Abstract
Werner syndrome (WS) is a premature aging disorder, displaying defects in DNA replication, recombination, repair, and transcription. It has been hypothesized that several WS phenotypes are secondary consequences of aberrant gene expression and that a transcription defect may be crucial to the development of the syndrome. We used cDNA microarrays to characterize the expression of 6,912 genes and ESTs across a panel of 15 primary human fibroblast cell lines derived from young donors, old donors, and WS patients. Of the analyzed genes, 6.3% displayed significant differences in expression when either WS or old donor cells were compared with young donor cells. This result demonstrates that the WS transcription defect is specific to certain genes. Transcription alterations in WS were strikingly similar to those in normal aging: 91% of annotated genes displayed similar expression changes in WS and in normal aging, 3% were unique to WS, and 6% were unique to normal aging. We propose that a defect in the transcription of the genes as identified in this study could produce many of the complex clinical features of WS. The remarkable similarity between WS and normal aging suggests that WS causes the acceleration of a normal aging mechanism. This finding supports the use of WS as an aging model and implies that the transcription alterations common to WS and normal aging represent general events in the aging process.
Werner syndrome (WS) is an autosomal recessive disease characterized by early onset of many signs of normal aging, such as graying of the hair, scleroderma-like skin changes, ocular cataracts, diabetes, degenerative vascular disease, osteoporosis, and high incidence of some types of cancers (1). As a segmental progeroid syndrome, WS does not exhibit all of the features of normal aging but nevertheless is a very useful model system for the molecular study of normal aging.
The molecular basis of WS is a single mutation in the WRN gene, resulting in a truncated WS protein (WRN) characterized by a loss of nuclear localization signal and protein function (2). WRN has been demonstrated to possess helicase and exonuclease activities (3, 4) and belongs to the RecQ family of helicases. Various defects in DNA replication, recombination, repair, and transcription are found in WS fibroblasts (reviewed in ref. 5). The mechanisms by which the biochemical deficiencies resulting from WRN mutations lead to the characteristic pathology of the syndrome are not yet understood. It has been hypothesized that several WS phenotypes are secondary consequences of aberrant gene expression (6) and that a transcription defect may be crucial to the development of the syndrome (7). Increasing evidence suggests that WRN has a role in transcription. Human WRN activates transcription in a yeast system (8), and recent studies from this laboratory demonstrated that RNA polymerase (pol) II transcription is reduced by 40–60% in WS cells, indicating a primary defect in transcription (7). Supporting this finding, we found that RNA pol II transcription is restored to normal levels by addition of wild-type WRN protein to WS cell extracts (7). So far, it has not been determined whether the WS transcription defect is global or localized to certain genes, and the roles for WRN in transcription remain elusive (9). This result prompted us to investigate the role of WRN in the differential expression of individual genes.
We used cDNA microarrays to study expression of 6,912 RNA pol II transcribed genes in a panel of 15 primary human fibroblast cell lines derived from normal young donors, normal old donors, and WS patients.
Materials and Methods
Cell Lines and Culture Conditions. Fifteen primary human skin fibroblast cell lines were obtained from Coriell Cell Repositories (Camden, NJ) and classified into three groups based on genotype as listed in Table 1: normal young (avg. 22.5 yr, n = 6), normal old (avg. 90 yr, n = 5), and WS (avg. 29 yr, n = 4). Cells were cultured in minimal essential medium supplemented with 10% FBS, 1% penicillin/streptomycin, 1% l-glutamine, and Geneticin G418 (400 μg/ml) (all components were from Life Technologies, Gaithersburg, MD).
Table 1. Cell lines used in this study.
| Coriell repository no. | Genotype | Donor phenotype | PDL | Age, yr |
|---|---|---|---|---|
| AG11747 | Normal young | Not clinically affected | 13 | 22 |
| AG10803 | Normal young | Not clinically affected | 9 | 22 |
| GM03440 | Normal young | Not clinically affected | ? | 20 |
| GM02937 | Normal young | Not clinically affected | ? | 22 |
| GM01891 | Normal young | Not clinically affected | ? | 24 |
| AG09975 | Normal young | Not clinically affected | 15 | 25 |
| AG10884 | Normal old | Not clinically affected | 10 | 87 |
