Abstract
During genome transcription in rotavirus, as with many segmented double-stranded RNA viruses, mRNA is transcribed within the intact subviral particle and translocated through specific channels in the capsid. To understand how the conformation of the capsid affects the efficiency of transcriptional events in the viral core, we carried out a series of comparative structural and biochemical studies to characterize four different structural forms of the virus exhibiting differing transcriptional behavior. Two of these were virus-antibody complexes having contrasting transcriptional capabilities, and two were variant structural forms of the virus that exist during the life cycle and also exhibit contrasting transcriptional behavior. Three-dimensional structural studies using electron cryomicroscopy showed that the binding of one Fab (8H2/G5) does not affect the conformation of the capsid, and the efficiency of mRNA production is similar to that of the native subviral particle. The other Fab (2A11/E9) introduces conformational changes in the capsid similar to those seen in the transcriptionally incompetent mature particle. In both of the transcriptionally incompetent particle types, mRNA synthesis was arrested after limited elongation with the resulting oligonucleotide transcripts remaining trapped inside the particles. Our results indicate that the continuous translocation of nascent mRNA through the capsid is critical for efficient transcript elongation and that the blockage of translocation causes premature termination of transcription.
Keywords: electron cryomicroscopy, virus structure, double-stranded RNA virus
After cell entry, a virus must present its genetic information to host cell ribosomes in a translatable form so that the virally encoded proteins needed for genome replication and the assembly of progeny virions may be synthesized. Viruses accomplish this critical step in a variety of different ways. Viruses in the family Reoviridae synthesize mRNA transcripts within structurally intact subviral particles by using a virally encoded endogenous transcriptase (1). Rotaviruses, which are prototypical members of this family, represent a useful system in which to investigate the mechanism of endogenous genome transcription in segmented double-stranded RNA (dsRNA) viruses.
Rotaviruses are large, complex icosahedral particles consisting of three concentric capsid protein layers enclosing a genome of 11 segments of dsRNA (reviewed in ref. 2). Rotaviruses attach to their host cells as triple-layered particles (TLPs) (2). Then, as a consequence of cell entry, the outermost capsid layer is lost, and the resulting double-layered particles (DLPs) become transcriptionally active. During this process, mRNA transcripts corresponding to the 11 dsRNA genome segments are synthesized by the concerted action of multiple enzymes within the viral core and then translocated 140 Å through a system of channels, penetrating the two capsid layers at the icosahedral 5-fold axes (3). Although the mRNA release channels have been identified, it is not understood how the architecture of the capsid places constraints upon the passage of mRNA through these channels nor how the efficiency of mRNA translocation in turn affects upstream events in the transcription process, i.e., transcript initiation and elongation.
To investigate the relationship between transcriptional events in the viral core and the efficiency of translocation through the capsid, we carried out a series of structural and biochemical studies to characterize two different virus-antibody complexes having contrasting transcriptional capabilities. A comparison of the transcriptional behavior of each DLP/Fab complex with the mode of Fab binding observed in the corresponding structure provides insight into the architectural requirements for efficient mRNA synthesis. Previous structural studies have used antibodies to characterize the functional domains of viral proteins involved in cell entry and neutralization (4–8). We present here a plausible mechanism by which antibodies can affect the structure of the viral capsid and prevent endogenous genome transcription. Our results also provide insight into the mechanism of transcriptional inhibition in the mature TLP.
METHODS
Preparation of Virus and Fabs.
DLPs from simian rotavirus strain SA11–4F were prepared as described (9). IgGs were purified from ascites (generous gift of R. L. Ward, Children’s Hospital Medical Center, Cincinnati, OH; ref. 10) by using a caprylic acid precipitation technique (11). Fab fragments were prepared by using a Pierce ImmunoPure Fab Preparation Kit, and the purity was confirmed by using SDS/PAGE analysis followed by silver staining.
In Vitro Transcription.
Virus/Fab complexes were prepared by incubating 0.2 μg of purified DLPs with 13.4 μg of Fab overnight at 4°C. At this stoichiometric ratio, the quantity of Fabs present in the reaction exceeds the number of VP6 epitopes, ensuring complete saturation of all available binding sites on the DLP. Virus/Fab complexes then were added to a standard rotavirus transcriptase reaction buffer (10) and incubated for 90 min at 37°C. Accumulation of radiolabeled mRNA was measured by using a trichloroacetic acid precipitation assay (12).
Electron Cryomicroscopy.
