Abstract
Hepatitis B virus (hepadnavirus) infections are maintained by the presence of a small and regulated number of episomal viral genomes [covalently closed circular DNA (cccDNA)] in the nuclei of infected cells. Although a number of studies have measured the mean copy number of cccDNA molecules in hepadnaviral-infected cells, the distribution of individual copy numbers have not been reported. Using a PCR-based assay, we examined the number of cccDNA molecules of the duck hepatitis B virus in single nuclei isolated from the liver of a chronically infected duck over the course of 131 days of infection. Nuclei were isolated from frozen serial biopsies and individually deposited into PCR microplates by flow sorting. Each nucleus was assayed by nested PCR for cccDNA and for cellular IFN-α genes as an internal control. We found that 90% of the nuclei assayed contained between 1 and 17 cccDNA molecules, with the remaining 10% containing more (90% confidence), and that differences in the mean number of copies and distribution of copy numbers occurred within the same animal at different times postinfection. Overall, the data suggest (i) that the number of cccDNA molecules per cell may fluctuate over time, and (ii) that, according to these fluctuations, a substantial fraction of cells may contain only one or a few copies. We infer from the results that infected hepatocytes express virus at different levels and that during cell division it is possible to segregate cells containing no cccDNA.
Hepadnaviruses are small, DNA-containing viruses that cause persistent infections of the liver. The prototype virus of this family is the human hepatitis B virus (HBV), a causative agent of chronic hepatitis and liver cancer in humans. Liver injury is not due to virus replication or toxic effects on the infected cell but to immunologic reactions against cells expressing viral antigens (1). Viral antigens are expressed from nuclear viral DNA, a covalently closed circular form (cccDNA), ≈3 kilobases in length. cccDNA is also responsible for the production of new viral DNA genomes through reverse transcription in the cytoplasm of genomic transcripts, the RNA pregenomes (2-5). The hepadnaviruses studied most frequently include human HBV and two experimental animal virus models, the woodchuck hepatitis virus (6) and the duck HBV (DHBV) (7).
The infecting genome is transported to the nucleus and converted to a cccDNA episome. Newly synthesized viral DNAs, produced from the initial episome by transcription and reverse transcription, are also transported into the nucleus and converted to cccDNA, a process we have called cccDNA amplification (4, 8). For DHBV it has been shown that the episomal copy number in the nucleus is eventually limited by production of viral envelope proteins (9-11), which direct newly synthesized DNA-containing viral cores into a virus assembly pathway in which they are ultimately secreted from the cells as enveloped virus. The importance of this control is demonstrated by the fact that mutational ablation of one of the DHBV envelope proteins, the pre-S protein, results in uncontrolled amplification and death of the infected cells (12-14).
Viral expression is controlled at one level by a gene dosage effect, according to the number of cccDNA copies in each nucleus (12). The average copy number has been reported to be between 6 and 60 copies per cell in human HBV-, woodchuck hepatitis virus-, or DHBV-infected liver cells (15-19). The range of copy numbers in different cells, however, has not been reported. In this study, we measured cccDNA copy numbers in single nuclei isolated from a DHBV-infected liver at different times after infection and during growth of the infected duckling. In the combined samples, we found a broad distribution of copy numbers between 1 and 17 per nucleus, suggesting that individual cells carry out different levels of virus gene expression and replication.
Methods
Animal. A White Pekin female duckling was obtained from Metzer Farms (Redlands, CA) and injected at 4 days of age with 2 × 107 DHBV DNA-containing particles. Blood was drawn at least weekly for the first 6 weeks and every other week thereafter. Liver biopsies were obtained surgically, under general anesthesia, by using aseptic technique at 11, 33, 66, 88, 109, and 131 days postinfection, and biopsy specimens were frozen at -80°C for later analysis. All animal procedures were conducted with the approval of the Institutional Animal Care and Use Committee. The facility is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
Isolation and Sorting of Nuclei. Approximately 5 mg of frozen liver tissue was homogenized in a 2-ml Dounce homogenizer with the loose-fitting ”A” pestle until the tissue was completely dispersed (≈20 strokes). The homogenization solution of 1 ml contained 10 mM Tris·HCl (pH 7.5), 3 mM MgCl2, 0.25 M sucrose, and 0.05% Triton X-100 (20). Nuclei were collected by low-speed centrifugation (2,000 rpm for 5 min in a Sorvall RT6000B), resuspended in 1 ml of homogenization buffer, and subjected to one additional round of centrifugation. A suspension of nuclei (≈106 per ml) was sorted (MoFlo cytometer and high speed cell sorter, Cytomation, Fort Collins, CO) into the individual wells of PCR plates (low binding, Fisher Scientific) containing 12 μl of a solution of 10 mM Tris·HCl (pH 7.5) and 0.1% Triton X-100, with proteinase K (200 μg/ml). Control experiments were performed in which nuclei were sorted into flat-bottom wells of a microtiter dish and observed microscopically to confirm that only a single nucleus was delivered to each well.
