Abstract
A well balanced body energy budget controlled by limitation of calorie uptake and/or increment of energy expenditure, which is typically achieved by proper physical exercise, is most effective against obesity and diabetes mellitus. Recently, peroxisome proliferator-activated receptor (PPAR) γ, a member of the nuclear receptor, and its cofactors have been shown to be involved in lipid metabolism and in the control of energy expenditure. Here we show that PPARγ coactivator 1 (PGC-1) β functions as ERRL1 (for ERR ligand 1), which can bind and activate orphan ERRs (estrogen receptor-related receptors) in vitro. Consistently, PGC-1β/ERRL1 transgenic mice exhibit increased expression of the medium-chain acyl CoA dehydrogenase, a known ERR target and a pivotal enzyme of mitochondrial β-oxidation in skeletal muscle. As a result, the PGC-1β/ERRL1 mice show a state similar to an athlete; namely, the mice are hyperphagic and of elevated energy expenditure and are resistant to obesity induced by a high-fat diet or by a genetic abnormality. These results demonstrate that PGC-1β/ERRL1 can function as a protein ligand of ERR, and that its level contributes to the control of energy balance in vivo, and provide a strategy for developing novel antiobesity drugs.
The most typical molecular mechanism that underlies gene expression regulated by nuclear receptors starts from the binding of their ligands, small lipophilic molecules such as steroids, retinoic acid, vitamin D3, and thyroid hormone (1). Endogenous levels of such ligands are strictly regulated by means of multiple enzymatic steps working toward their biogenesis and/or degradation, and these ligands are collectively called lipophilic ligands or lipophilic hormones of the endocrine system. By changing ligand levels, the endocrine system contributes to the adaptation to changes in the external or internal environment and thus to homeostasis (1). This system appears advantageous for slow and long-term adaptations but disadvantageous for quick responses because of the complicated regulation of ligand biogenesis. On the other hand, genome analysis has predicted the existence of numerous nuclear receptor-like molecules whose cognate lipophilic ligands are currently unknown, and these molecules are collectively called orphan receptors. The activation mechanisms of orphan receptors remain totally unknown. Estrogen receptor-related receptors 1 and 2 (ERR1 and -2) were the first identified orphan receptors (2, 3), and a third member (ERR3) has recently been isolated (4, 5). ERRs and estrogen receptors share significant structural similarity, but ERRs do not respond to estrogen. ERR1 has been proposed to act as a key transcriptional regulator of the gene encoding medium-chain acyl CoA dehydrogenase (MCAD), a pivotal enzyme in mitochondrial fatty acid β-oxidation (6, 7). These observations lead to the idea that ERR-mediated gene regulation plays important roles in the control of energy balance in the body by regulating fatty acid β-oxidation. However, because of the lack of knowledge on the precise mechanism of regulation of ERR-mediated gene expression, this idea remains to be tested.
Recent research in the nuclear receptor field has revealed several classes of cofactor proteins, e.g., the steroid receptor coactivator 1/p160 family, P/CAF, and CBP/p300, which play key roles in ligand-dependent transcriptional activation of the nuclear receptors (8, 9). These so-called coactivators are ubiquitously expressed, and their expression levels appear not to change during differentiation of cells or in response to changes in external and internal environments. More recently, a unique coactivator, termed peroxisome proliferator-activated receptor (PPAR) γ coactivator 1 (PGC-1) (10), was identified, which distinguishes itself from other coactivators by means of its tissue-specific and regulated expression. Namely, PGC-1 is expressed in different levels in brown adipose tissue (BAT), skeletal muscle, heart, kidney, and brain and is markedly up-regulated in BAT after acute exposure to cold stress (10). PGC-1 is also up-regulated in the liver and heart under fasting conditions (11, 12).
PPARγ is known to be the key regulator of adipogenesis (13), and its expression is augmented during the differentiation of adipocytes (13). However, we noticed that PGC-1 mRNA remains at very low levels during adipocyte differentiation of 3T3-L1 cells. We speculated that other yet-unidentified PGC-1-related molecules might exist and may function during adipocyte differentiation. We then searched for such potential PGC-1-related molecules among ESTs and indeed found ESTs showing significant homology to PGC-1, and we were able to isolate full-length cDNAs containing these ESTs. In this article, we report on the in vitro and in vivo characterization and functional analyses of the PGC-1-related protein, designated PGC-1β/ERR ligand 1 (ERRL1); it acts as a protein ligand of ERRs, and its expression levels control energy expenditure.
