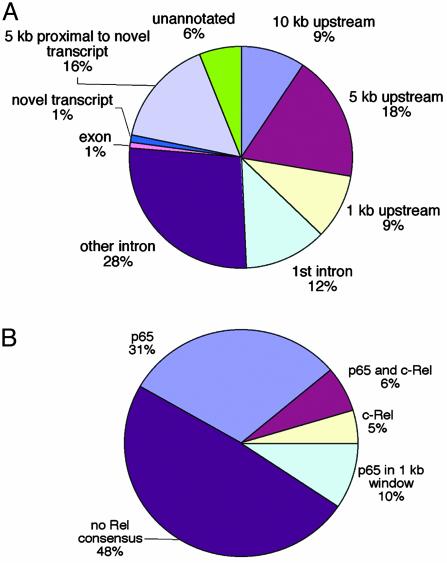
p65 Binds in Putative Promoters and Introns of Its Potential Targets. We analyzed the distribution of NF-κB relative to the 5′ ends (ATG) of annotated genes and mapped their location relative to introns, exons, and 3′ ends, and in unannotated intergenic regions (Fig. 4A). Nearly one-third (28%) of the NF-κB-bound fragments lie within 5 kb upstream of the 5′ end (ATG) of annotated genes, an enrichment of 3-fold over the whole chromosome. A significant portion of p65-binding sites were also present in the first intron (12%) or more internal introns (28%) of annotated genes, a finding consistent with NF-κB-binding intronic enhancer regions (22, 23). Thus, although many sites are present in the 5′ ends, NF-κB-binding sites are also located at other regions within a gene. One exception is exons, because only two p65-binding sites lie in fragments that contain only known exons. We also observed p65 binding proximal to and within pseudogenes, perhaps indicative of conserved regulatory sequences near these sequences.
Fig. 4.
Features of p65-binding sites on chromosome 22. (A) p65 binding relative to Sanger-annotated genes and hybridizing regions on chromosome 22 is illustrated. (B) Distribution of NF-κB consensus sequences in p65-binding sites. The sequences of p65-bound fragments on the microarray were searched for NF-κB consensus sites by using both an in-house chromosome annotation system and the TFSEARCH database (www.cbrc.jp/research/db/TFSEARCH.html).
p65 Binds to Consensus and Nonconsensus Sequences. The p65 targets on chromosome 22 were searched for NF-κB consensus sites (Fig. 4B). Thirty-five percent of the fragments contain a canonical p65 sequence (GGGRNNYCCC) (24), and closer inspection reveals that most of the remaining fragments contain sequences similar to the consensus. Interestingly, 65% of the binding regions 5 kb upstream of annotated genes contain at least one consensus site. In addition, 22% of the p65 target sequences have c-Rel recognition sites (GGGA/GNTTCC) either exclusively (11%) or in addition to a p65 site (11%) (24). Because p65 can function as a heterodimer with c-Rel, it might bind DNA in complex with c-Rel at those sites. Moreover, an additional 10% have a κB consensus site adjacent (within 500 bp) to the hybridizing fragment. Thus, it appears that 56% of the chromosome 22 binding sites contain or are next to a NF-κB site (Fig. 4B). These results are in agreement with studies of yeast in which factors bind regions that often do not have consensus sequences at a moderately high frequency (1) and illustrate the advantage of using an unbiased genomic DNA array for discovery of in vivo binding regions.
The p65 target sequences were searched for additional consensus sites in an attempt to reveal any evidence of cooperative binding based on an enrichment for other transcription factor binding sites. Two consensus sites that occurred with the highest frequency were those for GATA-3 (30%) and CRE-BP (46%) motifs. Recent genomewide computational analysis of transcription factor binding sites found GATA-3 and CRE-BP sites (in addition to others) lie near NF-κB sites (25). Our data are consistent with the hypothesis that these factors can potentially function together to regulate transcription.