| AG13208 | Normal old | Not clinically affected | 11 | 89 |
| AG13129 | Normal old | Not clinically affected | 11 | 89 |
| AG07725 | Normal old | Not clinically affected | 14 | 91 |
| AG08433 | Normal old | Not clinically affected | 17 | 94 |
| AG12795 | WS (mutation not identified) | Short stature, bird-like appearance, gray hair, juvenile bilateral cataracts, atrophic skin, and hypogonadism | 17 | 19 |
| AG12797 | WS (mutation not identified) | Short stature, bird-like appearance, gray hair, skin hyperpigmentation, juvenile bilateral cataracts, atrophic skin, diabetes, and hypogonadism | 10 | 36 |
| AG06300 | WS (F1074L replacement in the WRN protein) | Gray hair, muscle wasting, wrinkling of skin, dystrophic nails, high-pitched voice, hypogonadism, and a general aged appearance | 32 | 37 |
| AG12799 | WS (mutation not identified) | Short stature, gray hair, hyperpigmentation of skin, juvenile bilateral cataracts, atrophic skin, and hypogonadism | ? | 25 |
RNA Isolation and Microarray Hybridization. Single-channel labeling 33P nylon membrane-based cDNA microarrays containing 6,912 genes and ESTs were provided by the DNA Array Unit (DAU), National Institute on Aging, National Institutes of Health. Array hybridization and data analysis were supervised by the DAU and carried out as described (10, 11). Protocols on array printing, labeling, and hybridization, as well as information on software packages, are available at the DAU web site (www.grc.nia.nih.gov/branches/rrb/dna/dna.htm). Hybridization intensities were quantitated by using array-pro analysis software (Media Cybernetics, Silver Spring, MD), then stored as excel spreadsheets. See also Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Microarray Data Analysis and Significance Statistics. To eliminate noise from low-level expression, spots quantified at <10 were replaced by the value 10 (>25 data points). Nonspecific uniform background across entire arrays due to experimental variation was normalized in excel by using global normalization. The data value for each spot on each membrane was divided by the median intensity value of that membrane to obtain a normalized intensity value. Changes in gene expression between different RNA groups were then calculated by dividing the median of four replicate microarray measurements. The resulting value, referred to as the fold difference, was tested for significance following the guidelines suggested by Miller et al. (12) and others (13). Based on the variance between replicates, the number of replicates, and the number of genes analyzed, we determined the expected number of false positives for any given fold difference. At 1.5-fold difference, the two-tailed P value was 0.0006; thus, 0.06% of genes were likely to show a fold difference >1.5 by chance. Assaying 6,912 genes, that correlates to (6,912 × 0.0006) = 4.1 false positives. Four hundred thirty-five genes were different between groups by >1.5-fold, resulting in a low false discovery rate (false positives/true positives) of 4.1/435 = 0.9%. Genes were categorized as being similarly transcribed in WS and normal aging if expression correlated within 0.5-fold; that is, [(fold change WS/Young) — (fold change Old/Young)] < 0.5. Tables of differentially expressed genes were generated by using genespring software (Silicon Genetics, Redwood City, CA).
Verification of Microarray Data by Gene-Specific Relative RT-PCR. The RNA isolated to generate probes for the arrays (young and old donors) was used for RT-PCR with Ambion's RETROscript kit following the protocol provided by Ambion (Austin, TX). Genespecific primers were purchased from Ambion, primer sequences were obtained from Research Genetics (Huntsville, AL), and oligonucleotides were synthesized by Invitrogen.
Results
Supporting Information. The complete cDNA microarray data can be found in Tables 3–8, which are published as supporting information at the PNAS web site, www.pnas.org. The supporting information contains the raw data, normalization, analysis of intra- vs. inter-group variability, variability between replicates, and pooled vs. individual gene expression, with all genes identified in the study.
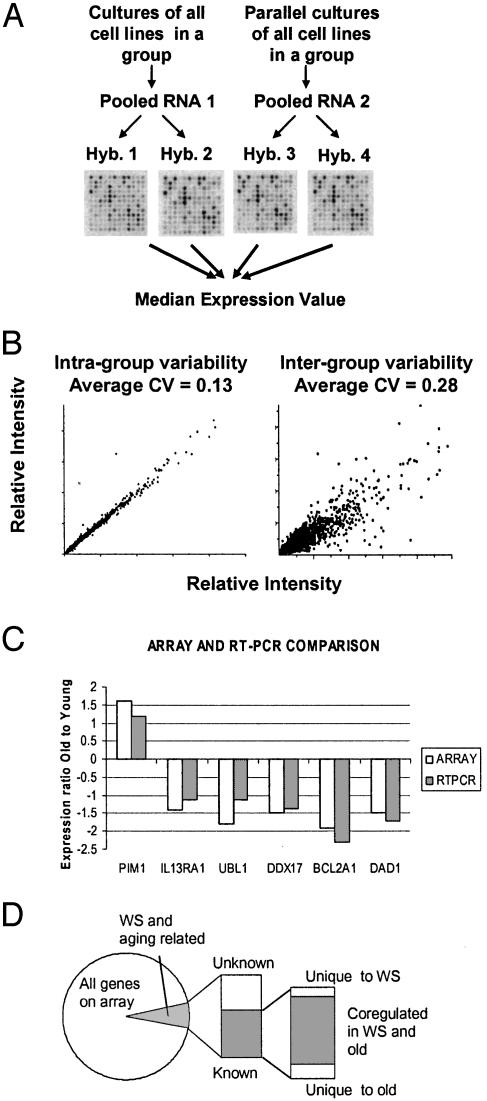
Verification of Microarray Data by Gene-Specific Relative RT-PCR. Expression levels of six randomly picked genes from Table 2 were measured by RT-PCR. As shown in Fig. 1C, RT-PCR analysis confirmed expression ratios measured by microarrays, correlating for both under- and overexpressed genes. The RT-PCR data, together with the statistics on variability, validate the reproducibility of our data. We have previously demonstrated that mRNA expression levels obtained by using this microarray platform correlate with Northern blotting (10).
