Abstract
Epigenetic silencing associated with aberrant methylation of promoter region CpG islands is one mechanism leading to loss of tumor suppressor function in human cancer. Profiling of CpG island methylation indicates that some genes are more frequently methylated than others, and that each tumor type is associated with a unique set of methylated genes. However, little is known about why certain genes succumb to this aberrant event. To address this question, we used Restriction Landmark Genome Scanning to analyze the susceptibility of 1,749 unselected CpG islands to de novo methylation driven by overexpression of DNA cytosine-5-methyltransferase 1 (DNMT1). We found that although the overall incidence of CpG island methylation was increased in cells overexpressing DNMT1, not all loci were equally affected. The majority of CpG islands (69.9%) were resistant to de novo methylation, regardless of DNMT1 overexpression. In contrast, we identified a subset of methylation-prone CpG islands (3.8%) that were consistently hypermethylated in multiple DNMT1 overexpressing clones. Methylation-prone and methylation-resistant CpG islands were not significantly different with respect to size, C+G content, CpG frequency, chromosomal location, or promoter association. We used DNA pattern recognition and supervised learning techniques to derive a classification function based on the frequency of seven novel sequence patterns that was capable of discriminating methylation-prone from methylation-resistant CpG islands with 82% accuracy. The data indicate that CpG islands differ in their intrinsic susceptibility to de novo methylation, and suggest that the propensity for a CpG island to become aberrantly methylated can be predicted based on its sequence context.
DNA methylation in mammals occurs at cytosines residues within the sequence context CpG. Although relatively rare (≈1 per 50–100 bp) throughout much of the human genome, CpGs are enriched in short stretches of CpG-dense DNA known as CpG islands (1). Recent estimates suggest that there are at least 29,000 such regions in the human genome, many of which surround the 5′ ends of genes (2). Whereas most CpG islands remain unmethylated in normal adult cells, they can become methylated de novo in human cancer cells. This aberrant methylation is accompanied by local changes in histone modification and chromatin structure, such that the CpG island and its embedded promoter take on a repressed conformation that is incompatible with gene transcription (3). This epigenetic alteration is thus associated with gene silencing, and together with point mutations and deletions, serves as one mechanism contributing to the inactivation of tumor suppressor and other critical genes in human cancers (4–6).
Despite numerous examples of methylation-associated gene silencing events in human cancer cells, it is not known why particular CpG islands succumb to aberrant methylation. Methylation “profiling” studies have shown that although there may be hundreds of different CpG islands methylated in any one tumor, some are methylated in multiple tumor types, whereas others are methylated in a tumor-type specific manner (7, 8). Moreover, each tumor type tends to exhibit a characteristic set of aberrantly methylated genes. A number of factors could contribute to the likelihood that a particular CpG island will be methylated in a given tumor. De novo methylation could occur in a stochastic manner, and the resulting spectrum of methylated loci could be a reflection of methylation-associated gene silencing events that confer a growth or survival advantage. The idea that CpG island methylation reflects events that are selected during tumor progression is supported by the observation that some genes (e.g., VHL, Rb) are methylated only in those tumor types in which they are also commonly mutated (9). Furthermore, methylation of tumor suppressor genes is generally observed only on the wild-type allele in cases where the other allele is mutated, whereas biallelic methylation is often observed when both alleles are wild type (10, 11).
Alternatively, there may be intrinsic differences in the susceptibility of individual CpG islands to de novo methylation. This latter hypothesis predicts that there are local features that influence the propensity for de novo methylation. For example, Alu and other repetitive elements have been suggested to serve as foci from which de novo methylation can spread (12, 13), whereas other elements have been suggested to provide a protective function (14, 15). The observation that DNA methyltransferases interact with transcription factors (16–18) has also led to the suggestion that de novo methylation may be “targeted” to particular regions through direct interaction with sequence-specific DNA-binding proteins.
The question of susceptibility is not easily addressed through studies of primary tumors because they represent the endpoint of an evolutionary process, a point in time long after the initial methylation events were incurred and the effects of selective pressures have been imposed. Therefore, to study the factors involved in aberrant methylation, we have used an in vitro model in which de novo methylation is evoked by overexpression of DNA cytosine-5-methyltransferase 1 (DNMT1). In previous studies, we showed that the overexpression of DNMT1 results in the progressive de novo methylation of endogenous CpG island sequences (13, 19). Based on the analysis of selected genes, we found that even in the context of increased de novo methylation capacity, only a subset succumbed to aberrant methylation. These findings led us to hypothesize that some CpG islands may be more prone to de novo methylation than others, and that intrinsic differences in methylation susceptibility might ultimately contribute to the nonrandom patterns of methylation observed in human tumors. An understanding of the nature of these differences could provide insight into the molecular basis for aberrant methylation.
In this study, we used restriction landmark genome scanning (RLGS) to analyze, on a genomewide basis, the methylation status of 1,749 unselected CpG islands in DNMT1 overexpressing cells relative to controls. We find that there are distinct classes of CpG islands with different propensities for de novo methylation. We used pattern recognition and machine learning techniques to identify sequence attributes and develop classification algorithms capable of discriminating between methylation-prone and methylation-resistant CpG islands. The data suggest that there is a sequence signature associated with aberrant methylation.