p65 Binds to Genes Whose Expression Is Regulated by TNF-α. We next examined the chromosomal distribution of p65 sites relative to genes whose expression is induced by proinflammatory stimuli. PolyA+ RNA was isolated from HeLaS3 cells treated with TNF-α for 90 min or 4 h and untreated cells, differentially labeled, and used to probe the chromosome 22 array. Three independent experiments were performed. Nearly identical results were found for the 90-min and 4-h treatments; thus, only the 90-min data are shown. The expression of 67 distinct genes was found to be altered on TNF-α treatment: 28 were up-regulated and 39 were down-regulated (Fig. 2 and Tables 2 and 3, which are published as supporting information on the PNAS web site, www.pnas.org). Interestingly, we found that the expression of 14 pseudogenes was also affected by TNF-α: nine were up-regulated and four were down-regulated, potentially because of cross-hybridization with functional paralogs.
Closer analysis of the p65-bound TNF-α-regulated genes revealed that 12 of the 28 up-regulated genes contain at least one and often multiple p65-binding sites nearby (Table 1). Two examples of up-regulated genes are phosphatidylinositol 4-kinase catalytic polypeptide α (PI4KCA) and neurofibromatosis 2 (NF2/merlin). The binding of p65 at the PI4KCA loci (Figs. 2B and 3A) suggests that p65 directly regulates PIK4 kinase gene expression. NF2 is a negative regulator of NF-κB signaling through its inhibition of IκBα degradation (26). The binding of p65 near the TNF-α-inducible NF2 gene suggests NF2s involvement in the autoregulatory loop whereby p65 activates an IκBα stabilizing protein in addition to IκBα, thereby enhancing the inhibition of NF-κB.
Table 1. Genes bound by p65 and regulated in response to TNF-α stimulation.
| Gene symbol | Description | p65-binding site (relative to ATG) |
|---|---|---|
| Up-regulated | ||
| ZBED4* | Zinc finger, BED domain 4 | +6.4 kb†, +28.6 kb†, +32.5 kb |
| ZNF378 | Zinc finger, DHHC domain 1 | +3.6 kb† |
| PIK4CA* | Type II phosphatidylinositol 4-kinase | -4.5 kb, -325 bp, and 8 internal |
| AP000557.3 | Similar to human PIK4CA | +543 bp, +5.4 kb |
| EWSR1* | Ewing sarcoma breakpoint region 1 | +1740 bp |
| SMC1L2 | Mitosis-specific chromosome segregation (SMC1)-like 2 | -5 kb, +30 kb |
| NF2* | Neurofibromatosis 2 | +61 kb |
| ARFGAP3* | ADP-ribosylation factor GTPase activating protein 3 | -10 kb |
| KIAA0542 | mRNA for KIAA0542 | +25, +32, +41 kb |
| AC007050.4* | POM121-like 1 | +697 bp |
| AC005005.6 | Homo sapiens cDNA: FLJ23382 | +32 kb |
| Down-regulated | ||
| BSAC/MKL1* | Megakaryoblastic leukemia 1 | -3 kb, -100 bp, +30 kb† MKL1* |
| SBF1* | SET binding factor 1 | +5.5 kb† |
| dJ756G23.3* | Similar to Drosophila Tr:Q24191 transcriptional repressor | +10 kb† |
| RABL2B | RAB—RAS family-like 2B | +15 kb |
| ADTB1 | Adaptin beta 1 (beta prime) | +4.8 kb† |
| bA247113.2 | Matches EST sequences | -18.5 kb, -4 kb |
p65 binding confirmed by PCR.
First intron of gene.
Unexpectedly, p65 binding was found to occur at 6 of the 38 genes down-regulated on TNF-α treatment (Table 1). Binding at four of these genes was confirmed by PCR. Other NF-κB family members play a role in transcriptional repression, but complexes containing p65 are thought to exclusively increase transcription (7, 12). These data raise the possibility that p65 might have an unappreciated role in the repression of gene expression.