DLP/Fab complexes were prepared as above and concentrated by using a MicroCon-100 microconcentrator. Specimens for microscopy were embedded in vitreous ice on holey carbon films by using standard procedures (13). DLP/Fab complexes imaged under transcription conditions were treated as described (3). Images were collected on a JEOL 1200EX electron cryomicroscope operated at ×30,000 magnification by using 100 keV electrons and a beam dosage of ≈5 electrons/Å2.
Three-Dimensional (3D) Structural Analysis.
Micrographs chosen for 3D structural analysis had a defocus value of ≈1.0 μm (≈2.2 μm for structures determined under transcription conditions) as estimated from the positions of the contrast transfer function (CTF) rings seen in the sum of individual particle image Fourier transforms (14). Micrographs were digitized by using a scanning interval corresponding to 5.33 Å in the object. 3D reconstructions were produced by using the software in the icos Toolkit suite (15) by using standard procedures (16–19). Reconstructions under nontranscribing conditions were computed to ≈21-Å resolution, and those under transcribing conditions to ≈30-Å resolution. CTF corrections were performed as described (14). For each reconstruction, the resolution was estimated by using Fourier ring correlation analysis (20), and 95% or more of the mean inverse eigenvalues (18) were less than 0.01, indicating that the orientation distribution and number of particles (ranging between 119 and 261) were sufficient. Surface representations of the reconstructions were displayed by using a contour level chosen to account for the expected number of VP6 molecules within the VP6 capsid region. The atomic coordinates of a Fab fragment from murine IgG2b (21) were modeled into an Fab region isolated from the vicinity of the 5-fold axis in each of the two structures. The atomic coordinates of the Fab were represented by using ribbons 2.85 (22) and then imported into explorer 3.5 for manual docking.
PAGE Analysis of Transcription Products.
DLPs, 2A11/E9 DLP/Fab complexes, or TLPs (10 μg per reaction) were suspended in 20 μl of transcription reaction buffer containing 100 mM Tris⋅HCl (pH 8), 10 mM MgOAc, 0.5 mM S-adenosyl methionine, 1 mM CaCl2, 1 mM ATP, 100 μM UTP and CTP, 99 μM GTP, and 60 μCi (1 μM) [α-32P]UTP (3,000 Ci/mmol, Amersham Pharmacia) and then incubated at ≈37°C for 30 min. Each reaction mixture then was adjusted to 30 μl with 10 mM CaCl2 and passed through a Micro Bio Spin P-30 spin column (Bio-Rad) to remove the majority of the unincorporated radionucleotides. The eluate was lyophilized and resuspended in buffer containing 7 M urea and 0.1% SDS. Samples were boiled for 5 min to disrupt the particles and denature the nucleic acid. Reaction products were electrophoresed on a 20% denaturing polyacrylamide gel and autoradiographed (23). RNA and DNA size markers were prepared by using the KinaseMax 5′ end labeling kit (Ambion) to radiolabel the internal standards provided with the kit.
RESULTS
Transcriptional Activity of DLP/Fab Complexes.
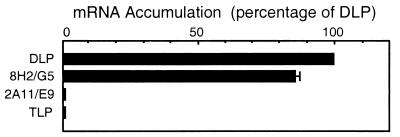
In our biochemical and structural studies, we made use of two Fab fragments, designated as 8H2/G5 and 2A11/E9, which were derived from IgG2b mAbs directed against the outer capsid protein VP6 in the DLP (10). In our studies, we chose to use monovalent Fab fragments instead of bivalent IgGs, as the ability of the intact antibody to crosslink its target molecules into large aggregates can complicate the interpretation of both biochemical and structural results. To determine the effect of these Fabs on the efficiency of genome transcription, we tested their ability to inhibit mRNA synthesis in rotavirus by using a standard in vitro transcription assay (Fig. 1). The antibody concentration was chosen to ensure that the number of Fab fragments was in excess of the number of VP6 epitopes (780 per particle) present in the reaction. Both the native DLP and the 8H2/G5 DLP/Fab complex were able to transcribe with essentially equal efficiency, whereas mRNA production nearly completely inhibited the 2A11/E9 DLP/Fab complex as well as the mature TLP, as has been reported (2).
Figure 1.
Comparison of the transcriptional efficiency among the native DLP, 8H2/G5 DLP/Fab complex, and the 2A11/E9 DLP/Fab complex. Each histogram represents the total mRNA yield, given as a percentage of that for the DLP. The transcriptional behavior of mature TLPs under similar conditions is shown for comparison purposes. Error bars indicate the distribution of multiple measurements when the range was greater than 1%.