Table 2. Gene expression in WS and normal old cells relative to normal young cells.
| GenBank | Old* | WS* | Gene name | Synonym |
|---|---|---|---|---|
| DNA/RNA metabolism and chromosomal processing | ||||
| AA459922 | 1.7 | 1.7 | Methyl-CpG binding domain protein 1 | MBD1 |
| AA450265 | 1.6 | 1.3 | Proliferating cell nuclear antigen | PCNA |
| AA029451 | 1.4 | 1.5 | Transcription factor 7-like 2 (T-cell specific, HMG-box) | TCF7L2 |
| AA485944 | -1.5 | -1.5 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 17 (72 kDa) | DDX17 |
| AA406285 | -1.8 | -2.0 | DR1-associated protein 1 (negative cofactor 2 alpha) | DRAP1 |
| AA136566 | -1.4 | -1.6 | Forkhead box M1 | FOXM1 |
| AA253413 | -1.4 | -1.6 | Friedreich ataxia | FRDA |
| AA608514 | -1.5 | -1.5 | H3 histone, family 3B (H3.3B) | H3F3B |
| AA291389 | -1.2 | -1.5 | Interferon-stimulated transcription factor 3, gamma (48 kDa) | ISGF3G |
| AA282537 | -1.6 | -1.7 | MADS box transcription enhancer factor 2, polypeptide B (myocyte enhancer factor 2B) | MEF2B |
| AA479052 | -2.0 | -2.1 | Polymerase (RNA) II (DNA directed) polypeptide A (220 kDa) | POLR2A |
| AA460830 | -1.4 | -1.6 | Polymerase (RNA) II (DNA directed) polypeptide J (13.3 kDa) | POLR2J |
| AA496809 | -1.5 | -1.8 | SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin | SMARCA1 |
| AA446018 | -1.4 | -1.5 | SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin | SMARCB1 |
| R91259 | -1.7 | -1.4 | Transcription factor BMAL2 | LOC56938 |
| AA465369 | -1.8 | -2.0 | Transcription factor Dp-2 (E2F dimerization partner 2) | TFDP2 |
| AA232647 | -1.9 | -1.6 | Zinc finger protein 161 | ZNF161 |
| AA252169 | -1.5 | -1.7 | Zinc finger protein 193 | ZNF193 |
| Cell growth | ||||
| AA262988 | -1.4 | -1.6 | Brain-derived neurotrophic factor | BDNF |
| AA480880 | -1.6 | -1.7 | Butyrate response factor 2 (EGF response factor 2) | BRF2 |
| AA458507 | -1.6 | -1.6 | Colony-stimulating factor 3 receptor (granulocyte) | CSF3R |
| AA443093 | -1.3 | -1.6 | Fbroblast growth factor receptor 2 | FGFR2 |
| AA001614 | -1.6 | -1.5 | Insulin receptor | INSR |
| N54596 | -1.7 | -1.7 | Insulin-like growth factor 2 (somatomedin A) | IGF2 |
| T62547 | -1.6 | -1.5 | Insulin-like growth factor 2 receptor | IGF2R |
| AA487034 | -1.5 | -1.5 | Transforming growth factor, beta receptor II (70-80 kDa) | TGFBR2 |
| Mitochondrial function | ||||
| W44701 | -1.7 | -1.8 | Solute carrier family 25 (mitochondrial carrier) | SLC25A6 |
| AA411554 | -1.8 | -1.8 | Solute carrier family 25 (mitochondrial carrier) | SLC25A16 |
| Cancer-related | ||||
| AA486403 | 1.7 | 2.1 | N-myc downstream-regulated | NDRG1 |
| AA447730 | 1.6 | 1.3 | Plm-1 oncogene | PIM1 |
| W72473 | 1.5 | 1.1 | Phosphoinositide 3-kinase, catalytic, alpha polypeptide | PIK3CA |
| H09066 | -1.6 | -1.7 | BRCA1-associated protein 1 (ubiquitin carboxy-terminal hydrolase) | BAP1 |
| AA425947 | -1.5 | -1.8 | Dickkopf (Xenopus laevis) homolog 3 | DKK3 |
| AA489246 | -1.9 | -2.0 | Suppression of tumorigenicity 14 (colon carcinoma, matriptase, epithin) | ST14 |
| AA261635 | -1.6 | -1.5 | Suppression of tumorigenicity 16 | ST16 |
| Stress response, DNA repair, and cell death/apoptosis | ||||
| AA598874 | 1.4 | 1.7 | Antioxidant protein 2 (non-selenium glutathione peroxidase, acidic calcium-independent phospholipase A2) | KIAA0106 |
| H11660 | 1.8 | 1.9 | p53-induced protein | PIG11 |
| AA453410 | 1.4 | 1.6 | Tumor necrosis factor receptor superfamily, member 10b | TNFRSF10B |
| AA459263 | -1.9 | -1.8 | BCL2-related protein A1 | BCL2A1 |
| AA427906 | -1.5 | -1.5 | Beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) | BECN1 |
| AA455281 | -1.5 | -1.6 | Defender against cell death 1 | DAD1 |
| AA281945 | -1.7 | -1.5 | Mitogen-activated protein kinase-activating death domain | MADD |
| AA293365 | -1.3 | -1.5 | Mitogen-activated protein kinase kinase 4 | MAP2K4 |
| R85257 | -1.8 | -1.9 | Protein tyrosine kinase 2 beta | PTK2B |
| AA496782 | -1.5 | -1.5 | Requiem, apoptosis response zinc finger gene | REQ |
| AA446223 | -1.9 | -1.7 | TGFB1-induced antiapoptotic factor 1 | TIAF1 |
| Cell cycle | ||||
| AA598974 | 1.7 | 1.6 | Cell division cycle 2, G1 to S and G2 to M | CDC2 |
| AA598776 | -1.8 | -1.7 | CDC20 (cell division cycle 20, S. cerevisiae, homolog) | CDC20 |
| T81764 | -1.5 | -1.6 | Cell division cycle 27 | CDC27 |
| Signal transduction | ||||
| AA432271 | 1.9 | 2.1 | Protein kinase, AMP-activated, beta 1 noncatalytic subunit | PRKAB1 |
| N78582 | 1.6 | 1.8 | Protein kinase, AMP-activated, beta 2 noncatalytic subunit | PRKAB2 |
| AA485366 | 1.2 | 1.5 | Protein kinase, cAMP-dependent, regulatory, type I, beta | PRKAR1B |
| AA125779 | -1.5 | -1.5 | Adenylate cyclase 7 | ADCY7 |
| H29256 | -1.6 | -1.6 | Calcium channel, voltage-dependent, L type, alpha 1D subunit | CACNA1D |
| AA456830 | -1.9 | -1.9 | Diacylglycerol kinase, alpha (80 kDa) | DGKA |
| AA450003 | -1.9 | -1.7 | Dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 4 | DYRK4 |
| AA521346 | -1.7 | -1.6 | Serine threonine protein kinase | NDR |
| Immune-related | ||||
| AA411324 | -1.4 | -1.6 | Interleukin 13 receptor, alpha 1 | IL13RA1 |
| AA057204 | -1.5 | -1.4 | Interleukin 2 receptor, beta | IL2RB |
| Protein metabolism | ||||
| AA450227 | -1.6 | -1.5 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | PSMD4 |