Digestion of Individual Nuclei. DNA was released from the individual nuclei by incubation with a proteinase K solution (200 μg/ml) for 60 min at 50°C, after which the proteinase K was inactivated by a further incubation at 75°C for 15 min. Cellular DNA and cccDNA were digested for 1 h at 37°C with a combination of EcoRI (5 units) and PstI (5 units) added in a total volume of 8 μl containing 2 μl of 10× PCR buffer (see below). EcoRI is a single cutter for cccDNA and PstI does not cut DHBV but cleaves once within each of the ≈10 individual copies of the duck IFN-α gene in the tandem array on the duck chromosome Z (21).
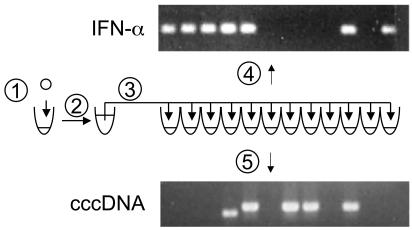
Measurement of cccDNA and IFN-α Copy Numbers in Each Nucleus. Total DNA released by proteinase K digestion and cut with EcoRI and PstI was diluted to 120 μl with a complete PCR reaction mixture containing, finally, 12 pmol each of primers CF1, CR1, IF1, and IR1 (Table 1), 4.2 units of Expand High Fidelity DNA polymerase (Roche Diagnostics), and 12 μl of the supplier's PCR buffer containing 15 mM MgCl2. The PCR mixture was distributed in 10-μl aliquots to 12 wells of a PCR microplate (Fig. 3) and subjected to the following amplification: denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 15 sec, 58°C for 20 sec, 72°C for 45 sec, and a final elongation step of 72°C for 4 min.
Table 1. PCR primers.
| Designation | Sequence, 5′-3′ |
|---|---|
| CF1 | TGT CCC GAG CAA ATA TAA TCC |
| CR1 | TGT GTA GTC TGC CAG AAG TCT TC |
| CF2 | TAT AAT CCT GCT GAC GGC C |
| IF1 | AAC GAC ACG CAG CAA GC |
| IR1 | GGA GGA AGT GTT GGA TGC |
| IF2 | CTC CAC CTC CTC CAA CAC |
| IR2 | TTG GAT GCA GCC GAA GTA |
Fig. 3.
Assay of a single nucleus for cccDNA and IFN-α genes. Single nuclei isolated from frozen biopsy specimens were deposited in the wells of a PCR microplate containing 7 μl of proteinase K (1). After digestion to release the DNA and heating to inactivate the proteinase K, the sample was diluted to 120 μl (2) with complete duplex PCR mixture containing cccDNA-specific primers, CF1 and CR1, IFN-α-specific primers, IF1 and IR1, and the restriction enzymes EcoRI and PstI. After incubation at 37°C for 1 h, the amplification reaction was distributed among 12 wells in a PCR microplate (3), and 35 cycles of amplifi-cation were carried out. Approximately 0.1 μl of each sample was transferred to two additional PCR microplates containing PCR mixes specific for amplifi-cation of cccDNA (4) or IFN-α (5) by nested primer sets CF2 and CR1, and IF2 and IR2, respectively. Amplification was carried out for an additional 35 cycles, and the products were analyzed by agarose gel electrophoresis. Products were visualized by ethidium bromide staining.
Approximately 0.1 μl of each reaction was transferred to each of two different PCR plates containing cccDNA- or IFN-α-specific nested primers. Seeding of the nested reactions was carried out by using a 96-well microplate pin replicator (Nalge Nunc). Each nested reaction consisted of a volume of 10 μl containing 4 pmol each of either CF2 and CR1 for cccDNA or IF2 and IR2 for IFN-α, 200 μM each dNTP, 0.5 units Taq DNA polymerase (Promega), and 1 μl of the supplier's 10× PCR mixture containing 15 mM MgCl2.