Electron Cryomicroscopy of DLP/Fab Complexes.
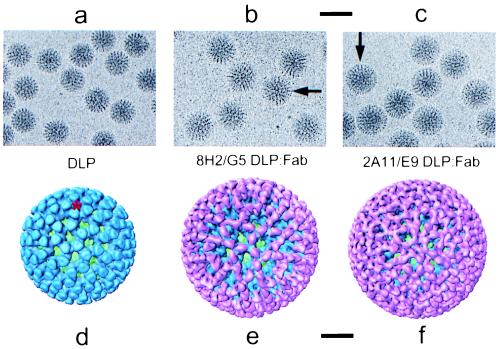
To visualize the complexes formed when these Fabs are bound to the DLP, the DLP/Fab complexes were imaged by using electron cryomicroscopy (Fig. 2 a-c). When compared with the native DLP (Fig. 2a), the DLP/Fab complexes (Fig. 2 b and c) each have a slightly greater diameter and a distinctly different appearance, suggesting that the Fabs are attached to epitopes on the exterior surface of VP6 and that each Fab has a different mode of binding.
Figure 2.
(a–c) Electron cryomicrograph images of native DLPs, 8H2/G5 DLP/Fab complexes, and 2A11/E9 DLP/Fab complexes. These images were recorded at a defocus of −2.2 μm. (Scale bar, 1,000 Å.) The binding of the Fabs form what appears to be an additional capsid layer on the DLP, visible in the images as a ring of mass density extending outward from a radius of 350 Å (arrows in b and c). (d–f) Surface representations of the 3D structures of the native DLP, 8H2/G5 DLP/Fab complex, and 2A11/E9 DLP/Fab complex, viewed along the icosahedral 3-fold axis. The inner capsid protein VP2 is shown in green, outer capsid protein VP6 in blue, and bound Fabs in pink. This color scheme is used in all figures. In the native DLP, the 780 molecules of VP6 are assembled as trimers organized on a T = 13 (levo) icosahedral lattice (9). One of the channels through which mRNA is released during transcription (3) is indicated with a red star. These channels lie along the icosahedral 5-fold axes. (Scale bar, 200 Å.)
3D Structural Analysis of DLP/Fab Complexes.
To visualize the interactions between these two Fabs and their epitopes on the VP6 surface, we determined the 3D structure of each DLP/Fab complex to a resolution of ≈21 Å from the corresponding electron micrographs (Fig. 2 d-f). In the 8H2/G5 DLP/Fab complex (Fig. 2e), the Fabs appear to bind in a vertical orientation with respect to the surface of the VP6 trimer. The top of the trimer approximates a triangular shape, and these epitopes are located along the edges of this triangle (see Fig. 3a). Because of the quasi-equivalent nature of the VP6 monomers in this T = 13 icosahedral lattice, the 260 trimers making up the capsid layer are not all spaced at equal distances from their neighbors. The consequence of this variable spacing is that some trimers are capable of binding more Fab molecules than others. At full saturation, a total of 510 of 780 epitopes are occupied by an 8H2/G5 Fab.
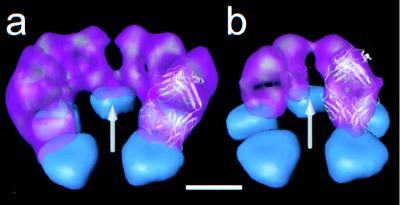
Figure 3.
Comparison of the orientation of Fab binding (pink) to VP6 (blue) around the mRNA release channel. (a) For 8H2/G5 Fab, eight of the 10 Fabs surrounding the channel are shown. (b) For 2A11/E9 Fab, four of the five interacting Fabs are shown. For reference, the uppermost 20 Å of the five VP6 trimers surrounding the channel in the native DLP are shown, and the icosahedral 5-fold axis passing through the center of the channel is indicated by an arrow (gray). The atomic structure of an Fab fragment from murine IgG2b (21) has been fitted into the molecular envelope (semitransparent) of one of the Fabs adjacent to the mRNA release channel to indicate that the Fabs have the expected morphology. (Scale bar, 50 Å.)
In the 2A11/E9 DLP/Fab complex (Fig. 2f), the Fabs interact with their epitopes in a 1:1 stoichiometry. These epitopes appear to be located near the vertices of the triangular-shaped distal portion of the VP6 trimer (see Fig. 3b). In contrast to 8H2/G5, these Fabs bind at a slight angle with respect to the surface of the trimer, forming a cage-like configuration that significantly narrows the opening of each channel in the VP6 capsid. Both the location of the epitope on the VP6 trimer and the geometry of the interaction between the Fab and VP6 surface are such that each trimer is able to accommodate the binding of three Fab molecules.