| AA598815 | -1.5 | -1.5 | Proteasome (prosome, macropain) subunit, alpha type, 5 | PSMA5 |
| T57841 | -1.6 | -1.7 | Ubiquitin fusion degradation 1-like | UFD1L |
| AA490124 | -1.8 | -1.7 | Ubiquitin-conjugating enzyme E2N (homologous to yeast UBC13) | UBE2N |
| AA488626 | -1.8 | -1.9 | Ubiquitin-like 1 (sentrin) | UBL1 |
Shown is a representative selection of the 435 genes differentially expressed when either WS or normal old cells were compared with normal young cells, including the 249 to which function was annotated.
Fold difference in gene expression relative to normal young.
Fig. 1.
(A) Replicate hybridizations. For each group of cell lines, two vials of RNA were isolated from identical cultures grown in parallel. Two independent probe preparations and hybridizations were made per pooled RNA sample, resulting in a total of four replicates. (B) Scatter plots of intra-group vs. inter-group variation. Based on three three-way comparisons, the average intra-group CV was 0.13 vs. an average inter-group CV of 0.28 based on 27 three-way comparisons. (C) Correlation between microarray data and RT-PCR. (D) Progression of gene expression analysis. Of the analyzed genes, 6.3% were differentially expressed in WS or aging. Of these, the genes with known function were categorized as being affected in WS only, aging only, or both.
Experimental Design and Reproducibility Statistics. The goal of this study was to determine gene expression changes in WS and old age. However, sources of gene expression changes that can complicate interpretation include biological and experimental variability, as well as genetic background differences between individuals. To minimize the effect of these variables, we used the following experimental strategy (Fig. 1 A), based on previously published designs (14–16). First, to verify that the growth conditions used in this study resulted in reproducible gene expression measurements, cells from each donor were grown up twice and RNA was isolated separately (biological replicates). The average coefficient of variation (CV = SD/mean) between biological replicates was 0.11. Variation due to technical issues was assessed by generating two independent probe preparations from each of these two RNA samples and hybridizing them to separate arrays, resulting in a CV of 0.12 (array replicates). Summarily, each age group was analyzed by four replicate microarray hybridizations, and, comparing all four replicates, the average CV was 0.16, demonstrating very high reproducibility. In a comparable study, the average CV was 0.21 after data filtering that removed most of the genes with high CVs (17). Array scans were inspected visually to confirm calculated ratios (Fig. 1 A). CV calculations for all genes are in Table 3.
Second, we designed the experiment to minimize bias due to individual polymorphism unrelated to the disease under study, thereby isolating changes specific to WS rather than individual patients. Age groups and WS patients were represented by an average of five cell lines, and RNA samples were pooled within each group before microarray hybridization. Preprofile mixing of RNA is a tested strategy (13, 16, 18), and recent data (14, 15) suggest that stringent yet robust data can be generated by mixing a small number of individuals with a defined condition (n = 5). These results confirm that preprofile mixing of RNA can effectively normalize both intra- and inter-patient sources of variation and that sample mixing results in relatively high sensitivity and specificity for gene expression changes that would be detected by many individual expression profiles. Pooling the RNA presupposes that variation within the group is smaller than variation between groups. To evaluate the validity of preprofile mixing of RNA in this study, in a preliminary experiment, RNA was isolated separately from three individual donors in each group and used to determine intra-group variability. Based on three three-way comparisons, the average intra-group CV was 0.13 vs. an average inter-group CV of 0.28 based on 27 three-way comparisons (Fig. 1B and Table 4). Grouping WS patients for gene expression analysis is consistent with the fact that different mutations found in WS all prevent nuclear localization of the protein that in turn loses its function (2). Thus, the biochemical WRN null phenotypes are expected to be identical between different WS patients. To further assess the effects of preprofile mixing of RNA, for each group in our study, we evaluated the difference between the pooled gene expression values and the median of three individually analyzed cell lines. The data are shown in Table 5, which also includes scatter plot representations of the correlation between pooled and individual gene expression. The average CVs were relatively low, indicating a good correlation: young, 0.32 (six pooled cell lines vs. median of three individual); old, 0.31 (five pooled cell lines vs. median of three individual); WS, 0.18 (four pooled cell lines vs. median of three individual). The CVs were highest for the young group, where only three of six donors were analyzed individually, supporting the hypothesis that multiple donors are needed to avoid bias from differences in genetic background.