p65 also Binds to Genes Whose Expression Is Not Regulated by TNF-α. A significant proportion of p65-binding sites (38%) lie near or within genes whose expression is not regulated by TNF-α (Fig. 2 A and D). Some of these genes are not expressed at detectable levels, but a majority are expressed, just not differentially, in response to the stimuli. Table 2 highlights several examples of genes bound by p65 that could potentially be regulated by TNF-α in other cell types or under stimuli other than TNF-α. p65 binding at many of these sites was confirmed by PCR, and Fig. 3A illustrates some of these examples, including thioredoxin (TXN2), target of myb (TOM), heparin cofactor 2 (HCF2), and glycoprotein Ib (platelet) beta polypeptide (GP1BP). HCF2 is an example of a nonregulated gene found on one strand of the chromosome while the up-regulated PI4KCA gene overlaps it on the other strand (Fig. 2B), raising the intriguing possibility that p65 regulates the expression of both of these genes. Regardless, these data suggest that, on entry to the nucleus, p65 binds a broader range of targets than those whose transcription is being regulated.
p65 Binding in Unannotated Regions. In addition to identifying binding sites proximal to known genes, this study revealed that 22% of the binding sites for p65 lie in unannotated regions (defined as >50 kb from any annotation) on chromosome 22. Two recent studies indicate that 50% of the expressed fragments of chromosome 22 do not contain annotated genes (19, 27). Searching for p65 binding 5 kb up- and downstream of the novel transcripts identified by Rinn et al. reveals 63 p65-bound regions, or 30% of the 209 p65-binding sites. Removing those within 50 kb of a known gene reduces this to 45 sites (22%; Fig. 4A). Thus, only 12 p65-binding sites (6%) are in regions completely devoid of any annotation or novel transcribed regions.
Discussion
This study demonstrates for the first time that it is possible to map transcription factor binding sites along an entire human chromosome. By mapping in vivo targets of p65 through ChIP, we found that p65 binds proximal to 5′ ends, as expected, but also with a high frequency at many other sites, including introns and sites distal to 5′ ends. Finding NF-κB-binding sites in introns is not unexpected because it was originally identified by its binding an enhancer element in the first intron of the κ light chain gene; since then, several examples of NF-κB to binding intronic sequences have emerged (22, 23, 28, 29). Our data also indicate that p65 not only binds genes whose expression is regulated by the proinflammatory cytokine TNF-α but also binds a large number of genomic loci whose expression is not regulated.
Unbiased Genomic DNA Arrays. Global analysis of protein–DNA interactions has been limited in mammalian cells by the types of microarrays available (2, 30). The present study uses an unbiased genomic DNA array of human chromosome 22, rather than arrays of fragments computed to contain known consensus sites or existing of known regulatory sequences or promoter proximal sequences (31, 32). Focused arrays based on consensus-site prediction will miss many targets that contain nonconsensus sequences (≈50% of the p65 targets in this study). Promoter-proximal arrays only identify those sites near the 5′ end. For p65, 90% of the targets do not lie in the 1-kb interval immediately upstream of the ATG. Thus, although chromosome arrays are expensive to construct and require more material to probe, they allow genomic sequences to be examined in an unbiased fashion and for the comprehensive identification of targets.
Genes Regulated by NF-κB. p65 was found to bind to sites within or near almost half of the genes up-regulated by TNF-α (Tables 1, 2, 3). Several potentially interesting and previously unidentified targets of p65 have emerged from this analysis (e.g., ZNF378, ZBED4, EWSR1, ARFGAP3, and PI4KCA). ZNF378 and ZBED4 are both uncharacterized zinc finger proteins, and their regulation by TNF-α suggests that they may function in mediating the downstream effects of this cytokine. Similarly, many of the other genes have not been previously shown to function in the TNF-α signaling pathway; hence, this study both adds to our knowledge of these individual genes and contributes to a greater understanding of TNF-α signaling events.
Surprisingly, we find p65 binding at a small fraction of the genes down-regulated on stimulation with TNF-α (Table 1), thereby revealing a previously uncharacterized aspect of p65 transcriptional regulation. One example is BSAC/MKL1 (Figs. 2C and 3A), an antiapoptotic transcriptional activator that has been shown to be down-regulated by TNF-α in murine embryonic fibroblasts (33). Fig. 2C illustrates that p65 binds two distinct sites at the 5′ end and one site in the first intron of the gene. To our knowledge, this is the first time p65 has been suggested to directly mediate the down-regulation of a gene in response to proinflammatory signals.