Materials and Methods
Plasmids. We used amino acids 1–147 of GAL4 that were fused to the ligand-binding domains of the following nuclear receptors (14): mouse androgen receptor (amino acids 607–899, GenBank accession no. X59592), human estrogen receptor α (251–595, X03635), human glucocorticoid receptor (489–777, M10901), human retinoic acid receptor (RAR) α (126–432, X06538), human RXRα (222–462, X52773), mouse PPARα (156–468, X57638), human PPARγ1 (176–478, L40904), human pregnane X receptor (110–434, AF084645), human ERR1 (147–422, L38487), human ERR2 (171–433, X51417), human ERR3 (173–436, AF058291), human hepatocyte nuclear factor 4α (125–465, X76930), human neuron-derived orphan receptor (361–626, D78579), human Nur-related factor 1 (264–535, S77154), human retinoid-related orphan receptor α1 (140–523, U04897), human steroidogenic factor 1 (64–461, U76388), human chicken ovalbumin upstream promoter (156–423, X12795), human testicular receptor 2-11 (149–603, M29960), and human RevErbA (199–614, M24898). Mouse PGC-1α cDNA was obtained by screening a mouse embryo cDNA library.
Transcriptional Activation Assays. Cells were maintained in DMEM supplemented with 10% serum. Transfections and reporter assays were performed as described (14).
Chromatin Immunoprecipitation Assay. Chromatin immunoprecipitation assay were carried out by using a chromatin immunoprecipitation assay kit (Upstate Biotechnology, Lake Placid, NY), following the supplier's instructions. The primers were designed to amplify the region -524 to -226 of the MCAD promoter containing an ERR-binding site, namely by using 5′-GATTTCCTTCTCAGTCTCCT-3′ (forward primer) and 5′-CGGAGAAGAAGACTGTGTGC-3′ (reverse primer).
High-Fat Diet. Mice were fed a regular diet (Oriental Yeast, Tokyo) or a high-fat diet, which contained casein (20% wt/wt), α-corn-starch (30.2%), sucrose (10%), lard (25%), corn oil (5%), minerals (3.5%), vitamins (1%), cellulose powder (5%), and D, l-methionine (0.3%).
Determination of Energy Expenditure. Oxygen consumption and carbon dioxide production were determined by using an indirect calorimeter system composed of a mass analyzer and a computer (15). Locomotor activity was recorded automatically every 10 min by an Animex-III (Shimadzu) laid under each respiration chamber. Energy expenditure was calculated as described (15).
Measurement of β-Oxidation. The rate of β-oxidation of fatty acid in skeletal muscle and liver of PGC-1β mice and age-matched control mice was examined as described (16), with slight modifications.
Statistical Analyses. Statistical comparisons of data from two experimental groups were made by using Student's t test. Comparison of data from multiple groups was made by ANOVA, and each group was compared with the others by the Fisher's protected least significant difference test (statview 4.0, Abacus Concepts, Berkeley, CA). Statistical significance was defined as P < 0.05 or P < 0.01.
Results
PGC-1β/ERRL1 Is Closely Related to PGC-1. The longest clone obtained by our cDNA screening for PGC-1-related molecules contained a 3.4-kb cDNA encoding an ORF of 1,014 aa with a predicted molecular mass of 112 kDa. We originally designated this protein as ERRL1 based on its properties as a protein ligand of ERRs (as shown later in detail), but we have used the name of PGC-1β throughout this article, because, during preparation of this article, Lin et al. (17) reported the cloning of a PGC-1 homologue, named PGC-1β, which turned out to only have one amino acid difference from ERRL1. PGC-1β and PGC-1 [renamed PGC-1α (17)] show a high degree of amino acid similarity except for the central extension unique to PGC-1β. The homologous region can be divided into five domains based on sequence similarity and predicted functional properties (Fig. 1A). The N-terminal region of PGC-1β (amino acids 1–282) contains two LXXLL motifs, which are a proposed binding motif to nuclear receptors (9), and has 41% identity with PGC-1α. The second region, which is unique to PGC-1β, contains E (glutamic acid) repeats and one LXXLL motif. The third region (amino acids 580–656) is highly conserved (47% amino acid identity). The fourth region (amino acids 657–882) is less conserved (22% amino acid identity) and contains a very short serine/arginine-rich (SR) domain (10, 17) in PGC-1β compared with the long SR domain in PGC-1α. The C-terminal domain (amino acids 883-1014) has a putative RNA-binding domain (10, 17) and is highly conserved (52% amino acid identity).