This study was designed in cooperation with the Statistics and Experimental Design Section (SEDS) and the DAU, both part of the Research Resources Branch at the National Institute on Aging, National Institutes of Health. The SEDS and DAU reviewed study design and analysis protocols to ensure that they were appropriate for the anticipated comparisons and statistical procedures.
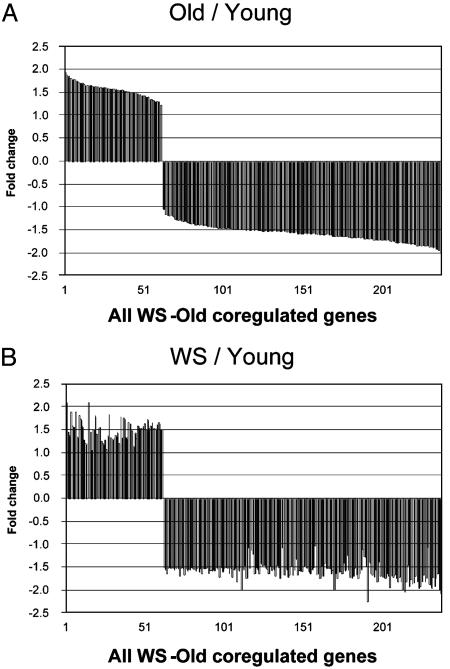
Gene Expression Analysis. We characterized gene expression patterns associated with WS and normal human aging by analyzing 6,912 RNA pol II transcribed genes across three groups of cell lines. When gene expression in normal old cells and WS cells was compared with the baseline gene expression of normal young cells, 6.3% (435) of the genes displayed >1.5-fold difference (Fig. 1D). Down-regulation (70%) dominated over upregulation (30%). Using the LocusLink database (www.ncbi.nlm.nih.gov/LocusLink//index.html), we annotated function to 249 of the 435 genes that had known function. A major result of our study is the finding that 91% of the genes with known function displayed similar expression changes (difference <0.5-fold) in WS and normal aging (Fig. 2). For these coregulated genes, the correlation coefficient between WS and old is 0.99, demonstrating an impressively similar regulation between WS and old on the level of individual genes. Further analysis was focused on genes with known function, and a representative selection of the coregulated genes is shown in Table 2 (all coregulated genes are in Table 6, part A). Three percent were differentially expressed in WS only (Table 6, part B) and 6% were differentially expressed in normal old only (Table 6, part C). A full list of all 435 genes and ESTs is shown in Table 7; the complete list of expression ratios for the 6,912 genes assayed is shown in Table 8.
Fig. 2.
Visual representation of gene expression changes associated with WS and normal aging. Shown are column charts illustrating the similar transcription profiles of old (A) and WS (B) cells when compared with young cells. Genes are plotted in the same order in A and B and are listed after expression level in old relative to young. All genes in this figure are listed in Table 6, and select examples are listed in Table 2.
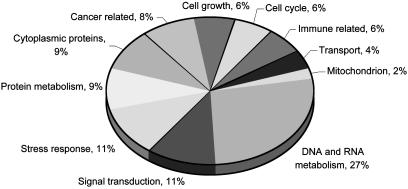
Gene Expression Changes Common to WS and Normal Aging. Functional categorization of the coregulated genes enabled us to speculate on the pathways affected in premature and normal aging. Fig. 3 gives the percentage of genes in each functional group relative to all genes coregulated in WS and normal old. DNA/RNA metabolism and chromosomal recombination. The largest functional group consisted of genes related to DNA and RNA processing. Seventy-five percent of these were down-regulated. The gene displaying the largest decrease in expression was RNA pol II polypeptide A (POLR2A). Another RNA pol II subunit, polypeptide J (POLR2J), was also down-regulated. The RNA pol II complex transcribes protein-encoding genes and interacts with the promoter regions as well as with a variety of transcription factors, thereby controlling a wide range of cellular processes. We have shown that WS cells are deficient in pol II transcription (3). In addition to RNA pol II itself, a number of RNA pol II transcription factors were less expressed with age. Transcription factor Dp-2 (TFDP2) is a component of the drtf1/e2f transcription factor complex and regulates genes encoding proteins required for the progression of S-phase during the cell cycle. Reduced expression of the FOXM1 (HFH-11) transcription factor gene corresponded to previous findings in elderly patients and patients with Hutchinson–Gilford progeria (19). The down-regulated SMARCA1 and SMARCB1 products are part of a complex that opens the chromatin to facilitate the transcriptional machinery to access their targets (20).
Fig. 3.
Functional categories of gene expression changes associated with WS and normal aging. Percentage is given relative to all WS-old coregulated genes assigned to functional categories.
Cell growth, cell cycle, and oncogenes. The growth-deficient phenotype of WS cells correlates with the repressed expression of 12 genes related to cell growth (Table 3). Diminished capability in responding to growth factors was indicated by the reduced expression of BRF2, encoding a putative nuclear transcription factor regulating the response to growth factors. Gene down-regulation included CSF3R, which encodes a receptor that transduces signals regulating the proliferation, differentiation, and survival of myeloid cells. Reduced expression of the insulin receptor gene INSR, insulin-like growth factor 2 gene (IGF2), and the IGF2 receptor (IGF2R) may provide insights into the increased incidence of diabetes mellitus in WS.