Competitive PCR and Primer Extension. Competitive PCR was carried out with a genetically marked template containing a 4-bp insertion at the XhoI site in the duck IFN-α coding sequence. DNA template was prepared from a mixture of duck erythrocyte nuclei and a 10-fold molecular excess of plasmid competitor DNA, linearized with BamHI. Nuclei (107) and competitor plasmid (108) molecules were mixed in a volume of 0.5 ml of 10 mM Tris·HCl buffer (pH 8.0) containing 0.15 M NaCl, 0.5% SDS, and 0.5 mg/ml pronase. Digestion was carried out at 37°C for 3 h, and the DNA was purified by phenol extraction and ethanol precipitation. The DNA was digested with EcoRI and PstI, and an amount equivalent to ≈1,000 cells was subjected to 35 cycles of PCR amplification with the primers IF1 and 5′biotinylated IR1 and Expand High Fidelity DNA polymerase (Roche Diagnostics) conditions. The 5′-biotinylated strand of the product was isolated on streptavidin-coated magnetic beads (Dynal) and analyzed by primer extension.
Primer extension was carried out in a volume of 10 μl containing template plus 200 μM dNTP, 1 pmol 5′ 32P-IF1 primer, 0.5 units of Taq DNA polymerase (Promega), and 1 μl of the supplier's 10× PCR buffer containing 15 mM MgCl2. The reaction conditions were 94°C for 4 min, followed by 25 cycles of 94°C for 15 sec, 58°C for 20 sec, and 72°C for 45 sec, and a final 4-min elongation at 72°C. The 32P-labeled primer extension products of 250 and 254 nucleotides were resolved by electrophoresis through an 8% polyacrylamide sequencing gel and quantified by phosphorimaging.
Analysis of the Data. The data from the assays of single nuclei for IFN-α genes and cccDNAs were analyzed by using computational methods to describe the random sorting of individual templates into 12 wells of a PCR microplate, and incorporated a fixed probability that each molecule would be detected by the nested PCR assay. The probability of observing k (k = 1-12) PCR-positive wells when r templates were uniformly distributed into 12 wells was computed. For each observed k, the r that maximized this probability was determined. Accordingly, each observed value of k corresponds to a maximum likelihood estimate (mle) of r. Means and standard deviations for the number of IFN-α genes and cccDNAs for each set of nuclei assayed were calculated after converting the values of k for the data set to the corresponding mles.
The lower bound of the range of the distribution of cccDNA copies per nucleus was defined as one copy per nucleus, and the fraction of nuclei containing only one copy was estimated by a statistical analysis. The 90% upper bound of the distribution, rM, was defined as that number which, for at least 90% of all nuclei, r < rM. These statistical derivations are described in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
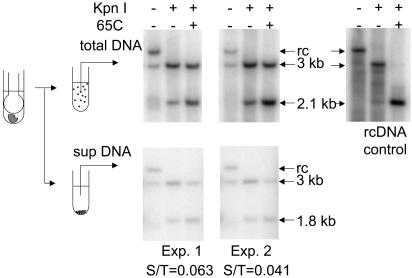
Retention of cccDNA in Nuclei Obtained from Frozen Tissue. We wanted to use frozen liver samples to determine the distribution of cccDNA copy numbers in hepatocyte nuclei. Therefore, we tested how well cccDNA would be retained in the nucleus during the thawing, homogenization, and nuclear isolation. Frozen liver tissue was homogenized and divided into two parts. One part was incubated on ice while the second part was subjected to low-speed centrifugation to remove the nuclei, which were discarded. Both samples were adjusted to 1% SDS and the protein-DNA-detergent complexes were precipitated by the addition of 0.5 M KCl. The supernatants containing cccDNA were collected after microcentrifugation and extracted with phenol to remove remaining proteins and replicative intermediates, which are covalently bound to protein. The aqueous phase was precipitated with two volumes of ethanol, the precipitate was dissolved, and the amount of cccDNA was determined by Southern blot hybridization. The design of the experiment and the results are shown in Fig. 1.
Fig. 1.
Retention of cccDNA in the nucleus during nuclear isolation. Approximately 5 mg of frozen liver was homogenized, and the homogenate was assayed directly for cccDNA (upper left) or after centrifugation to remove the nuclei (lower). DNA was purified by phenol extraction, to remove most of the replicative intermediates covalently bound by protein, followed by ethanol precipitation. A sample of replicative intermediates isolated after digestion with pronase was used as a control for specificity of the assay (rcDNA control). DNA samples from equivalent amounts of homogenate were loaded after the indicated treatments. The fraction of cccDNA not retained in the nuclear fraction was calculated by comparing the signal of 3-kb linear DNA in the supernatant fraction with that in the total DNA sample after KpnI digestion and heating. The results of two experiments are shown as the fraction of cccDNA in the supernatant (S/T).