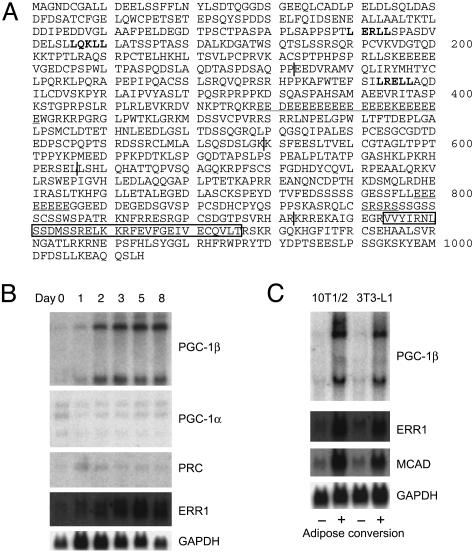
Fig. 1.
Amino acid sequence and the expression profile of ERRL1/PGC-1β.(A) LXXLL motifs are shown in bold. Glutamic acid (E) repeats and serine/arginine (SR)-rich regions are underlined. A putative RNA-binding motif is boxed. Vertical lines show tentative domain borders. A splice variant of ERRL1/PGC-1β was identified, which lacks 39 aa (from the 156th leucine to the 194th lysine). The 260th leucine is a proline in reported PGC-1β (17). (B) 3T3-L1 cells were induced to differentiate into adipocytes by treatment with dexamethasone, 1-methyl-3-isobutylxanthine, and insulin (day 0). RNAs were isolated on the days indicated, and Northern blot analysis was performed. The membrane was sequentially hybridized with the probes indicated. (C) Expression of PGC-1β, ERR1, MCAD, and a control GAPDH mRNA in 3T3-L1 and 10T1/2 cells. -, preadipocytes; +, mature adipocytes.
Similar Expression Patterns Between PGC-1β and ERR. We then examined the expression patterns of PGC-1β in different tissues from adult mice. Two PGC-1β mRNAs ≈10 kb and 4 kb in length were observed. PGC-1β mRNAs were abundant in brain, BAT, heart, kidney, and skeletal muscle, and were also detected in stomach and white adipose tissue (WAT) (data not shown and ref. 17). Consistent with a previous report (10), PGC-1α expression was elevated in mouse BAT after exposure to cold stress, but PGC-1β mRNAs were only marginally up-regulated by this stress. We noticed here that the expression pattern of PGC-1β closely resembles that of ERR1 (6), with both mRNAs being highly expressed in tissues that can use lipids as a source of cellular energy, e.g., BAT, heart, skeletal muscle, and kidney. We could not detect ERR2 mRNA expression in any of the tissues by the Northern blot analysis, which is consistent with a previous study (2).
PGC-1β mRNAs were present at very low levels in 3T3-L1 preadipocytes and were markedly induced during adipocyte differentiation (Fig. 1B). In contrast, mRNAs of PGC-1α and PGC-1-related coactivator (18), another PGC-1α-related molecule, remained at low levels during this adipogenesis (Fig. 1B). Similar augmentation of PGC-1β mRNAs was observed in mature adipocytes differentiated from 10T1/2 cells (Fig. 1C). Again, expression profiles of ERR1 and its target MCAD were quite similar to those of PGC-1β (Fig. 1 B and C); ERR2 and ERR3 mRNA expression was not detected in these cells.