Based on the reduced replicative potential of WS cells and cells from normal old donors, we expected to see differential regulation of genes associated with cell cycle progression. Of the 11 genes related to cell cycle regulation, 7 were down-regulated (Table 6). Our findings support previous observations that mitotic misregulation is likely to play a role in the aging process (19). Correlating with the increased occurrence of cancers in old age and in WS, we found up-regulation of oncogenes (NDRG1, PIM1, RAB11A, and PIK3CA) and down-regulation of tumor suppressor genes (ST14, ST16, DKK3, and BAP1).
Stress response, DNA repair, and apoptosis. Transcriptional regulation of genes involved in apoptosis was ambiguous, possibly indicating a preference for specific apoptotic pathways in WS and normal aging. Four negative regulators of apoptosis (TIAF1, DAD1, BCL2A1, and BECN1) were down-regulated in both WS and normal aging, suggesting an increased susceptibility to apoptosis with age. This finding is supported by increased expression of TNFRSF10B that encodes a tumor necrosis factor (TNF) receptor. Expression was decreased for UBL1, which encodes a Rad51/Rad52-interacting protein (21) that functions in recombinational repair and protects against anti-Fas/APO-1 and TNF-induced cell death. At the same time, two proapoptotic genes are down-regulated, REQ encodes a protein that functions as a transcription factor required for the induction of apoptosis, and mitogen-activated protein kinase-activating death domain is a propagator of the apoptotic signal. MAP2K4, also down-regulated, is found to be consistently inactivated in many types of tumors (22). Decreased expression of PTK2B, whose product is involved in activation of the mitogen-activated protein kinase signaling pathway, supports an age-related mitogen-activated protein kinase pathway deficiency.
Gene Expression Changes Specific to WS or Normal Aging. Fourteen (6%) of the differentially expressed genes were specific to normal old, and seven genes (3%) were specific to WS (Table 6). The genes specific to WS or normal old did not offer any apparent evidence for the phenotypic differences between the premature aging in WS and normal aging. A possible explanation for this finding is that differences will appear only under certain conditions, e.g., after challenging the cells. Further studies are needed to address this question and build on the work presented here.
Discussion
This report compares global genome expression patterns in WS patients with normal human aging. We used cDNA microarrays to characterize expression of 6,912 genes and ESTs across a panel of 15 human skin fibroblast cell lines from young, old, and WS donors. Postadolescent young controls, rather than children, were used to separate age-related changes from developmental changes (23). Of the 6,912 genes assayed, 6.3% (435) displayed significant differences in expression, when cells from either WS patients or old donors were compared with cells from young donors. Among the 435 genes generated by the two comparisons, 91% of those with known function overlapped between the two, and their correlation coefficient was 0.99. This result suggests that not only the phenotypes but also the pathways involved in generating WS and aging are exceedingly similar. Our findings underscore the relevance of using WS as an aging model and represent a further step toward the desired goal of understanding the processes behind normal aging. Identifying genes whose expression changes by as little as 1.5-fold is particularly relevant because, in terms of normal physiology, age-associated gene expression changes are thought to be small (24). Current data indicate that almost all changes in gene expression that are age-related occur in the 30% to 3-fold range (reviewed in ref. 25). As a result of high reproducibility across replicate experiments, we were thus able to identify a large number of genes not previously known to be regulated during the aging process.
Gene expression changes associated with aging have been reported. A study by Ly et al. (19) using human cell lines compared genomic expression between children and elders. However, their setup did not discriminate between developmental changes through puberty and age-associated changes (23). Since then, a number of studies in non-human species (24, 26–29) have revealed complex aging-related expression patterns that thus far point to no single mechanism explaining the aging process. Welle et al. (30) found that only around one-third of age-related gene expression changes correlated between mice and humans, emphasizing the necessity of exploratory studies in human model systems. Existing information on gene expression in WS cells is sparse. In 1991, Murano et al. (31) published nine overexpressed extracellular matrix-related genes in one WS cell line. Protein expression in WS fibroblasts vs. normal was analyzed by Toda et al. (32), who found that 12 proteins were differentially expressed. Their threshold of detection was 3-fold, and it is reasonable to assume that many proteins are differentially expressed at a lower level. It has been suggested that, in addition to RNA pol II, RNA pol I activity is affected in WS cells (33). Shiratori et al. (34) recently proposed a role for WRN in rRNA unwinding and synthesis rather than mRNA synthesis. However, they also found that total RNA synthesis by WS fibroblasts was 43–90% of that in normal fibroblasts, an effect unlikely to be caused by defects in rRNA transcription alone. The microarrays in our study did not contain rRNA; thus, RNA pol I-dependent transcription was not addressed. Further work is needed to resolve the extent of change in RNA pol I and II activity in WS cells. In summary, little if anything is known regarding the role of WRN in the transcription of specific genes, and to our knowledge there have been no studies of global genome expression comparing WS and normal cells.