Because the samples contained small amounts of residual relaxed circular DNA (rcDNA), a procedure was performed to distinguish the cytoplasmic rcDNA from cccDNA that might have been released from nuclei as a nicked form, which would comigrate with rcDNA. Two-thirds of each DNA sample was digested with KpnI, an enzyme that cut both rcDNA and nicked cccDNA at one site to produce 3,021-bp linear DNAs. After cutting, one-half of the digested sample was heated to 65°C for 5 min to denature the 40- to 52-bp cohesive region, found only in rcDNA. Denaturation of the cohesive region resulted in the production of discrete fragments of ≈1.8 kb and 1.2 kb (Fig. 1, rcDNA control, 1.2-kb band not seen), whereas BamHI-digested cccDNA remained intact as a 3,021-bp linear DNA. Thus, the 3,021-bp linear DNA represented the total amount of intact and nicked cccDNA in the original sample. Phosphorimage analysis of the Southern blot of two different liver samples revealed that 94% and 96% of all cccDNA in the tissues was removed with the nuclear fraction, indicating that an average of 95% of the cccDNA remained associated with the nuclei during the major steps of nuclear isolation from frozen tissue.
Measurement of IFN-α Gene Copy Number. In assaying for the copy number of cccDNA molecules, we concurrently assayed each nucleus for the number of copies of a multicopy cellular gene, IFN-α, as an internal control for recovery of nuclear DNA, efficiency of detection of templates, and overall accuracy of the method. To interpret the results of these controls, we needed to know the actual number of IFN-α templates per cell that could be detected by our IFN-α-specific primers.
It has been reported that ≈10 copies of the duck IFN-α gene are found in each female duck and that 20 copies are found in each male duck (21). The difference in copy numbers between females and males exists because IFN-α genes are found in a tandem array near the one end of the Z sex chromosome. Karyotypically, male birds are ZZ and females are XZ. For the production of a genetically marked competitor template, the IFN-α product amplified from duck DNA was cloned into plasmid pSP65 by blunt-end ligation, and the single XhoI site in the coding sequence was destroyed by cleavage, extension of the 3′ recessed ends by Klenow fragment of DNA polymerase I, and blunt-end ligation. This genetically marked template thus contained a 4-bp insertion.
A known number of female duck erythrocyte nuclei was mixed with a 10-fold excess of plasmid DNA molecules containing the competitor IFN-α template to simulate the reported copy number of IFN-α genes. Total DNA was extracted from the mixture and the copurified DNAs, equivalent to ≈1,000 cells, were digested with EcoRI and PstI (see Methods) and used as a template for PCR with one biotinylated primer and one nonbiotinylated primer. The product of this reaction was then subjected to 25 cycles of primer extension with the 5′ 32P-labeled nonbiotinylated primer after purification of the biotinylated strand on streptavidin-coated magnetic beads. Finally, separation of the labeled primer extension products on a sequencing gel revealed the ratio of the endogenous product and the competitor (+4) product, which was quantified by phosphorimaging (Fig. 2). This assay was performed seven times on each of six different DNA preparations, and the copy number per cell of IFN-α genes was calculated for each reaction. The mean copy number per nucleus of the 42 assays was 9.64 ± 1.46. This value agrees well with the published value of 10 copies per cell.
Fig. 2.
Assay for IFN-α copy number by competitive PCR. Erythrocyte nuclei purified from the infected female duck used in the study were counted in a hemocytometer and mixed with a 10-fold excess of plasmids containing a genetically marked IFN-α competitor template. DNA was isolated by proteinase K digestion, phenol extraction, and ethanol precipitation. DNA equivalent to ≈1,000 nuclei (3 ng) was used in each of seven amplification reactions with the IFN-α-specific primers, IF1 and IR1, where IR1 was 5′-biotinylated. The 5′-biotinylated strand from the product of this reaction was isolated on streptavidin-coated magnetic beads and subjected to 25 cycles of primer extension with 5′ 32P-labeled IF1 primer. The primer extension products were separated on a sequencing gel and quantified by phosphorimaging. The entire procedure was carried out on six independently isolated DNAs, yielding a total of 42 assays.
Infection. Table 2 summarizes the virological data on the infected duckling used in this study. As can be seen, a chronic infection was maintained throughout the study, as judged by the presence of a viremia. Body-weight measurements show that the study spanned the phase of rapid growth through attainment of the adult body mass. Histological examination of serial biopsies indicated that mild inflammation occurred at 66, 88, and 109 days postinfection that was not observed in the other biopsy specimens.