The Fabs in both DLP/Fab complexes have the expected morphology, as indicated by the placement of the atomic structure of an Fab molecule (21) within the Fab molecular envelopes computationally isolated from the two structures (Fig. 3). The slight merging of mass density seen among some adjacent Fab molecules is likely the result of antibody flexibility or perhaps is caused by the close proximity of binding as it would appear in a structure at this resolution.
Comparison of the Capsid Region in the Transcriptionally Competent and Incompetent Structures.
The attachment of 8H2/G5 Fab to one site on the capsid surface has very little effect on the efficiency of genome transcription, whereas the attachment of either 2A11/E9 Fab or VP7 to nearby locations on VP6 correlates with a nearly complete inhibition of mRNA production. To investigate the structural basis for these effects of ligand attachment, we compared the protein regions surrounding the mRNA release channels in the two transcriptionally competent structures with the corresponding regions in the two incompetent structures.
The mRNA release channels are formed by five trimers of VP6 surrounding the icosahedral 5-fold axes (Fig. 4a). In the transcriptionally competent 8H2/G5 DLP/Fab complex, the Fabs lining the channel opening are attached to the top of VP6 in an orientation that tilts the main axis of each antibody away from the icosahedral 5-fold axis (Fig. 3a). The 3D structure of the 8H2/G5 DLP/Fab complex under transcription conditions shows a narrow plug of mass density corresponding to nascent mRNA in the center of the mRNA release channel (data not shown), as was previously seen in the structure of the actively transcribing native DLP (3).
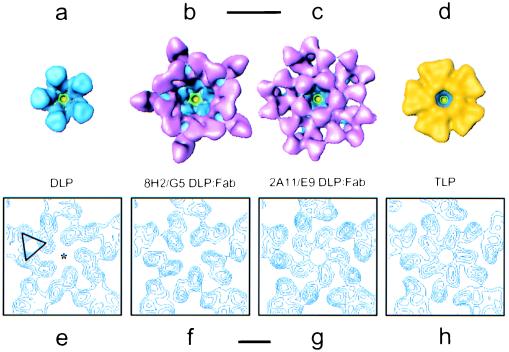
Figure 4.
(a–d) Comparison of the mRNA release channel architecture in the native DLP, 8H2/G5 DLP/Fab complex, 2A11/E9 DLP/Fab complex, and mature TLP, illustrating the degree to which additional protein on the VP6 surface affects the dimensions of the channel opening. Protein is colored according to the same scheme used in Fig. 2, except that VP7 in the TLP is indicated in yellow. (Scale bar, 100 Å.) (e–h) Representative contoured cross-sections perpendicular to the 5-fold axis (star in e), illustrating subtle conformational changes occurring in the VP6 trimers lining the channel at a particle radius of ≈293 Å. These maps have been represented in terms of variance and scaled to have the same minima and maxima, and each contour represents an interval of ≈0.37 σ. One of the five trimers in the DLP (e) is marked with a triangle. (Scale bar, 50 Å.)
In the transcriptionally incompetent 2A11/E9 DLP/Fab complex, three Fabs are bound to each trimer surrounding the channel (Fig. 4c). The five Fabs facing the channel are bound such that the main axis of the antibody is oriented toward the icosahedral 5-fold axis, narrowing the diameter of the channel to ≈35 Å in the vicinity of the elbow region of the Fab (Fig. 3b). As expected, the structure of the complex under transcription conditions does not show any structural evidence of mRNA translocation through the channels (data not shown). The absence of visible mRNA in the structure under transcription conditions does not indicate that transcript initiation or even limited elongation is completely blocked but rather that uninterrupted translocation, as is needed to visualize mRNA clearly in the 3D structure, does not occur. The mRNA release channel also is partially closed in the mature TLP, where each trimer of VP6 is bound by three molecules of VP7 that extend partially over the mouth, also reducing the diameter to ≈35 Å (Fig. 4d).