Gene expression patterns in senescent WS cells overlap with those in normal strains (35), and senescence seems to be a p53-dependent event mediated via pathways identical to those in normal cells (36). We saw no induction of p53 in either old or WS cells (Table 6), indicating that the gene profiles reflected early passage rather than cellular senescence. Also, the gene profile of replicative senescence (early passage <25 population doublings (PDL), late passage >60 PDL) has previously been shown by Park et al. (37) to be different from progeria or elderly donor, suggesting that the gene profiles presented here are specific to normal aging and WS.
We found a strikingly high degree of similarity between transcription patterns in normal aging and WS. It has been hypothesized that several WS phenotypes are secondary consequences of aberrant gene expression (6), and our results suggest that the same is the case for normal aging. The transcription process requires unwinding of the DNA duplex, and involvement of helicases in transcription is exemplified by the observation that one of the basal transcription factors, TFIIH, exhibits DNA unwinding activity (38). In addition to WS, six other segmental progeroid syndromes are caused by mutations in helicase genes, which implies contribution of aberrant helicases to phenotypes in normal aging (6). One of these is Cockayne syndrome (CS), in which we have demonstrated a transcription defect after hydrogen peroxide exposure (10). CS has been speculated to be a “transcription syndrome” (39), and the present work supports the hypothesis that altered transcription may also be central to the defect in WS. If aging pathways exist, these may involve helicases needed to unwind the DNA as well as proteins and regulatory factors required for transcription.
Genes in the DNA and RNA metabolism category accounted for 27% of shared transcription alterations (Fig. 3), indicating that aberrations in DNA and RNA pathways are closely associated with WS and aging. This result is supported by previous findings that, in addition to its role in transcription, WRN copurifies with a 17S DNA replication complex (40) and participates in numerous protein–protein interactions, mainly with proteins related to DNA and RNA metabolism, including DNA repair (reviewed in ref. 9). Although their biological relevance remains to be understood, protein–protein interactions are thought to contribute to the WS phenotype.
We propose a model in which WRN protein, by virtue of its helicase and transcription-activating activities, as well as its protein interactions, is directly or indirectly involved in the transcription of genes upstream in the network of “aging pathways.” Reduced expression of RNA pol II and associated transcription factors may directly affect the expression of many genes. Hyporecombination in WS could be related to suppressed expression of recombination proteins, such as UBL1, that interact with Rad51 and Rad52. Extended S-phase and reduced replicative potential in WS cells could be explained by the aberrant gene expression related to cell cycle progression. Growth-related genes seem to be uniformly down-regulated, correlating with the reduced growth potential of WS cells. Up-regulation of oncogenes and down-regulation of tumor suppressor genes correlate with cancer predisposition in WS patients and elders. Thus, defects in the transcription of the genes identified in this study could result in many of the complex clinical features of WS. The remarkable similarity between WS and normal aging suggests that WS causes the acceleration of a normal aging mechanism. On a broader level, our results support the use of WS as an aging model and imply that transcription alterations affecting specific pathways are also a major part of the normal aging process.
Supplementary Material
Acknowledgments
We thank members of the Laboratory of Molecular Gerontology for reading and comments. We thank Dr. Kevin Becker (National Institute on Aging) for biostatistical advice. This work was supported by the Danish Center for Molecular Gerontology, the Danish Medical Research Council (9902876), Aarhus Universitets Forskningsfond (E-2002SUN-3-11), and Lundbeckfonden (117/02).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: WS, Werner syndrome; WRN, WS protein; pol, polymerase; CV, coefficient of variation; DAU, DNA Array Unit.
References
- 1.Martin, G. M., Sprague, C. A. & Epstein, C. J. (1970) Lab. Invest. 23, 86–92. [PubMed] [Google Scholar]
- 2.Matsumoto, T., Shimamoto, A., Goto, M. & Furuichi, Y. (1997) Nat. Genet. 16, 335–336. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki, N., Shimamoto, A., Imamura, O., Kuromitsu, J., Kitao, S., Goto, M. & Furuichi, Y. (1997) Nucleic Acids Res. 25, 2973–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, S., Li, B., Gray, M. D., Oshima, J., Mian, I. S. & Campisi, J. (1998) Nat. Genet. 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opresko, P. L., Cheng, W. H., von Kobbe, C., Harrigan, J. A. & Bohr, V. A. (2003) Carcinogenesis 24, 791–802. [DOI] [PubMed] [Google Scholar]
- 6.Nakura, J., Ye, L., Morishima, A., Kohara, K. & Miki, T. (2000) Cell Mol. Life Sci. 57, 716–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balajee, A. S., Machwe, A., May, A., Gray, M. D., Oshima, J., Martin, G. M., Nehlin, J. O., Brosh, R., Orren, D. K. & Bohr, V. A. (1999) Mol. Biol. Cell 10, 2655–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye, L., Nakura, J., Morishima, A. & Miki, T. (1998) Exp. Gerontol. 33, 805–812. [DOI] [PubMed] [Google Scholar]
- 9.Fry, M (2002) SAGE KE, 10.1126/sageke.2002.13.re2.