Table 2. Virological and histological evaluation of an infected duckling.
| Days postinfection | Body weight, g | Viremia* | Histologic observations |
|---|---|---|---|
| 11 | 430 | 7 × 109 | Normal |
| 33 | 1,700 | 2 × 1010 | Normal |
| 66 | 2,360 | 8 × 109 | Mild portal inflammation |
| 88 | 2,350 | 6 × 109 | Mild portal inflammation |
| 109 | 2,200 | 1 × 109 | Mild portal inflammation |
| 131 | 2,200 | 1 × 109 | Normal |
DNA-containing particles per milliliter
Concurrent Assays of Single Nuclei for cccDNA and IFN-α The analysis protocol for determination of both cccDNA and IFN-α copy numbers on each individual nucleus is shown in Fig. 3. Nested PCR was aided by a microplate pin replicator, which was used to transfer a small amount of product from the first duplex PCR to each of the second DHBV- or IFN-α-specific reactions. The presence of a specific product was determined by agarose gel electrophoresis. Occasionally, smaller products of PCR were seen in the cccDNA amplification reactions, as seen in the example in Fig. 3. These products were probably derived from deleted cccDNA templates produced from linear DNA, as we described (22, 23).
Data from 648 assays were obtained. The raw data are presented in matrix form for each biopsy in Fig. 6, which is published as supporting information on the PNAS web site, and are summarized here. Only 312 nuclei assayed were positive for DHBV cccDNA, despite the fact that all hepatocytes were infected, as judged by immunohistochemical detection of viral antigens (data not shown). Presumably, not all liver nuclei were derived from hepatocytes. Nonhepatocytes found in the livers of ducks would include erythrocytes and mononuclear cells in addition to the various nonparenchymal cells. The failure to detect any IFN-α genes in 58 wells was presumed to be due to failure of the flow sorter to deliver a nucleus to that well. Such failure rates were confirmed by direct microscopic visual inspection of wells for a nucleus, enhanced by staining with ethidium bromide. From this visual examination it was apparent that the sorter had delivered a droplet but that the droplet did not contain a nucleus. In the assays of these 58 IFN-α-negative samples, no cccDNA templates were detected, confirming that cccDNA templates detected were associated with nuclei and not free in solution.
There was low correlation between the number of reactions positive for cccDNA per nucleus and those positive for IFN-α (ρ = 0.08 for the combined data; see Table 3). Had the low levels of cccDNA been due to poor recovery or detection of DNA templates in some nuclei, we would have expected to see low numbers of IFN-α-positive reactions preferentially associated with those nuclei. The low correlation indicates that the number of cccDNA-positive reactions per nucleus was not a result of failure of the assay for that nucleus.
Table 3. cccDNA and IFN-α copy numbers in different biopsies.
| cccDNA*
|
|||||
|---|---|---|---|---|---|
| Days postinfection | IFN-α mean | Mean | Fraction of nuclei with one copy | 90% upper bound | IFN-α and cccDNA correlation p |
| 11 | 9.2 ± 3.2 | 4.3 ± 3.8 | >0.17 | <21 | 0.00 |
| 33 | 9.4 ± 3.7 | 4.7 ± 3.1 | >0.06 | <18 | 0.23 |
| 66 | 8.5 ± 3.4 | 8.6 ± 5.8 | >0.01 | <36 | -0.09 |
| 88 | 8.4 ± 3.1 | 2.9 ± 2.2 | >0.18 | <10 | 0.04 |
| 109 | 9.5 ± 3.4 | 4.3 ± 4.8 | >0.15 | <30 | 0.16 |
| 131 | 10.0 ± 2.9 | 7.1 ± 4.3 | >0.02 | <22 | 0.16 |
| Combined | 9.1 ± 3.3 | 5.2 ± 4.4 | >0.13 | <17 | 0.08 |
For positive nuclei only
Efficiency of Detection of Nuclear Templates. The efficiency of detection of nuclear DNA template was examined by converting the data for the distribution of IFN-α-positive reactions into corresponding mle (see Methods) and determining the mean mle for 590 positive nuclei assayed. This calculation yielded a mean mle of 9.1, a value that agrees well with the copy number 9.64 determined by competitive PCR. Therefore, assuming that the efficiency of detection of nuclear DNA was close to 100%, we omitted this variable from our subsequent calculations.