Materials and Methods
Cell Lines. The generation and characterization of matched clonal cell lines expressing 50-fold increased levels of DNMT1 (HMT.17, HMT.19, HMT.1E1) or the empty vector (NEO.1, NEO.4, NEO.20) from simian virus 40 (SV40)-transformed human fibroblasts has been described (19). Cells were maintained in Eagle's minimal essential medium (EMEM) containing 10% FCS, 1 mM glutamine, and 400 μg/ml G418.
RLGS. RLGS was performed as described (8). Briefly, DNA (5 μg) was digested with NotI, end labeled, digested with EcoRV, and separated in the first dimension on a 0.8% agarose tube gel. DNA was subjected to in-gel digestion with HinfI and separated in the second dimension on a 5% polyacrylamide gel. Dried gels were exposed to x-ray film. A fragment was called methylated if there was a visually apparent decrease in spot intensity relative to surrounding spots, and to the spots in the same neighborhood on the other profiles. Based on Southern blot analyses, the distinction between unmethylated and methylated was most reliable when methylation exceeded ≈40%.
Southern Analysis. Genomic DNA (10 μg) was digested overnight with NotI and EcoRV, separated on a 1% agarose gel, transferred to a nylon filter (ZetaProbe, Bio-Rad), and hybridized with a random-prime-labeled DNA probe. Blots were washed to a final stringency of 0.1% SDS/0.1× SSC at 65°C, and exposed to x-ray film. Probes were generated from the corresponding genomic NotI/EcoRV boundary library clone or amplified by PCR from a human fibroblast DNA template (see Supporting Text and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org).
Statistical Analyses and Simulations. One-dimensional, average-linked, hierarchical cluster analysis was performed by using an agglomerative clustering algorithm (cluster) and visualized by using treeview software (http://rana.lbl.gov/EisenSoftware.htm).
For the tests of randomness of methylation in the HMT clones, we performed simulations based on the frequency of actual methylation events observed in each clone (363 of 1,749, 194 of 1,749, and 375 of 1,749 in HMT.17, HMT.19, and HMT.1E1, respectively). Simulations were performed in which exactly 363 (or 194, or 375) entries were selected from a total of 1,749 possibilities and the number of times each entry was selected over 1,000 simulations was recorded. Empirical results indicated that each CpG island has a 20%, 10%, and 20% chance to be methylated by each of the HMT clones, respectively. Therefore, given an equal probability of methylation, no CpG island should be methylated in all three clones with a chance higher than 0.2 × 0.1 × 0.2 = 0.004, and the probability of 66 being methylated in all three clones is 0.004/66 = 0.00006.
Sequence Analysis. Genomic clones corresponding to various RLGS spots were obtained from a human NotI/EcoRV boundary library (20). In other cases, genomic sequence was derived from RLGS verified, “in silico” NotI/EcoRV/HinfI digests of the human genome draft sequence (21). Genomic regions surrounding the corresponding NotI sites were extracted from the Human Genome (http://genome.ucsc.edu) and Celera databases. CpG islands were identified by using the cpgreport algorithm (www.ebi.ac.uk/cpg), and 4 kb of sequence centered on this region was extracted. CpG island attributes were determined by using the newcpgreport program (www.uk.embnet.org/Software/EMBOSS) with a sliding window of 200 bp and a step increment of 1.
Supervised and Unsupervised Classification. We developed a method for classifying CpG islands into response categories (e.g., methylation-prone or methylation-resistant) based on DNA sequence. This method consisted of three steps: (i) the identification of exact match sequence strings in the 4-kb CpG island centered sequences from the training set, (ii) selection of the optimal set of sequence patterns that discriminate the training set by using a multistep tree classification approach, and (iii) class prediction based on the frequency of occurrence of the optimal set of sequence patterns. The accuracy of the resulting classifier was estimated by 10-fold cross validation. For details, see Supporting Text.
Results
Genomewide Methylation Analyses. In previous studies we showed that overexpression of DNMT1 results in the progressive de novo methylation of selected gene targets (19). Here we took a genomewide approach to determine whether, and to what degree, CpG islands differ in their susceptibility to aberrant methylation. RLGS allows for the unbiased analysis of >1,000 CpG islands without prior knowledge of sequence or selection of specific gene targets. This method is based on the two-dimensional separation of end-labeled fragments generated by digestion with the methylation-sensitive enzyme, NotI. Such sites occur on average every 100 kb in the human genome and are confined almost exclusively to CpG islands (22). Methylation blocks NotI digestion, resulting in the loss of a spot on the two-dimensional profile. A high degree of reproducibility in the two-dimensional profiles has been demonstrated. This technique has been useful in the identification of novel methylation, amplification, and deletion events in human and mouse tumors (23).
RLGS was performed on six independent human fibroblast cell clones derived from a common parent that differed only in their levels of DNMT1 expression (19). We previously described the generation of single cell clones stably overexpressing human DNMT1 or the empty vector. Southern blot analysis for the integrated construct confirmed that each clone was derived from an independent integration event. We compared three clones expressing ≈50-fold increased levels of DNA methyltransferase activity (HMT.17, HMT.19, and HMT.1E1) to three vector-only controls (NEO.1, NEO.4, and NEO.20). To control for methylation events that might continue to accumulate over time, DNA for the RLGS analysis was collected from all clones at passage 10, or ≈30 population doublings after the original transfection with DNMT1. The two-dimensional pattern of radiolabelled NotI fragments was visually compared between the six profiles as well as to that of a control peripheral blood lymphocyte profile, and an “address” was assigned to each spot (Fig. 4, which is published as supporting information on the PNAS web site). Radioactive spots that were >≈70% decreased in intensity relative to surrounding spots on the same profile, as well as by comparison to the other profiles was recorded as a loss. A total of 1,749 NotI fragments were analyzed in each of six profiles, for a total of 10,494 potential CpG island methylation events.