Within the upper portion of the VP6 region, beyond the particle radius ≈310 Å, the trimers are well separated from one another and the monomers are observed to form only intra-trimer interactions. At the present resolution, the distribution of mass appears to be quite similar throughout this region in all four of the structures. Toward the base of the mRNA release channel, the VP6 trimers form a network of trimer–trimer interactions extending between the radii ≈275 Å and ≈310 Å. Throughout this region, the trimers in the two transcriptionally incompetent structures appear to have a slightly different conformation from those in the two transcriptionally competent structures. One indicator of these structural rearrangements is that the mRNA release channel appears to undergo a subtle change in topology. The changes in this region are illustrated in representative contoured cross-sections of the channels corresponding to a radius of ≈293 Å, where changes in the structure of VP6 are most evident (Fig. 4 e–h). In the two transcriptionally competent structures, the channel is comparatively wider in this region, whereas in the two transcriptionally incompetent structures, there is a slight narrowing of the channel by ≈8 Å. No other significant structural differences are evident in the VP6 layer below the region of inter-trimer interactions or in the VP2 layer surrounding the mRNA release channels at the present resolution. In all likelihood, the structural rearrangements seen in the region of trimer–trimer interactions near the base of the VP6 capsid result from small conformational changes, which originate at the point of ligand attachment and then are transmitted inside; however, because of the uncertainty with which the ligand-VP6 boundary can be delineated at the present resolution, conformational changes in the upper region of VP6 are difficult to detect.
The structural changes observed in the VP6 capsid region are small and must be interpreted with caution, as such variations can easily arise from differences in imaging conditions and the choice of contour levels. The contour levels used with these reconstructions were chosen in each case to account for the expected 780 molecules of VP6 within the VP6 capsid region. Because of the consistent trend that is observed, with the two transcriptionally competent structures exhibiting one conformation of VP6 and the two incompetent structures exhibiting an alternate conformation, it is unlikely that these differences are simply the result of variations in imaging conditions.
Analysis of Transcription Products in Transcriptionally Incompetent Particles.
To determine whether the binding of 2A11/E9 Fab or VP7 to the particle interrupts the transcription process at the stage of initiation or during a downstream event, particles were incubated with the necessary precursors for mRNA synthesis, and [α-32P]GTP was included in the reaction mixture to label any polymerized products. After disruption of the particles, the products of the reaction were resolved on a denaturing 20% polyacrylamide gel (23). An autoradiograph produced from this gel indicates that initiation and limited elongation occurred in both the 2A11/E9 DLP/Fab complex and mature TLP, although no full-length transcripts could be detected (Fig. 5).
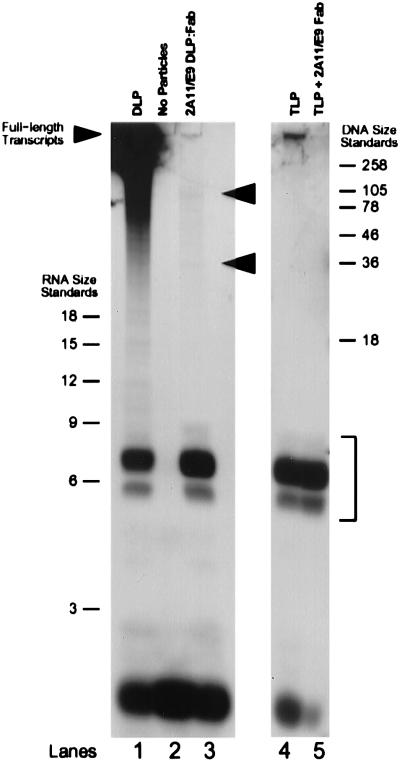
Figure 5.
Denaturing PAGE analysis of transcription products in the two transcriptionally incompetent particle types. The position of the short oligonucleotides between five and seven bases in length is indicated with a bracket. Arrowheads (lane 3) indicate the positions of several higher molecular weight RNA species produced in the 2A11/E9 DLP/Fab complex. The band at the bottom of the gel in each lane consists of residual unincorporated radiolabeled GTP not completely removed from the reaction mixture during spin column centrifugation. The intensity of this band is not precisely the same in each lane because of slight differences in the efficiency with which the spin column removed unincorporated radionucleotides after transcription.
In the native DLP reaction (Fig. 5, lane 1), the transcription products ranged in size from full-length mRNAs to shorter transcripts in the process of being elongated. The reaction products included a number of oligonucleotides that appeared to range between five and seven bases in length. In the 2A11/E9 DLP/Fab complexes (Fig. 5, lane 3), nearly all of the reaction products were between 5 and 7 nt in length, although a small number of transcripts were observed to be as long as ≈90 nt. The length distribution of these reaction products was identical after incubation of the reaction for 30 min, 2 hr, and 4 hr (data not shown), suggesting that transcriptional inhibition is not affected by transient dissociation and reassociation of Fab fragments. In the TLP reaction (Fig. 5, lane 4), the reaction products also consisted almost entirely of short oligonucleotides 5–7 bases long. There was a small quantity of high molecular weight transcripts produced as well, but these were absent when 2A11/E9 Fabs were included in the reaction mixture at 50 μg/ml (Fig. 5, lane 5), indicating that these longer transcripts most likely were produced by a small number of contaminating DLPs also present in the TLP reaction mixture.