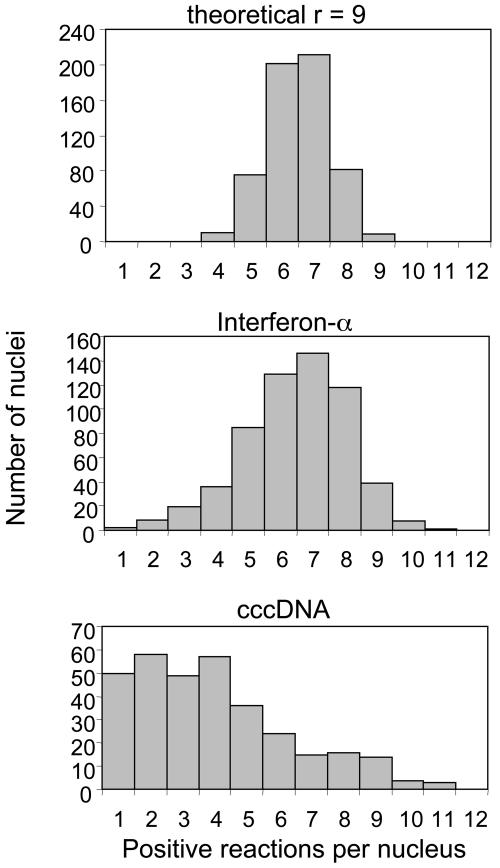
Distribution of cccDNA Copy Number. The distribution of cccDNA-positive reactions in all samples was clearly broader than that obtained for IFN-α genes, which are assumed to be present at a unique value of 9 or 10 copies in all cells (Fig. 4). This apparent difference suggests that the copy number of cccDNA per nucleus is not a unique value but a distribution of values. Because randomly delivering a fixed number of molecules in 12 reactions can, by chance, result in different numbers of positive wells, as seen in the theoretical distribution in Fig. 4, the distribution of positive wells for all nuclei does not define a unique distribution of cccDNAs. However, it was possible to estimate the range of the distribution of cccDNA molecules that gives rise to the data for the combined 312 DHBV-positive nuclei (shown in Fig. 4). On the low end, our data indicate that at least 13% of all nuclei analyzed contained exactly one cccDNA molecule, with a confidence level of 90%. On the upper end of the range, we have calculated that no more than 10% of nuclei contain >17 cccDNA molecules, with a confidence level of 90%. It is not possible from these data to distinguish among a variety of cccDNA copy number distributions within these limits; that is, the data do not exclude that specific numbers of copies of cccDNA were absent from the population of nuclei analyzed, or that only certain copy numbers within the range were present. Nevertheless, the data support the conclusion that the copy number per nucleus in the population was variable within broad limits.
Fig. 4.
Distribution of positive reactions for all nuclei. (Top) The theoretical distribution of 9 templates among 12 reactions, assuming 100% detection of each template. (Middle and Bottom) The distributions for the number of positive reactions per nucleus for the combined data from six biopsies. Only nuclei showing at least one positive reaction for IFN-α (Middle) or cccDNA (Bottom) are included in the distributions.
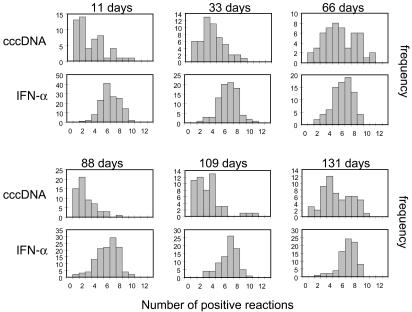
Comparison of Different Biopsies. Whereas the assays for IFN-α generated similar distributions of positive reactions per nucleus for each biopsy, different distributions of cccDNA-positive reactions were seen (Fig. 5). All distributions were rather broad, but in general the number of positive reactions increased successively in biopsies taken up to 66 days, decreased at 88 days, and increased in the subsequent biopsies at 109 and 131 days. The statistical data summarizing these assays are shown in Table 3. Changes in the mean number of positive reactions and their distribution were specific to the cccDNA assays and were comparatively minor for the IFN-α assays.
Fig. 5.
Distribution of positive reactions for each biopsy. The separate distributions for IFN-α- and cccDNA-positive reactions are shown for each biopsy. Only assays yielding at least one positive reaction for IFN-α or cccDNA are included in the respective distributions.