The total burden of CpG island methylation was estimated from the number of spots present (e.g., unmethylated) on each RLGS profile. Of the 1,749 possible fragments, the NEO control clones retained an average of 1,691 (range 1,676–1,705), whereas the HMT clones retained an average of 1,438 fragments (range 1,374–1,555). This corresponds to a significantly greater number of potential methylation events in the HMT as compared with the NEO clones (310 versus 58, respectively; P = 0.01, Student's t test). These data indicate that overexpression of DNMT1 is associated with a genomewide increase in CpG island methylation. Assuming 29,000 CpG islands per haploid genome (2) and an estimated 90% concordance between NotI sites and CpG islands (22), we estimate that the HMT clones undergo an average of 4,672 CpG island methylation events.
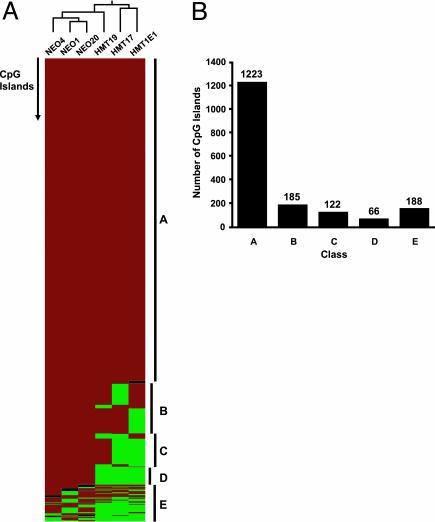
Hierarchical cluster analysis was used to assess the overall degree of similarity between the clones (Fig. 1). The NEO and HMT clones clustered into separate groups, suggesting that the clones can be separated based on their methylation profiles. Several classes of CpG islands with distinct epigenetic behavior were observed. Strikingly, most of the NotI fragments (1,223 of 1,749, 69.9%) were retained in all six RLGS profiles, suggesting that the vast majority of CpG islands are unaffected by DNMT1 overexpression. An additional 373 (21%) exhibited DNMT1-specific “hits” in that they were retained in all of the NEO clones and were lost only in one or more of the HMT clones. A subset of these (66 of 1,749, 3.8%) were retained in all three NEO clones and lost in all three HMT clones. The analysis of multiple independent control (NEO) clones allowed an estimation of the degree of interclonal heterogeneity in RLGS profiles. Only 153 of the 1,749 fragments analyzed (9%) varied in status among the NEO clones. All but five of these were also variably methylated in the HMT clones. These data suggest that there is <10% clonal heterogeneity in global CpG island methylation in the absence of added DNMT1. These data are similar to those of Zhu et al. (24), who showed an ≈10% heterogeneity (156 of 1,068 fragments analyzed) in global RLGS profiles among T cell clones derived from a single patient.
Fig. 1.
Methylation events at 1,749 CpG island fragments as determined by RLGS analysis. (A) Hierarchical cluster analysis examining the overall similarity in RLGS patterns between HMT and NEO clones. Each row represents 1 of 1,749 CpG island fragments, and each column represents a different clone. Relatedness of the clones is represented by a tree (top) whose branch lengths represent the degree of similarity (red, fragment retained; green, fragment lost; black, undetermined). Several distinct classes of CpG islands were observed: those retained in all six clones (A); those retained in all Neo clones but lost in one (B), two (C) or three (D) HMT clones; and those exhibiting frequent loss in both NEO and HMT clones (E). (B) The number of fragments in each class.
Considering the estimate of 9% frequency of loss of any one fragment among clones in the absence of DNMT1, it seems unlikely that 66 CpG islands could be concurrently methylated in any three clones solely by chance. We ran simulations based on the observed frequency of CpG island methylation in each of the HMT clones (363 of 1,749, 194 of 1749, 375/1749 in HMT.17, HMT.19, and HMT.1E1, respectively) to test the probability that 66 CpG islands could be concurrently methylated in the HMT clones. Results of 1,000 simulations showed that each CpG island has a 20%, 10%, and 20% chance to be methylated in each of the HMT clones. Therefore, if all CpG islands have an equal chance of being methylated, none should be methylated in all three clones with a probability greater than 0.2 × 0.1 × 0.2 = 0.004, and the chance that 66 would be concurrently methylated is 0.004/66 or 0.00006. Taken together, these data indicate that methylation is nonrandom, and that given equal methylation potential, CpG islands differ in their intrinsic susceptibility to de novo methylation. We defined those CpG islands that remained unmethylated in all six clones as “methylation-resistant” and the 66 CpG islands that were consistently hypermethylated in response to DNMT1 as “methylation-prone.”