We also detect p65 binding at several genes that are not regulated by TNF-α. One specific example of NF-κB binding in the absence of gene activation occurs at the Igλ locus (IGL). Although NF-κB was originally identified as a regulator of an Igκ intronic enhancer, it has also recently been implicated in the regulation of Igλ expression (31, 34). NF-κB binding is found at several sites in the locus, particularly upstream of the IGLV1 and IGLV3 clusters (Table 3). Taken together, these data indicate that NF-κB binding occurs at a large number of target sites including those that may be functionally significant in other cell types or under different conditions. Our data indicate that NF-κB binding in and of itself is not sufficient to drive altered expression, suggesting additional mechanisms for gene activation.
We did not find p65 binding at several of the TNF-α-responsive genes. p65 may function at an earlier time point than that analyzed (90 min). Another possibility is that TNF-α may regulate these genes through other transcription factors (such as AP1) (35) or indirect effects such as a downstream transcriptional cascade or posttranscriptional mechanisms. We found p65 bound to two transcription factors that are down-regulated (Table 1): a protein similar to a Drosophila transcriptional repressor and BSAC/MKL1. Down-regulation of either of these genes could alter the expression profile on exposure to TNF-α.
p65 Binds in Unannotated Regions. p65 binds in unannotated regions; however, many of these binding sites (22%) lie near previously uncharacterized transcriptionally active regions, or TARs, as identified by Rinn et al. (19), raising the possibility that p65 might regulate the expression of these sequences. In addition, 6% of p65-binding sites are in completely unannotated regions. Such binding sites might function as distal enhancer regions and regulate genes at a distance, perhaps by mechanisms such as chromosomal looping (36). It is also possible that unidentified genes lie in these regions.
Characterization of the human genome not only requires the identification of coding segments of genes and the mapping of novel transcripts but also how individual genes are separately regulated. These studies demonstrate that it is possible to map the binding sites of a DNA-binding protein across an entire chromosome in multicellular organisms. It should now be possible to extend this approach to an entire mammalian genome.
Supplementary Material
Acknowledgments
We thank F. Sayward for valuable assistance, and S. Vidan, M. Smith, and C. Horak for helpful discussions and critical comments on the manuscript. R.M. is supported by a National Institutes of Health predoctoral training grant, and G.E. is supported by National Institutes of Health Postdoctoral Fellowship F32 HG02446-01. This work was supported by National Institutes of Health Grant HG02357.
Abbreviations: ChIP, chromatin immunoprecipitation; Ct, critical threshold; ICAM, intercellular adhesion molecule; TNF, tumor necrosis factor.
References
- 1.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 2.Horak, C. E., Luscombe, N. M., Qian, J., Bertone, P., Piccirrillo, S., Gerstein, M. & Snyder, M. (2002) Genes Dev. 16, 3017–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 4.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells, J., Graveel, C. R., Bartley, S. M., Madore, S. J. & Farnham, P. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horak, C. E., Mahajan, M. C., Luscombe, N. M., Gerstein, M., Weissman, S. M. & Snyder, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May, M. J. & Ghosh, S. (1998) Immunol. Today 19, 80–88. [DOI] [PubMed] [Google Scholar]
- 8.Gugasyan, R., Grumont, R., Grossmann, M., Nakamura, Y., Pohl, T., Nesic, D. & Gerondakis, S. (2000) Immunol. Rev. 176, 134–140. [DOI] [PubMed] [Google Scholar]