Discussion
Our study established that the mean cccDNA copy numbers per nucleus in hepatocytes of a DHBV-infected duck ranged between 2.9 and 8.6 in biopsies taken over the course of 131 days of chronic infection. A similar range of low copy numbers (1.2-6.9) was obtained by a more conventional Southern blot analysis (Fig. 7, which is published as supporting information on the PNAS web site). At all times the distribution was broad, with the 90% upper bound of the distribution varying among the different time points between 10 and 36 copies per cell. Moreover, a significant fraction of nuclei in each biopsy contained only one or a few molecules. Neither the basis for the broad distribution at any time nor for the variation from biopsy to biopsy is known. Although some variation in copy numbers may be due to differences among cells in the extent of the initial cccDNA amplification, changes in the overall distribution over time suggest that fluctuations in the pool size of cccDNA may occur within single cells or single cell lineages.
Such changes might be related to the physiological state of the liver: for example, the fractional increase in liver mass with time is greatest in Pekin ducks at early ages and declines until around the age of 45 days, when maximum liver weight is attained (24). High liver growth rates might result in cccDNA copy number dilution when cells divide. In addition, mild inflammatory changes in the liver (Table 2) may have had some influence on cccDNA copy numbers through a variety of mechanisms that are not understood. Whether these fluctuations in cccDNA copy numbers were actually related to growth, inflammation, or regeneration in the liver is speculative, and establishing such a relationship would require much more observation.
The cccDNA form of the hepadnavirus genome is one of a large variety of nonessential episomal genetic elements that are hosted by prokaryotic and eukaryotic organisms. Among these are the various circular DNA plasmids found in bacteria and fungi, and the viral episomal DNA forms that persist in higher eukaryotes, namely those of herpesviruses, papillomaviruses, and hepadnaviruses. In bacteria, plasmid maintenance during cell growth is generally a result of sufficient plasmid replication to ensure a high probability of distribution to both daughter cells, and the copy number control is a result of negative feedback mechanisms that suppress runaway plasmid replication (for review, see ref. 25). In such systems, the copy number per cell has been shown to be a narrow Gaussian distribution due to the unstable nature of the negative regulator molecules. In papillomavirus-infected cells episomal maintenance is conferred by the tethering of episomes to mitotic chromosomes by the transcriptional activator E2-TA (26, 27). Evidence suggests that the copy number of episomes stably maintained in cells depends on the number of available E2-TA molecules, possibly to provide tethering sites (28). Latent episomes of the herpesvirus Epstein-Barr virus (EBV) are also probably maintained by tethering of the DNA to mitotic chromosomes by EBNA-1 (29, 30). In addition, the replication of EBV episomes is coupled to the replication of cellular DNA through association of the chromosome replication factors ORC and MCM with oriP, the episomal replication origin (31-33). Actual copy-number distributions for herpesvirus and papillomavirus stable plasmids have not been determined. Among the animal viruses, the episomal maintenance of hepadnaviruses most resembles that of high-copy bacterial plasmids, with negative feedback by a product (the large envelope protein) preventing runaway replication by inhibiting the conversion of rcDNA to cccDNA. However, the broad distribution of cccDNA copy numbers suggests that the large envelope protein is relatively stable compared with the negative regulators of bacterial plasmids, and therefore its level in the cell does not decrease rapidly with the cccDNA copy number. No evidence for tethering of hepadnavirus cccDNA to cellular chromosomes has been reported.
Our results suggest that hepadnaviruses can support stable maintenance of the infected state with small numbers of cccDNAs, which apparently can be as low as one molecule per cell. One may speculate that some infected cells may contain no cccDNAs because they were lost through cell division or some other process. We did not test this idea directly. However, in contrast to bacterial plasmids and other viral episomes, the loss of the last remaining episome from the nucleus is not an irreversible event in the state of infection of the cell because many copies of cccDNA precursors, i.e., DNA- or RNA-containing capsids and pregenomes, are present in the cytoplasm and can replenish the pool of cccDNA under normal conditions of chronic infection (34). Finally, the existence of a large fraction of hepatocytes with only one or a few cccDNA molecules (or possibly no molecules) per nucleus may allow the rapid segregation of uninfected cells when cell proliferation occurs under conditions in which viral DNA synthesis is inhibited during, for example, immune clearance or antiviral therapy.
Supplementary Material
Acknowledgments
We thank Daniel D. Loeb for a critical reading of the manuscript. This work was supported by National Cancer Institute Grant CA84017.
Abbreviations: cccDNA, covalently closed circular DNA; DHBV, duck hepatitis B virus; mle, maximum likelihood estimate; rcDNA, relaxed circular DNA.