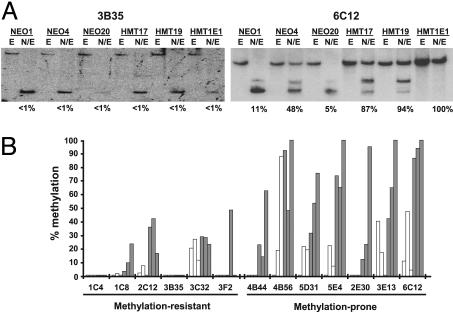
The methylation status determined by RLGS was verified by Southern blot analysis on selected CpG islands from the methylation-prone (4B44, 4B56, 5D31, 5E4, 2E30, 3E13, 6C12) and methylation-resistant (1C4, 1C8, 2C12, 3B35, 3C32, 3F2) groups (Fig. 2). Analysis of 13 CpG islands showed that those in the methylation-resistant group were either completely unmethylated in all clones, or in cases where there was some low-level methylation, there was little difference between the HMT and the NEO clones (Fig. 2). In contrast, there was a general trend for increased methylation in the HMT clones relative to the NEO clones in the methylation-prone group. In general, the RLGS “call” and the actual methylation status were most consistent when methylation exceeded 40%, consistent with previous studies (8). These data also confirm that the loss of a spot on the RLGS profile caused by methylation of the NotI site and not a deletion or recombination event.
Fig. 2.
Methylation analysis of representative RLGS fragments. (A) Genomic DNA from DNMT1-overexpressing (HMT) and vector-only control clones (NEO) was digested with EcoRV (E) or NotI plus EcoRV (N/E) and subjected to Southern blot analysis using probes specific to the indicated RLGS fragment. Representative blots of fragments 3B35 and 6C12 are shown. (B) The percent methylation was determined from scanned images of the resulting autoradiographs and quantified as the relative intensities of the completely digested band to larger methylated band(s). Twelve representative RLGS fragments from the methylation-prone and methylation-resistant groups were analyzed. Open bars, NEO clones; filled bars, HMT clones.
CpG Island Characteristics. The development of an arrayed human genomic NotI-EcoRV boundary library (20) and in silico NotI/EcoRVI/HinfI digests of the completed segments of the human genome project (21), have allowed the assignment of specific sequences to many, but not all, spots on the two-dimensional array. We obtained sequence information from 15 of the methylation-prone and 47 of the methylation-resistant NotI fragments. All 62 fragments analyzed were found to lie within a CpG island according to established definitions (25), and all but five also conformed to the more stringent definition of Takai and Jones (26). Overall, the CpG islands ranged in length from 228 bp to 3.1 kb and had an average C+G content of 69% (range 57.6–78.8%), and an average ratio of observed over expected CpG frequency of 0.85 (range 0.66–1.01). As expected, most (92%) of the CpG island sequences were found to be gene-associated in that they overlapped with either the exon of a known gene or with spliced ESTs (Table 3, which is published as supporting information on the PNAS web site). There did not appear to be any evidence for the methylation-prone CpG islands to cluster at any particular chromosomal locus, although 2 of 15 (3E55 and 3D70) included exons for adjacent homeobox genes that lie ≈40 kb apart on chromosome 13q12 (IPF1 and CDX2, respectively). Likewise, for gene associated CpG islands, there was no relationship between its location within the gene (promoter versus nonpromoter) and methylation class.
Discriminating Methylation-Prone and Methylation-Resistant CpG Islands. The hypothesis that CpG islands differ in their inherent susceptibility to aberrant methylation presupposes that there are cis-acting features that distinguish methylation-prone and methylation-resistant CpG islands. To address this question, we first compared general CpG island characteristics, including size, G+C content, and the ratio of observed to expected CG frequency (CGo/CGe) between the two groups (Table 1). There were no statistically significant differences in these three attributes.
Table 1. General CpG island attributes.
| Methylation-prone (n = 15)
|
Methylation-resistant (n = 47)
|
||||
|---|---|---|---|---|---|
| Attribute | Mean ± SD* | Range | Mean ± SD* | Range | P value* |
| Length, bp | 1,098.4 ± 586.0 | 473-2718 | 1,098.4 ± 630.7 | 228-3182 | 1.00 |
| %C+G | 69.6 ± 4.31 | 61.7-75.8 | 68.8 ± 4.99 | 57.7-78.9 | 0.57 |
| CGo/CGe | 0.87 ± 0.08 | 0.77-1.01 | 0.85 ± 0.08 | 0.66-1.01 | 0.29 |
| Sp1 sites† | 0.47 ± 0.64 | 0-2 | 0.70 ± 0.91 | 0-3 | 0.36 |
Sequences were analyzed by using the NEWCPGREPORT algorithm (http://www.uk.embnet.org/Software/EMBOSS) by using a sliding window of 200 bp and a step increment of 1.
Groups were compared by Student's t test.
Number of occurrences per sequence based on the consensus GRGGCRGGGW.