PGC-1α as a Broad-Range Nuclear Receptor Agonist. We next examined whether PGC-1β can function as a coactivator of PPARγ, using PGC-1α as a control coactivator. Contrary to our expectations, we were unable to observe any evidence indicating the stimulation of PPARγ-mediated transcription by PGC-1β (data not shown). In the control experiments, we noticed that the expression of PGC-1α on its own was able to drastically stimulate PPARγ-mediated transcription without the addition of any exogenous lipophilic ligands. PGC-1α is known to physically interact not only with PPARγ but also with several other nuclear receptors (10, 11, 19). Furthermore, PGC-1α has been shown to be induced by certain environmental changes such as cold exposure (10) and fasting (11, 12). These observations raise the possibility that the expression of PGC-1α alone can stimulate multiple nuclear receptor pathways. This putative character is analogous to classical lipophilic ligands of the nuclear receptors. To address this possibility, we used nuclear receptors whose DNA-binding domains were replaced by the GAL4 DNA-binding domain, which allows the comparison of transcriptional activation profiles of various molecules with the same reporter such as (UAS)4-Luc. Indeed, PGC-1α was able to activate transcription by means of multiple nuclear receptors, including orphan HNF-4α, SF1, and ERRs (Fig. 2 A and B). Among these, PGC-1α most strongly activated HNF-4α, which is consistent with a recent report (11), followed by estrogen receptor α, SF1, ERRs, PPARs, pregnane X receptor, RARα, and RXRα (Fig. 2 A and B). These observations indicate that PGC-1α can function as a broad-range nuclear receptor agonist.
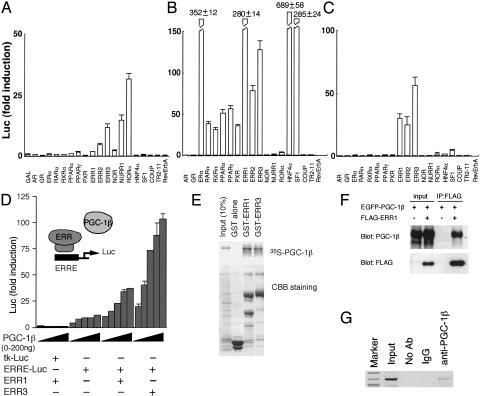
Fig. 2.
Profiles of PGC-1α and PGC-1β as protein ligands on specific nuclear receptors. (A–C) Transcriptional activation of various GAL4-fused nuclear receptors was examined in a transient transfection assay in the absence (A) or presence of PGC-1α (B) and PGC-1β (C). Mean values of triplicate experiments are shown as fold induction, where the Luc activity of nonfused GAL4 protein (GAL) as reference value (A), or where the Luc activity of GAL4-fused nuclear receptor in the absence of PGC-1α or PGC-1β serves as the reference values, which were obtained in the experiment shown in A (B and C). (Error bars, standard deviation.) (D) Dose-dependent activation of full-length ERR-mediated transcription via ERRE by PGC-1β. Mean values from triplicate experiments are shown as fold induction, where the Luc activity of TK-Luc in the absence of PGC-1β serves as the reference value. (Error bars, standard deviation.) (E) In vitro synthesized 35S-labeled PGC-1β shows strong interaction with GST-ERR1 and -ERR3. (F) Immunoprecipitation of PGC-1β with ERR1. HEK293T cells were transfected with pEGFP-PGC-1β with or without pCMX-FLAG-ERR1, cell lysates were immunoprecipitated with an anti-FLAG antibody, and immunoprecipitates were analyzed by Western blotting with anti-PGC-1β and anti-FLAG antibodies. (G) Chromatin immunoprecipitation assay on the MCAD promoter in 3T3-L1 cells using an anti-PGC-1β antibody. The chromatin from 3T3-L1 cells was cross-linked with formaldehyde and was immunoprecipitated with an anti-PGC-1β antibody. IgG was used as a nonrelated antibody for a negative control. The precipitated DNA was amplified by PCR by using the primers that cover the promoter region of the MCAD gene containing an ERR-responsive element.
PGC-1β as an ERR Protein Ligand. Using the same GAL4-fused nuclear receptor set, we next searched for potential partners of PGC-1β and observed that PGC-1β specifically activated ERR-mediated transcription: ERR3 was most strongly activated by PGC-1β, followed by ERR1 and ERR2 (Fig. 2 A and C). In this assay, we used strictly stripped serum. This might be a reason of loss of PGC-1β effect on glucocorticoid receptor-, TR-, and RAR-mediated transcription; PGC-1β has been recently reported to interact with glucocorticoid receptor, TR, and RAR in a ligand-dependent manner (17). We then tested the transcriptional activation properties of PGC-1β on full-length ERRs. Consistent with the experiments above, PGC-1β dose-dependently stimulated full-length ERR3- and full-length ERR1-mediated transcription by means of the ERR responsive element (ERRE) from the MCAD gene promoter (6, 7) (Fig. 2D).