- 9.Locksley, R. M., Killeen, N. & Lenardo, M. J. (2001) Cell 104, 487–501. [DOI] [PubMed] [Google Scholar]
- 10.Ainbinder, E., Revach, M., Wolstein, O., Moshonov, S., Diamant, N. & Dikstein, R. (2002) Mol. Cell. Biol. 22, 6354–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pahl, H. L. (1999) Oncogene 18, 6853–6866. [DOI] [PubMed] [Google Scholar]
- 12.Rothwarf, D. M. & Karin, M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 13.Weinmann, A. S. & Farnham, P. J. (2002) Methods 26, 37–47. [DOI] [PubMed] [Google Scholar]
- 14.Luscombe, N. M., Royce, T., Bertone, P., Echols, N., Horak, C. E., Chang, J. T., Snyder, M. & Gerstein, M. (2003) Nucleic Acids Res. 31, 3477–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quackenbush, J. (2002) Nat. Genet. 32, Suppl., 496–501. [DOI] [PubMed] [Google Scholar]
- 16.van de Stolpe, A., Caldenhoven, E., Stade, B. G., Koenderman, L., Raaijmakers, J. A., Johnson, J. P. & van der Saag, P. T. (1994) J. Biol. Chem. 269, 6185–6192. [PubMed] [Google Scholar]
- 17.Kunsch, C. & Rosen, C. A. (1993) Mol. Cell. Biol. 13, 6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner, B. P., Westerheide, S. D. & Baldwin, A. S., Jr. (2001) Mol. Cell. Biol. 21, 7065–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn, J. L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N. M., Hartman, S., Harrison, P. M., Nelson, F. K., Miller, P., Gerstein, M., et al. (2003) Genes Dev. 17, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins, J. E., Goward, M. E., Cole, C. G., Smink, L. J., Huckle, E. J., Knowles, S., Bye, J. M., Beare, D. M. & Dunham, I. (2003) Genome Res. 13, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison, P. M., Hegyi, H., Balasubramanian, S., Luscombe, N. M., Bertone, P., Echols, N., Johnson, T. & Gerstein, M. (2002) Genome Res. 12, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schjerven, H., Brandtzaeg, P. & Johansen, F. E. (2001) J. Immunol. 167, 6412–6420. [DOI] [PubMed] [Google Scholar]
- 23.Maehara, K., Hasegawa, T. & Isobe, K. I. (2000) J. Cell. Biochem. 77, 474–486. [DOI] [PubMed] [Google Scholar]
- 24.Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A., Podkolodnaya, O. A., Kolpakov, F. A., et al. (1998) Nucleic Acids Res. 26, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannenhalli, S. & Levy, S. (2002) Nucleic Acids Res. 30, 4278–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J. Y., Kim, H., Jeun, S. S., Rha, S. J., Kim, Y. H., Ko, Y. J., Won, J., Lee, K. H., Rha, H. K. & Wang, Y. P. (2002) Biochem. Biophys. Res. Commun. 296, 1295–1302. [DOI] [PubMed] [Google Scholar]
- 27.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 28.Bendall, H. H., Sikes, M. L. & Oltz, E. M. (2001) J. Immunol. 167, 264–269. [DOI] [PubMed] [Google Scholar]
- 29.Guo, Z., Boekhoudt, G. H. & Boss, J. M. (2003) J. Biol. Chem. 278, 23570–23578. [DOI] [PubMed] [Google Scholar]
- 30.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 31.Wells, J., Yan, P. S., Cechvala, M., Huang, T. & Farnham, P. J. (2003) Oncogene 22, 1445–1460. [DOI] [PubMed] [Google Scholar]
- 32.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A. & Dynlacht, B. D. (2002) Genes Dev. 16, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasazuki, T., Sawada, T., Sakon, S., Kitamura, T., Kishi, T., Okazaki, T., Katano, M., Tanaka, M., Watanabe, M., Yagita, H., Okumura, K. & Nakano, H. (2002) J. Biol. Chem. 277, 28853–28860. [DOI] [PubMed] [Google Scholar]
- 34.Combriato, G. & Klobeck, H. G. (2002) J. Immunol. 168, 1259–1266. [DOI] [PubMed] [Google Scholar]
- 35.Ventura, J. J., Kennedy, N. J., Lamb, J. A., Flavell, R. A. & Davis, R. J. (2003) Mol. Cell. Biol. 23, 2871–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. (2002) Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.