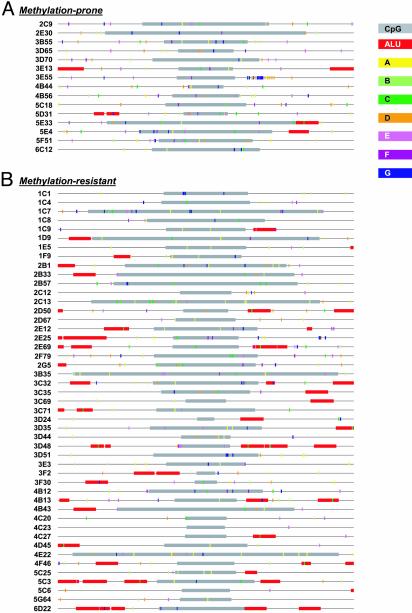
We next used pattern recognition and a supervised learning strategy to identify more complex sequence attributes that might discriminate between the two classes of CpG islands. To take an unbiased approach to pattern discovery, we normalized the sequence data by determining the center of each CpG island and extracting 2 kb of sequence information in either direction. This size was chosen because the CpG islands in the study ranged in size from 0.2 to 3.2 kb and because cis-acting features might lie within or flanking the CpG island. Raw sequence (e.g., unmasked) from both strands was used so that the analysis would be independent of orientation. The training set consisted of the 4-kb centered sequences from nine methylation-prone and nine methylation-resistant CpG islands. Our approach involved three steps. First, an in-house DNA pattern discovery tool (27) was used to identify fixed-length DNA patterns. The algorithm used differs from similarity-based motif discovery tools in that it uses a combinatorial approach to identify the longest common exact match sequence string among a group of DNA sequences. The algorithm allows for nonconsecutive sequence strings (e.g., the use of N for any base) and other physical constraints (27). The pattern recognition procedure identified >100 patterns of length >7 bp in the training set. Next, a multistep decision-tree analysis (28, 29) was performed on the training set CpG islands to rank the patterns whose frequency of occurrence best discriminated the two classes. Seven DNA patterns (TCCCCCNC; TTTCCTNC; TCCNCCNCCC; GGAGNAAG; GAGANAAG; GCCACCCC; and GAGGAGGNNG) were selected as having the highest discrimination potential. The frequency of occurrence of each of the patterns in the CpG islands is listed in Table 4, which is published as supporting information on the PNAS web site, and their spatial distribution shown in Fig. 3.
Fig. 3.
Sequence features associated with methylation-prone (A) and methylation-resistant (B) CpG islands. Map of various sequence features in the 62 CpG islands used in this study. Each line represents a 4-kb centered sequence used for pattern recognition and discriminant analysis. CpG islands (gray) were defined by the cpgreport algorithm; Alu sequences (red) were detected by repeatmasker. The seven novel patterns are also shown: A, yellow; B, light green; C, dark green; D, orange; E, pink; F, magenta; G, dark blue.
We next derived a classification function based on the selected DNA patterns. A numeric vector representing the frequency of occurrence of each of the seven DNA patterns was constructed for each sequence in the training set and used as input for discriminant analysis to develop a predictive rule. The training set consisted of the same 18 CpG islands (nine methylationprone, nine methylation-resistant) used for the pattern recognition module, and the classification algorithm was a linear program optimization-based discriminant analysis method (30, 31). The accuracy of the resulting classifier was estimated by 10-fold cross-validation. Results of the cross validation tests indicated that the methylation-prone CpG islands were classified with 100% accuracy (zero of nine misclassified) and the methylation resistant CpG islands were classified with 78% accuracy (two of nine misclassified) for an overall rate of correct classification of 89%. Successive iterations of the pattern selection and two-stage discriminant analysis that incorporated more sequence patterns (e.g., the 13 best, 20 best, etc.) in the frequency vector did not improve the accuracy of the function.
To validate the classification function, we next tested it against the remaining 44 CpG islands whose class was known from the RLGS analysis, but whose sequences were not used in the pattern recognition or training modules (e.g., unseen data). In this “blind” test, methylation-prone and methylation-resistant CpG islands were correctly classified at a rate of 83% (one of six misclassified) and 78% (7 of 38 misclassified), respectively. Thus, the classification function derived from short, exact match sequence patterns using the optimization-based discrimnant analysis model was able to “predict” the status of a series of unknown CpG islands with an overall correct classification rate of 82%.
We also tested the utility of the sequence patterns in an unsupervised learning strategy. In contrast to supervised methods, unsupervised learning methods do not use a training set to derive prediction rules. Attribute vectors based on the frequency of the seven DNA patterns were determined for all 62 CpG islands, and served as direct input for cluster analysis using PAM (partitioning around medoids) (32). In this experiment, the correct classification rate was 87% (2 of 15 misclassified) for the methylation-prone, and 57% (20 of 47 misclassified) for the methylation-resistant sequences. Compared with the supervised method, the classification of methylation-resistant sequences in the unsupervised method was somewhat inferior. The results illustrate both the usefulness of sequence information for prediction of the methylation status of CpG islands and the potential of machine learning techniques, especially supervised learning, in extracting important patterns and developing reasonably accurate prediction functions.
Discussion
We have taken a functional genomics approach to ask why certain genes are targets of aberrant methylation in human cancers. Our data show that although increased expression of DNMT1 results in an overall increase in CpG island methylation, not all CpG islands are equally affected. Although most CpG islands were resistant to de novo methylation, we defined a subset that was highly susceptible to aberrant methylation. We developed a classifier based on the frequency of seven novel sequence patterns that was capable of discriminating methylation-prone and methylation-resistant CpG islands with 82% accuracy. The implications of our studies are twofold. First, the data indicate that CpG islands differ in their innate susceptibility to aberrant methylation. Such differences could contribute to the propensity of some genes to de novo methylation in human cancers. Secondly, we provide evidence that the epigenetic state of a CpG island can be predicted based on its sequence context, suggesting that there are local features that contribute to the risk of aberrant methylation.