We next tested whether PGC-1β physically interacts with ERRs. Matrix-bound GST-ERR1 and -ERR3, but not GST alone, retained radiolabeled PGC-1β efficiently (Fig. 2E). As reported previously (6, 7), we observed that ERRs could bind radiolabeled MCAD ERRE oligonucleotides in a gel mobility shift assay, and that the ERR-DNA complexes were supershifted by the addition of the PGC-1β protein (data not shown). Given that lipophilic hormones were not added either in the synthesis of the proteins or in the binding reactions, these results indicate that PGC-1β and ERRs can directly interact on the ERR target promoter DNAs without the help of lipophilic ligands. The interaction between PGC-1β and ERRs in the cultured cells was confirmed by the immunoprecipitation assay (Fig. 2F) and the chromatin immunoprecipitation assay (Fig. 2G). Similar results have recently demonstrated by Hentschke et al. (19). These results collectively demonstrate that PGC-1β can function as a protein ligand of ERRs and activate ERR-mediated transcription, at least in cultured cells.
Creation of PGC-1β Mice. To examine the effects of increased PGC-1β expression in vivo, we created PGC-1β transgenic mice, which we expected would exhibit phenotypes that mimic those induced by activated ERR-mediated transcription. The chicken β-actin promoter with the cytomegalovirus immediate early enhancer (the CAG promoter) (20) was used to drive the expression of the mouse PGC-1β transgene in mice (Fig. 3 A and B).
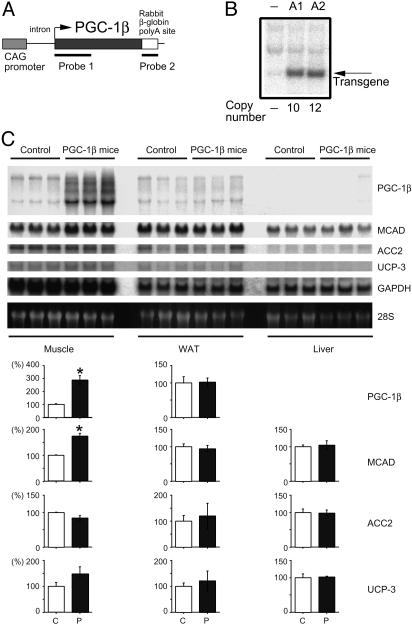
Fig. 3.
Creation of PGC-1β mice. (A) A schematic drawing of the PGC-1β transgene and the positions of probes used for Southern (probe 1) and Northern (probe 1 and 2) blots are shown. (B) Two transgenic lines (A1 and A2) were established, containing 10 and 12 copies of the transgene, respectively. (C) Northern blot analysis on the skeletal muscle, WAT, and liver from 8-week-old PGC-1β (line A1) and control mice. Each lane contained 20μg of total RNA. mRNA expression of PGC-1β, MCAD, ACC2, UCP-3, and control GAPDH were examined. Three age-matched male mice in each group were used. Quantification of Northern signals (control as 100%) is shown under the panels. Data are represented as the mean ± SEM (*, P < 0.05). C, control mice; P, PGC-1β mice.
Expression of the PGC-1β transgene was evaluated by Northern blot analysis. When a transgene-specific probe (Fig. 3A, Probe 2) was used, only a single 4-kb band was detected (data not shown). The CAG promoter has been reported to be strongly active in ubiquitous tissues (20). Unexpectedly, however, the use of this promoter resulted in high expression levels of the PGC-1β transgene in several restricted tissues such as brain, BAT, heart, skeletal muscle, and kidney, but a low-level of expression in liver, lung, and white adipose tissue (Fig. 3C and data not shown), although we observed a high expression in cultured mature adipocytes (Fig. 1 B and C). These expression profiles corresponded relatively well to the endogenous PGC-1β expression patterns, suggesting that tissue-specific regulatory cis-elements crucial for PGC-1β expression may reside in the PGC-1β cDNA used in this construct. This possibility remains to be clarified. The expression pattern of PGC-1α was not affected in PGC-1β mice (data not shown). As predicted from the in vitro data, the increased expression of PGC-1β enhanced MCAD mRNA expression in vivo, as shown in the skeletal muscle of PGC-1β mice (Fig. 3C). The increased expression of PGC-1β did not affect the expression of the ACC2 and the UCP-3 genes in the skeletal muscle (Fig. 3C), showing specificity.