- 10.Kyng, K. J., May, A., Brosh, R. M., Jr., Cheng, W. H., Chen, C., Becker, K. G. & Bohr, V. A. (2003) Oncogene 22, 1135–1149. [DOI] [PubMed] [Google Scholar]
- 11.Fan, J., Yang, X., Wang, W., Wood, W. H., III, Becker, K. G. & Gorospe, M. (2002) Proc. Natl. Acad. Sci. USA 99, 10611–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, R. A., Galecki, A. & Shmookler-Reis, R. J. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56, B52–B57. [DOI] [PubMed] [Google Scholar]
- 13.Lawrance, I. C., Fiocchi, C. & Chakravarti, S. (2001) Hum. Mol. Genet. 10, 445–456. [DOI] [PubMed] [Google Scholar]
- 14.Bakay, M., Chen, Y. W., Borup, R., Zhao, P., Nagaraju, K. & Hoffman, E. P. (2002) BMC Bioinformatics 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Y. W., Zhao, P., Borup, R. & Hoffman, E. P. (2000) J. Cell Biol. 151, 1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostermeier, G. C., Dix, D. J., Miller, D., Khatri, P. & Krawetz, S. A. (2002) Lancet 360, 772–777. [DOI] [PubMed] [Google Scholar]
- 17.Weindruch, R., Kayo, T., Lee, C. K. & Prolla, T. A. (2002) Mech. Ageing Dev. 123, 177–193. [DOI] [PubMed] [Google Scholar]
- 18.Hilsenbeck, S. G., Friedrichs, W. E., Schiff, R., O'Connell, P., Hansen, R. K., Osborne, C. K. & Fuqua, S. A. (1999) J. Natl. Cancer Inst. 91, 453–459. [DOI] [PubMed] [Google Scholar]
- 19.Ly, D. H., Lockhart, D. J., Lerner, R. A. & Schultz, P. G. (2000) Science 287, 2486–2492. [DOI] [PubMed] [Google Scholar]
- 20.Versteege, I., Sevenet, N., Lange, J., Rousseau-Merck, M. F., Ambros, P., Handgretinger, R., Aurias, A. & Delattre, O. (1998) Nature 394, 203–206. [DOI] [PubMed] [Google Scholar]
- 21.Shen, Z., Pardington-Purtymun, P. E., Comeaux, J. C., Moyzis, R. K. & Chen, D. J. (1996) Genomics 36, 271–279. [DOI] [PubMed] [Google Scholar]
- 22.Su, G. H., Song, J. J., Repasky, E. A., Schutte, M. & Kern, S. E. (2002) Hum. Mutat. 19, 81. [DOI] [PubMed] [Google Scholar]
- 23.Cristofalo, V. J. (2000) Nat. Med. 6, 507. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. K., Klopp, R. G., Weindruch, R. & Prolla, T. A. (1999) Science 285, 1390–1393. [DOI] [PubMed] [Google Scholar]
- 25.Han, E. & Hilsenbeck, S. G. (2001) Mech. Ageing Dev. 122, 999–1018. [DOI] [PubMed] [Google Scholar]
- 26.Kayo, T., Allison, D. B., Weindruch, R. & Prolla, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. K., Weindruch, R. & Prolla, T. A. (2000) Nat. Genet. 25, 294–297. [DOI] [PubMed] [Google Scholar]
- 28.Weindruch, R., Kayo, T., Lee, C. K. & Prolla, T. A. (2001) J. Nutr. 131, 918S–923S. [DOI] [PubMed] [Google Scholar]
- 29.Zou, S., Meadows, S., Sharp, L., Jan, L. Y. & Jan, Y. N. (2000) Proc. Natl. Acad. Sci. USA 97, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welle, S., Brooks, A. & Thornton, C. A. (2001) Physiol. Genomics 5, 67–73. [DOI] [PubMed] [Google Scholar]
- 31.Murano, S., Thweatt, R., Shmookler-Reis, R. J., Jones, R. A., Moerman, E. J. & Goldstein, S. (1991) Mol. Cell. Biol. 11, 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toda, T., Satoh, M., Sugimoto, M., Goto, M., Furuichi, Y. & Kimura, N. (1998) Mech. Ageing Dev. 100, 133–143. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. K., Johnson, R. E., Yu, S. L., Prakash, L. & Prakash, S. (1999) Science 286, 2339–2342. [DOI] [PubMed] [Google Scholar]
- 34.Shiratori, M., Suzuki, T., Itoh, C., Goto, M., Furuichi, Y. & Matsumoto, T. (2002) Oncogene 21, 2447–2454. [DOI] [PubMed] [Google Scholar]
- 35.Choi, D., Whittier, P. S., Oshima, J. & Funk, W. D. (2001) FASEB J. 15, 1014–1020. [DOI] [PubMed] [Google Scholar]
- 36.Davis, T., Singhrao, S. K., Wyllie, F. S., Haughton, M. F., Smith, P. J., Wiltshire, M., Wynford-Thomas, D., Jones, C. J., Faragher, R. G. & Kipling, D. (2003) J. Cell Sci. 116, 1349–1357. [DOI] [PubMed] [Google Scholar]
- 37.Park, W. Y., Hwang, C. I., Kang, M. J., Seo, J. Y., Chung, J. H., Kim, Y. S., Lee, J. H., Kim, H., Kim, K. A., Yoo, H. J., et al. (2001) Biochem. Biophys. Res. Commun. 282, 934–939. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffer, L., Roy, R., Humbert, S., Moncollin, V., Vermeulen, W., Hoeijmakers, J. H., Chambon, P. & Egly, J. M. (1993) Science 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 39.Bootsma, D. & Hoeijmakers, J. H. (1993) Nature 363, 114–115. [DOI] [PubMed] [Google Scholar]
- 40.Lebel, M., Spillare, E. A., Harris, C. C. & Leder, P. (1999) J. Biol. Chem. 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.