At present, the factors that contribute to methylation-susceptibility are not known. We identified 66 CpG islands that were highly susceptible to de novo methylation in that they were consistently hypermethylated in multiple independent clones with elevated levels of DNMT1. Biniszkiewicz et al. (33) also showed that among selected imprinted genes, the H19/Igf2 locus was de novo methylated in response to moderately elevated levels of DNMT1, whereas several others remained unaffected. Interestingly, GRB10, a maternally imprinted gene (34) was among the methylation-prone CpG islands identified here. GRB10 is also deregulated in ES cells lacking DNMT1 (35), suggesting that this locus may be exquisitely sensitive to alterations in DNMT1 levels. The inherent susceptibility of some loci to de novo methylation could contribute to their aberrant methylation in human cancers. Two of the methylation-prone CpG islands are associated with genes that encode homeobox transcription factors (CDX2 and IPF1). As a family, the homeobox genes are frequently down-regulated in association with aberrant methylation in human cancer cells (36, 37), and the HOX gene clusters are a hotspot of de novo methylation in lung cancers (38).
Although not as dramatic as the levels achieved here, increased expression of DNMT1 has been observed in human tumors (reviewed in ref. 9), and the levels of DNMT1 can impact on transformation potential and tumor formation in mouse models (39–41). The precise role for DNMTs in the aberrant methylation that accompanies tumorigenesis remains controversial, however, because there appears to be no direct relationship between DNMT expression and the methylation of specific tumor suppressor genes (e.g., ref. 42). Our data suggest that although increased expression of DNMT1 may increase the cellular capacity for aberrant methylation, the propensity for any one locus to succumb to this event is dictated by local factors. In human tumors, the final profile of methylated genes will be further shaped over time by the pressures encountered during tumor progression and the selection for methylation-associated gene silencing events that confer a growth or survival advantage. This would explain why numerous attempts to correlate DNMT expression with the methylation status of particular gene targets in primary tumors have met with little success.
The finding that the vast majority of CpG islands remained unmethylated despite overexpression of DNMT1 suggests that for most genes there is a strong protective mechanism against de novo methylation. The molecular basis of such protection is not well understood. An element containing Sp1 sites in the 5′ end of Aprt is necessary to protect the CpG island from de novo methylation (15). Interestingly, we found that the frequency of Sp1 sites did not significantly differ between methylation-prone and the methylation-resistant CpG islands (Table 1). Moreover, the addition of the Sp1 consensus as an additional attribute in discriminant analysis reduced the accuracy of the resulting classifier, both in cross validation tests (from 89% to 83%) and blind predictions (from 82% to 65%) (Supporting Text). These data suggest that the presence of Sp1 sites has little bearing on the susceptibility of CpG islands to aberrant methylation, at least in the DNMT1 overexpression model. It also has been proposed that ongoing transcription may indirectly protect CpG islands from methylation (9, 43). This hypothesis predicts that genes with a restricted expression pattern might be more prone to aberrant methylation than widely expressed genes. Indeed, many of the methylation-resistant CpG islands were associated with genes with housekeeping functions or with broad expression patterns, including MYC, FOS, CDK6, MBD1, and SF3A1 (Table 3) (40). Likewise, many of the methylation-prone CpG islands were associated with tissue-specific genes, such as TBR1, SIM2, GRM6, and XT3 (40). However, there were also examples in which the opposite was true (e.g., WNT10B, NPTXR, POP3). Thus, although there was a tendency for differential representation of widely expressed versus tissue-specific genes between the two groups, the correlation was not absolute.
We describe a multistage discriminant analysis approach that can predict the epigenetic state of different genomic regions based on sequence context. Discriminant analysis and machine learning techniques have been used to identify functional components of the genome based on sequence attributes. For example, Ioshikhes et al. (44) showed that certain CpG island attributes (e.g., C+G content, CpG observed/expected) could be used to develop a function capable of discriminating transcription start site-associated CpG islands from nontranscription start site-associated CpG islands. Similarly, DNA sequence attributes and mRNA characteristics have been used to develop discriminant functions that predict 5′-flanking regions/first exons and internal protein coding regions in genomic DNA (45). Here we have added a pattern recognition component to first identify potential sequence patterns and then used these as attributes to develop a classification function capable of discriminating methylation-prone and methylation-resistant CpG islands.
The reasonable accuracy with which the classifier discriminates methylation-prone and methylation-resistant CpG islands suggests that there is a sequence “signature” associated with susceptibility to, or protection from, aberrant methylation. However, it does not necessarily imply that the sequence patterns themselves have a biological function. Nevertheless, it is interesting to note that two of these patterns (e.g., F and G) can be found in a subset of Alu sequences. Alu and other endogenous retroelements are heavily methylated in the human genome (46) and have been suggested to act as foci from which methylation can spread (12). Notably, we found no particular association of bona fide Alu sequences with the methylation-prone CpG islands. In fact, Alu elements tended to be more frequently associated with the methylation-resistant group (Fig. 3), suggesting that it is the patterns themselves that are important rather than their association with Alus. Likewise, several patterns are homopurine/homopyrimidine stretches. These sequences form triple helical structures under superhelical tension, and have been reported to occur near matrix attachment regions, replication origins, recombination hot spots and in the promoters/enhancers of genes (ref. 47 and references therein). Such alternative DNA structures are suggested to serve as targets for de novo methylation (48).