DISCUSSION
Endogenous genome transcription is an important event in the life cycle of segmented dsRNA viruses. In rotaviruses, transcription occurs within structurally intact subviral double-layered particles, the structural form of the virus, which results from the removal of the outer capsid layer at cell entry. Nascent mRNA is observed to exit the particle through channels penetrating the VP6 capsid layer at the icosahedral vertices, although the precise pathway through the VP2 layer is not known. To gain insight into the molecular mechanisms of genome transcription, we carried out comparative biochemical and structural analyses of four particle forms exhibiting differing transcriptional capabilities. Both the native DLP and the 8H2/G5 DLP/Fab complex were able to produce mRNA transcripts with essentially equal efficiency, whereas the 2A11/E9 DLP/Fab complex and the mature TLP were unable to produce full-length transcripts.
Despite significant differences in transcriptional capabilities, the proteins present in the DLP portion of each structure appear to be essentially identical at the present resolution. The readily observable architectural difference among these structures is the degree to which the presence of additional protein on the VP6 surface reduces the dimensions of the mRNA release channels in the capsid. In the two transcriptionally incompetent structures, the binding of 2A11/E9 Fab or VP7 to the surface of VP6 reduces the effective opening of the channels at the particle periphery to approximately 50% of their normal width. Furthermore, as a consequence of ligand attachment in the two transcriptionally incompetent structures, small but consistent conformational changes are observed to occur in the inner region of the VP6 capsid layer near the VP6-VP2 interface. These structural rearrangements appear to alter trimer–trimer interactions and, as a consequence, also reduce the effective diameter of the base of the mRNA release channel by ≈20%.
Structural changes in the capsid could conceivably affect the transcription process in several ways. Protein attachment to the viral surface could transmit structural rearrangements into the particle interior, disrupting the proper alignment of the transcription enzymes and the dsRNA template and hindering the efficient movement of the dsRNA with respect to the polymerase during nucleotidyl transfer. Under these conditions, transcript initiation potentially could be inhibited. Alternatively, the attachment of protein to specific locations on VP6 could either directly or indirectly prevent the efficient translocation of nascent transcripts through the exit channels. Such a mechanism of inhibition likely would give rise to a series of oligonucleotides having discrete lengths determined primarily by the point at which translocation is blocked.
To differentiate between these potential mechanisms of ligand-induced transcription inhibition, we carried out electrophoretic analysis of the transcription reaction products generated in the two transcriptionally incompetent particles. In the native DLPs, full-length transcripts were produced as expected, although interestingly, a population of short oligonucleotides ranging in length from five to seven bases also was observed. In the two transcriptionally incompetent structures, however, nearly all of the transcripts remained as short oligonucleotides ranging in length from five to seven bases. In the 2A11/E9 DLP/Fab complex, a small number of oligonucleotides were elongated beyond seven bases, with two predominant forms appearing at ≈35 bases and ≈90 bases, whereas in the mature TLP, no elongation beyond seven bases was observed. These observations indicate that, in the two transcriptionally incompetent structures, transcription is not inhibited before initiation but rather during elongation.
Under transcription conditions, the native DLP is able to produce full-length transcripts because the conformation of the capsid facilitates the continuous elongation and translocation of the nascent mRNA out of the particle. The short oligonucleotides observed in the native DLP may represent a small population of initiated transcripts that failed to be elongated because they could not be successfully translocated into the mRNA release channels. In this respect, the proper threading of an initiated transcript through the exit channel is perhaps the critical event required for the transition from initiation to sustained elongation. Even in the native DLP, it is conceivable that not all initiated transcripts necessarily would make this transition successfully. A similar phenomenon has been observed in mammalian orthoreovirus, where subviral particles analogous to the rotavirus DLP produce both full-length transcripts as well as short oligonucleotides during transcription (24–26), whereas mature particles, analogous to the rotavirus TLP, produce only short oligonucleotides (27).