References
- 1.Rapicetta, M., Ferrari, C. & Levrero, M. (2002) J. Med. Virol. 67, 454-457. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Opazo, N., Chakraborty, P. R. & Shafritz, D. A. (1982) Cell 29, 129-136. [DOI] [PubMed] [Google Scholar]
- 3.Mason, W. S., Aldrich, C., Summers, J. & Taylor, J. M. (1982) Proc. Natl. Acad. Sci. USA 79, 3997-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuttleman, J., Pourcel, C. & Summers, J. (1986) Cell 47, 451-460. [DOI] [PubMed] [Google Scholar]
- 5.Summers, J. & Mason, W. S. (1982) Cell 29, 403-415. [DOI] [PubMed] [Google Scholar]
- 6.Summers, J., Smolec, J. M. & Snyder, R. (1978) Proc. Natl. Acad. Sci. USA 75, 4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason, W. S., Seal, G. & Summers, J. (1980) J. Virol. 36, 829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, T.-T., Coates, L., Aldrich, C. E., Summers, J. & Mason, W. S. (1990) Virology 175, 255-261. [DOI] [PubMed] [Google Scholar]
- 9.Summers, J., Smith, P. & Horwich, A. L. (1990) J. Virol. 64, 2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summers, J., Smith, P., Huang, M. & Yu, M. (1991) J. Virol. 65, 1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenhoff, R. & Summers, J. (1994) J. Virol. 68, 4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenhoff, R. & Summers, J. (1994) J. Virol. 68, 5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenhoff, R. L., Luscombe, C. A. & Summers, J. (1998) Virology 251, 85-96. [DOI] [PubMed] [Google Scholar]
- 14.Lenhoff, R. L., Luscombe, C. A. & Summers, J. (1999) Hepatology 29, 563-571. [DOI] [PubMed] [Google Scholar]
- 15.Jilbert, A. R., Wu, T. T., England, J. M., Hall, P. M., Carp, N. Z., O'Connell, A. P. & Mason, W. S. (1992) J. Virol. 66, 1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addison, W. R., Wong, W. W., Fischer, K. P. & Tyrrell, D. L. (2000) Antiviral Res. 48, 27-37. [DOI] [PubMed] [Google Scholar]
- 17.Dandri, M., Burda, M. R., Will, H. & Petersen, J. (2000) Hepatology 32, 139-146. [DOI] [PubMed] [Google Scholar]
- 18.Zhu, Y., Yamamoto, T., Cullen, J., Saputelli, J., Aldrich, C., Miller, D., Litwin, S., Furman, P., Jilbert, A. & Mason, W. (2001) J. Virol. 75, 311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajino, K., Jilbert, A. R., Saputelli, J., Aldrich, C. E., Cullen, J. & Mason, W. S. (1994) J. Virol. 68, 5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newbold, J. E., Xin, H., Tencza, M., Sherman, G., Dean, J., Bowden, S. & Locarnini, S. (1995) J. Virol. 69, 3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nanda, I., Sick, C., Munster, U., Kaspers, B., Schartl, M., Staeheli, P. & Schmid, M. (1998) Chromosoma 107, 204-210. [DOI] [PubMed] [Google Scholar]
- 22.Yang, W. & Summers, J. (1995) J. Virol. 69, 4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, W. & Summers, J. (1998) J. Virol. 72, 8710-8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, Y.-Y. & Summers, J. (2000) J. Virol. 74, 5257-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.del Solar, G. & Espinosa, M. (2000) Mol. Microbiol. 37, 492-500. [DOI] [PubMed] [Google Scholar]
- 26.Lehman, C. & Botchan, M. (1998) Proc. Natl. Acad. Sci. USA 95, 4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voitenleitner, C. & Botchan, M. (2002) J. Virol. 76, 3440-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penrose, K. J. & McBride, A. A. (2000) J. Virol. 74, 6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middleton, T. & Sugden, B. (1994) J. Virol. 68, 4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda, T., Otter, M. & Wahl, G. M. (2001) Mol. Cell. Biol. 21, 3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schepers, A., Ritzi, M., Bousset, K., Kremmer, E., Yates, J. L., Harwood, J., Diffley, J. F. & Hammerschmidt, W. (2001) EMBO J. 20, 4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhuri, B., Xu, H., Todorov, I., Dutta, A. & Yates, J. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirai, K. & Shirakata, M. (2001) Curr. Top. Microbiol. Immunol. 258, 13-33. [DOI] [PubMed] [Google Scholar]
- 34.Huang, M. & Summers, J. (1991) J. Virol. 65, 5435-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.