This study serves as a proof-of-principle that the epigenetic state of a genomic region can be predicted based on underlying sequence context. Although the sequence patterns and the classifier developed here are likely to be specific for methylation occurring in response to overexpression of DNMT1, the methods developed should be more broadly applicable to de novo methylation as it occurs in human cancers. Extensive CpG island methylation profiling data, stemming from RLGS analyses or new microarray-based techniques, already exist for a number of human tumor types (8, 23, 49, 50). Thus, a similar strategy could be applied to derive tumor type-specific DNA patterns and classification algorithms. Moreover, the development of reasonably accurate classification models such as those described here could allow one to predict the methylation status of CpG islands across the genome based on the analysis of a reasonable subset.
Supplementary Material
Acknowledgments
We thank Drs. Paul Wade and Maureen Powers for their thoughtful review of the manuscript. This work was supported by National Cancer Institute Grant CA077337 (to P.M.V.) and National Science Foundation Grant CCR-9721402 (to E.K.L.). P.M.V. is supported in part by funds from the Avon Foundation. F.A.F. was supported by a American Cancer Society Fellowship PF-GMC-102929.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RLGS, restriction landmark genome scanning; DNMT1, DNA cytosine-5-methyltransferase-1.
References
- 1.Bird, A. P. (1986) Nature 321, 209–213. [DOI] [PubMed] [Google Scholar]
- 2.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Antequera, F., Boyes, J. & Bird, A. (1990) Cell 62, 503–514. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., Herman, J. G., Graff, J. R., Vertino, P. M. & Issa, J. P. (1998) Adv. Cancer Res. 72, 141–196. [PubMed] [Google Scholar]
- 5.Costello, J. F. & Plass, C. (2001) J. Med. Genet. 38, 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 7.Esteller, M., Corn, P. G., Baylin, S. B. & Herman, J. G. (2001) Cancer Res. 61, 3225–3229. [PubMed] [Google Scholar]
- 8.Costello, J. F., Fruhwald, M. C., Smiraglia, D. J., Rush, L. J., Robertson, G. P., Gao, X., Wright, F. A., Feramisco, J. D., Peltomaki, P., Lang, J. C., et al. (2000) Nat. Genet. 24, 132–138. [DOI] [PubMed] [Google Scholar]
- 9.Clark, S. J. & Melki, J. (2002) Oncogene 21, 5380–5387. [DOI] [PubMed] [Google Scholar]
- 10.Esteller, M., Fraga, M. F., Guo, M., Garcia-Foncillas, J., Hedenfalk, I., Godwin, A. K., Trojan, J., Vaurs-Barriere, C., Bignon, Y. J., Ramus, S., et al. (2001) Hum. Mol. Genet. 10, 3001–3007. [DOI] [PubMed] [Google Scholar]
- 11.Myohanen, S. K., Baylin, S. B. & Herman, J. G. (1998) Cancer Res. 58, 591–593. [PubMed] [Google Scholar]
- 12.Yates, P. A., Burman, R. W., Mummaneni, P., Krussel, S. & Turker, M. S. (1999) J. Biol. Chem. 274, 36357–36361. [DOI] [PubMed] [Google Scholar]
- 13.Graff, J. R., Herman, J. G., Myohanen, S., Baylin, S. B. & Vertino, P. M. (1997) J. Biol. Chem. 272, 22322–22329. [DOI] [PubMed] [Google Scholar]
- 14.Millar, D. S., Paul, C. L., Molloy, P. L. & Clark, S. J. (2000) J. Biol. Chem. 275, 24893–24899. [DOI] [PubMed] [Google Scholar]
- 15.Turker, M. S. (1999) Semin. Cancer Biol. 9, 329–337. [DOI] [PubMed] [Google Scholar]
- 16.Robertson, K. D., Ait, S. A., Yokochi, T., Wade, P. A., Jones, P. L. & Wolffe, A. P. (2000) Nat. Genet. 25, 338–342. [DOI] [PubMed] [Google Scholar]
- 17.Fuks, F., Burgers, W. A., Godin, N., Kasai, M. & Kouzarides, T. (2001) EMBO J. 20, 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Croce, L., Raker, V. A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., Fuks, F., Lo, C. F., Kouzarides, T., Nervi, C., et al. (2002) Science 295, 1079–1082. [DOI] [PubMed] [Google Scholar]
- 19.Vertino, P. M., Yen, R. W., Gao, J. & Baylin, S. B. (1996) Mol. Cell. Biol. 16, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plass, C., Weichenhan, D., Catanese, J., Costello, J. F., Yu, F., Yu, L., Smiraglia, D., Cavenee, W. K., Caligiuri, M. A., deJong, P. & Held, W. A. (1997) DNA Res. 4, 253–255. [DOI] [PubMed] [Google Scholar]
- 21.Zardo, G., Tiirikainen, M. I., Hong, C., Misra, A., Feuerstein, B. G., Volik, S., Collins, C. C., Lamborn, K. R., Bollen, A., Pinkel, D., et al. (2002) Nat. Genet. 32, 453–458. [DOI] [PubMed] [Google Scholar]
- 22.Lindsay, S. & Bird, A. P. (1987) Nature 327, 336–338. [DOI] [PubMed] [Google Scholar]
- 23.Costello, J. F., Smiraglia, D. J. & Plass, C. (2002) Methods 27, 144–149. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, X., Deng, C., Kuick, R., Yung, R., Lamb, B., Neel, J. V., Richardson, B. & Hanash, S. (1999) Proc. Natl. Acad. Sci. USA 96, 8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardiner-Garden, M. & Frommer, M. (1987) J. Mol. Biol. 196, 261–282. [DOI] [PubMed] [Google Scholar]
- 26.Takai, D. & Jones, P. A. (2002) Proc. Natl. Acad. Sci. USA 99, 3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, E. K., Easton, E. L. & Johnson, E. L. (2001) Graphs Combinatorics, in press.