In the 2A11/E9 DLP/Fab complex, as in the native DLP, the shorter oligonucleotides likely represent aborted transcripts that failed to be properly translocated out of the core. The longer mRNA species may represent partially elongated transcripts, which, though successfully translocated into the mRNA release channel system, nonetheless were unable to pass through the mouth of the channel at the particle periphery because of the presence of bound Fabs narrowing the opening. In the VP6 capsid region, the mRNA release channel approximates a conical shape measuring ≈65 Å across at the mouth and nearly 70 Å in length. A transcript passing through this relatively wide region may begin to form small elements of secondary structure, which are too bulky to move out of the obstructed opening in the 2A11/E9 DLP/Fab complex. Indeed, the 5′ untranslated regions of the 11 gene segments are proposed to form secondary structures involving stem loops and hairpins (28), all of which would likely be too large to pass efficiently through the constricted mouth of the mRNA release channel. The point at which secondary structure becomes bulky enough to reduce the efficiency of translocation is likely to be somewhat different for each segment, the result being that elongated transcripts arrested in the mRNA release channel are not all the same length. The reason for which the length distribution of transcription products in the 2A11/E9 DLP/Fab complex is different from that observed in the native DLP is perhaps caused by subtle ligand-induced conformational changes in the capsid that decrease the likelihood that an initiated transcript will properly exit the core, leading to a predominance of short oligonucleotides.
In contrast to the 2A11/E9 DLP/Fab complex, elongation beyond seven bases is not observed in the mature TLPs, suggesting that the binding of VP7 introduces conformational changes in the capsid architecture, which effectively prevent transcripts from being elongated beyond this point. The lack of longer oligonucleotides clearly indicates that obstructions in the VP6 region of the channels do not play a role in the transcriptional inhibition observed in the TLP. Two possible explanations may account for this difference between the 2A11/E9 DLP/Fab complex and the mature TLP. First, the difference in behavior may be the result of subtle variations in the degree to which bound ligands affect the conformation of the capsid and thus hinder the translocation of mRNA. Second, this difference may arise from the fact that the mature TLP and the 2A11/E9 DLP/Fab complex are not formed in the same way. 2A11/E9 DLP/Fab complexes are prepared in vitro by combining transcriptionally competent DLPs with 2A11 Fabs, whereas mature TLPs are synthesized in vivo by the transcapsidation of DLP-like assembly intermediates during the particle maturation phase of the replication cycle. It is presently unknown whether in vivo DLP-like assembly intermediates are transcriptionally competent.
A comparison of the transcriptional behavior among the three ligand-bound structures suggests that the principal determinant of transcription inhibition is likely to be the location of the ligand attachment site on VP6. In the 8H2/G5 DLP/Fab complex, location of the epitope is such that only 510 Fab molecules can bind without steric hindrance and, at the present resolution, essentially no conformational changes are seen in the body of the trimer upon Fab binding. In the two transcriptionally incompetent structures, however, the location of the binding sites are such that, not only is each VP6 molecule bound by a ligand, but subtle structural changes also appear to be transmitted into the interior of the VP6 layer, affecting the architecture of the capsid in the vicinity of the VP2-VP6 interface. These changes in the body of the VP6 trimers perhaps are required to accommodate bound protein (2A11/E9 Fab or VP7) in a 1:1 stoichiometry or perhaps are triggered by the attachment of these proteins to specific locations on the top of the trimer.
Based on the comparative structural and biochemical analyses of the transcriptionally competent and incompetent structures presented here, the following conclusions can be made. In the native DLP, the three stages of transcription, namely initiation, elongation, and translocation, generally occur in succession without interruption, as the capsid is designed to facilitate the continuous elongation and translocation of nascent mRNA out of the core during transcription. The transcriptionally competent 8H2/G5 DLP/Fab complex also is able to produce full-length transcripts because the configuration of the capsid, being unaffected by the manner in which the Fabs are attached to the particle, likewise readily accommodates the continuous translocation of nascent mRNA. In the transcriptionally incompetent 2A11/E9 DLP/Fab complex, however, our observations suggest that transcription is inhibited because the binding of 2A11/E9 Fab to a specific epitope on VP6 alters the state of the capsid such that most transcripts cannot be translocated successfully out of the core. Likewise, in the mature TLP, the binding of VP7 alters the architecture of the particle such that the capsid is no longer able to facilitate the translocation of mRNA in the presence of the outer capsid layer. In this regard, transcription inhibition in the mature TLP may resemble that of the mature orthoreovirion, where conformational changes accompanying the loss of the outer capsid proteins enable the particle to become transcriptionally competent (27, 29). Comparative structural studies at higher resolution will further our understanding of the way in which small conformational changes in the VP6 capsid layer can perturb the function of the endogenous transcription machinery in the core.