- 28.Breiman, L., Friedman, J. H., Olshen, R. A. & Stone, C. J. (1984) Classification and Regression Trees (Wadsworth and Brooks/Cole, New York).
- 29.Mathsoft (2001) S-Plus 6 User Manual (Mathsoft, Inc., Seattle).
- 30.Lee, E. K., Gallagher, R. J. & Patterson, D. (2003) INFORMS J. Comput. 15, 23–41. [Google Scholar]
- 31.Gallagher, R. J., Lee, E. K. & Patterson, D. (1997) Ann. Op. Res. 74, 65–88. [Google Scholar]
- 32.Kaufman, L. & Rousseeuw, P. J. (1990) Finding Groups in Data: An Introduction to Cluster Analysis (Wiley, New York).
- 33.Biniszkiewicz, D., Gribnau, J., Ramsahoye, B., Gaudet, F., Eggan, K., Humpherys, D., Mastrangelo, M. A., Jun, Z., Walter, J. & Jaenisch, R. (2002) Mol. Cell. Biol. 22, 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyoshi, N., Kuroiwa, Y., Kohda, T., Shitara, H., Yonekawa, H., Kawabe, T., Hasegawa, H., Barton, S. C., Surani, M. A., Kaneko-Ishino, T. & Ishino, F. (1998) Proc. Natl. Acad. Sci. USA 95, 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson-Grusby, L., Beard, C., Possemato, R., Tudor, M., Fambrough, D., Csankovszki, G., Dausman, J., Lee, P., Wilson, C., Lander, E. & Jaenisch, R. (2001) Nat. Genet. 27, 31–39. [DOI] [PubMed] [Google Scholar]
- 36.Raman, V., Martensen, S. A., Reisman, D., Evron, E., Odenwald, W. F., Jaffee, E., Marks, J. & Sukumar, S. (2000) Nature 405, 974–978. [DOI] [PubMed] [Google Scholar]
- 37.Suh, E. R., Ha, C. S., Rankin, E. B., Toyota, M. & Traber, P. G. (2002) J. Biol. Chem. 277, 35795–35800. [DOI] [PubMed] [Google Scholar]
- 38.Shiraishi, M., Sekiguchi, A., Oates, A. J., Terry, M. J. & Miyamoto, Y. (2002) Oncogene 21, 3659–3662. [DOI] [PubMed] [Google Scholar]
- 39.Wu, J., Issa, J. P., Herman, J., Bassett, D. E. J., Nelkin, B. D. & Baylin, S. B. (1993) Proc. Natl. Acad. Sci. USA 90, 8891–8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakin, A. V. & Curran, T. (1999) Science 283, 387–390. [DOI] [PubMed] [Google Scholar]
- 41.Eads, C. A., Nickel, A. E. & Laird, P. W. (2002) Cancer Res. 62, 1296–1299. [PubMed] [Google Scholar]
- 42.Eads, C. A., Danenberg, K. D., Kawakami, K., Saltz, L. B., Danenberg, P. V. & Laird, P. W. (1999) Cancer Res. 59, 2302–2306. [PubMed] [Google Scholar]
- 43.Bird, A. (2002) Genes Dev. 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 44.Ioshikhes, I. P. & Zhang, M. Q. (2000) Nat. Genet. 26, 61–63. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, M. Q. (2002) Nat. Rev. Genet. 3, 698–709. [DOI] [PubMed] [Google Scholar]
- 46.Yoder, J. A., Walsh, C. P. & Bestor, T. H. (1997) Trends Genet. 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 47.Li, G., Tolstonog, G. V. & Traub, P. (2002) DNA Cell Biol. 21, 163–188. [DOI] [PubMed] [Google Scholar]
- 48.Vertino, P. M. (1999) in Eukaryotic DNA Methyltransferases, eds. Cheng, X. & Blumenthal, R. M. (World Scientific, River Edge, NJ), pp. 341–372.
- 49.Smiraglia, D. J. & Plass, C. (2002) Oncogene 21, 5414–5426. [DOI] [PubMed] [Google Scholar]
- 50.Yan, P. S., Chen, C. M., Shi, H., Rahmatpanah, F., Wei, S. H., Caldwell, C. W. & Huang, T. H. (2001) Cancer Res. 61, 8375–8380. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.