Acknowledgments
We are indebted to Richard Ward (Children’s Hospital Medical Center, Cincinnati, OH) for generously providing the ascites from which these antibodies were purified. We thank S. Crawford for helpful technical advice and Dr. R.F. Ramig for helpful comments. This research was supported by National Institutes of Health Grants AI-36040 (B.V.V.P.) and DK-30144 (M.K.E.) and by the R. Welch Foundation. J.A.L. acknowledges the support of a training grant GM-08280 from the National Institute of General Medical Sciences. We also acknowledge use of the Biomedical Computation and Visualization Laboratory at Baylor College of Medicine, supported by a grant from the National Science Foundation.
Footnotes
Abbreviations: dsRNA, double-stranded RNA; TLP, triple-layered particle; DLP, double-layered particle; 3D, three-dimensional.
References
- 1.Fields B N. In: Virology. Fields B N, Knipe D M, Chanock R M, Melnick J L, Roizman B, Hope R E, editors. New York: Raven; 1996. pp. 1553–1555. [Google Scholar]
- 2.Prasad B V V, Estes M K. In: Structural Biology of Viruses. Chiu W, Burnett R, Garcia R, editors. New York: Oxford Univ. Press; 1997. pp. 239–268. [Google Scholar]
- 3.Lawton J A, Estes M K, Prasad B V V. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 4.Prasad B V V, Burns J W, Marietta E, Estes M K, Chiu W. Nature (London) 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 5.Wikoff W R, Wang G, Parrish C R, Cheng R H, Strassheim M L, Baker T S, Rossmann M G. Structure (London) 1994;2:595–607. doi: 10.1016/s0969-2126(00)00062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wien M W, Filman D J, Stura E A, Guillot S, Delpeyroux F, Crainic R, Hogle J M. Nat Struct Biol. 1995;2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]
- 7.Hewat E A, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, Newman J, Domingo E, Mateu M G, Stuart D I. EMBO J. 1997;16:1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Che Z, Olson N H, Leippe D, Lee W M, Mosser A G, Rueckert R R, Baker T S, Smith T J. J Virol. 1998;72:4610–4622. doi: 10.1128/jvi.72.6.4610-4622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad B V V, Wang G J, Clerx J P M, Chiu W. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 10.Ginn D I, Ward R L, Hamparian V V, Hughes J H. J Gen Virol. 1992;73:3017–3022. doi: 10.1099/0022-1317-73-11-3017. [DOI] [PubMed] [Google Scholar]
- 11.McKinney M M, Parkinson A. J Immunol Methods. 1987;96:271–278. doi: 10.1016/0022-1759(87)90324-3. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. E18–E19. [Google Scholar]
- 13.Dubochet J, Adrian M, Chang J J, Homo J C, LePault J, McDowall A, Schultz P. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 15.Lawton J A, Prasad B V V. J Struc Biol. 1996;116:209–215. doi: 10.1006/jsbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 16.Olson N H, Baker T S. Ultramicroscopy. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowther R A. Philos Trans R Soc London B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 18.Crowther R A, DeRosier D J, Klug A. Proc R Soc London Ser A. 1970;317:319–340. [Google Scholar]
- 19.Fuller S D. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 20.van Heel M. Ultramicroscopy. 1987;21:95–99. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 21.Guddat L W, Shan L, Anchin J M, Linthicum D S, Edmundson A B. J Mol Biol. 1994;236:247–274. doi: 10.1006/jmbi.1994.1133. [DOI] [PubMed] [Google Scholar]
- 22.Carson M. J Appl Crystrallogr. 1991;24:958–961. [Google Scholar]
- 23.Donis-Keller H, Maxam A M, Gilbert W. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamakawa M, Furuichi Y, Nakashima K, LaFiandra A J, Shatkin A J. J Biol Chem. 1981;256:6507–6514. [PubMed] [Google Scholar]
- 25.Zarbl H, Hastings K E M, Millward S. Arch Biochem Biophys. 1980;202:348–360. doi: 10.1016/0003-9861(80)90437-3. [DOI] [PubMed] [Google Scholar]
- 26.Bellamy A R, Nichols J L, Joklik W K. Nat New Biol. 1972;238:49–51. doi: 10.1038/newbio238049a0. [DOI] [PubMed] [Google Scholar]
- 27.Yamakawa M, Furuichi Y, Shatkin A J. Virology. 1982;118:157–168. doi: 10.1016/0042-6822(82)90329-4. [DOI] [PubMed] [Google Scholar]
- 28.Patton J T, Wentz M, Xiaobo J, Ramig R F. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]