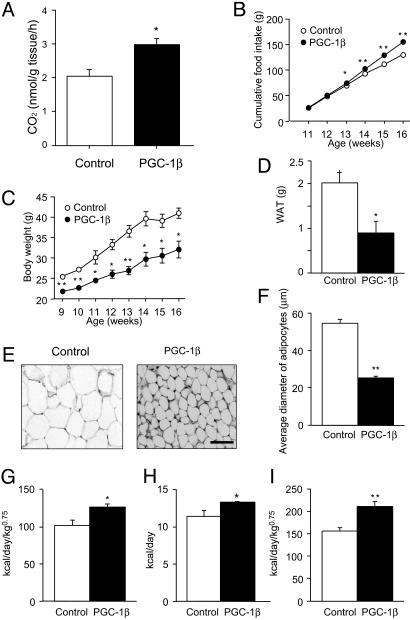
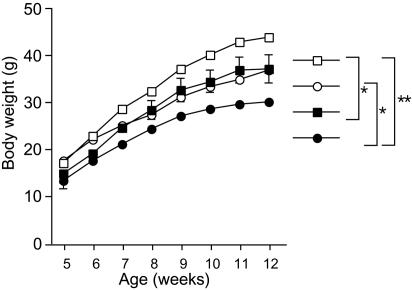
PGC-1β Mice Are Hyperphagic but Lean. The apparent phenotype observed in PGC-1β mice was leanness, which agreed well with the phenotype predicted from increased β-oxidation of fatty acids (Fig. 4A). This phenotype was more prominent when mice were fed on a high-fat diet. Groups of six male wild-type control mice and six male PGC-1β mice, at 9 weeks of age, were freely fed on a high-fat diet, and the food consumption and weight of each mouse were measured every week. PGC-1β mice consumed significantly more food than the control mice (Fig. 4B). However, the transgenic mice weighed 15–25% less than the control mice before and throughout the feeding periods (Fig. 4C), and they accumulated significantly less fat in their adipose tissues (data not shown). The epididymal WAT in the PGC-1β mice weighed only 0.92 ± 0.28 g, compared with 2.0 ± 0.30 g in control mice (Fig. 4D and Table 1). In contrast, liver weights were not significantly different (PGC-1β mice, 1.23 ± 0.06 g; control mice, 1.4 ± 0.08 g). The adipose cells of transgenic mice were smaller than those of the control mice (average diameter of adipocytes in PGC-1β mice, 25.1 ± 1.1 μm; control mice, 54.5 ± 2.2 μm; Fig. 4 E and F). We also observed significant reductions in body weight and adipose tissue mass in the A2 line of PGC-1β mice (data not shown).
Fig. 4.
Phenotypes of PGC-1β mice. (A) Increased β-oxidation in the skeletal muscle of PGC-1β mice. No increase of β-oxidation was observed in the liver of PGC-1β mice (data not shown). (B) Food intake was measured every week and expressed as cumulative food intake per mouse during the indicated period. Data are means ± SEM (n = 6 per animal group; error bars are smaller than the symbols; *, P < 0.05; **, P < 0.01). (C) PGC-1β mice (•) weighed significantly less than their wild-type controls (○). Values represent mean body weight ± SEM (n = 6 per animal group; *, P < 0.05; **, P < 0.01). For some data points, the error bars are smaller than the symbols. (D) Comparison of epididymal WAT weight between PGC-1β mice and wild-type control mice. Columns represent mean values of WAT weight ± SEM (n = 6 per animal group; *, P < 0.05). (E) Comparison of the morphology of WAT between PGC-1β mice and littermate wild-type control mice. (Scale bar = 50 μm.) (F) Average diameter of adipose cells. The diameters of the cells were measured from the sections shown in E (n = 20, **, P < 0.01). (G–I) Resting (G) and total (H and I) energy expenditures were significantly higher in PGC-1β mice than in control mice at 12 weeks of age. Columns represent mean values of energy expenditure ± SEM (n = 6 per animal group; *, P < 0.05; **, P < 0.01).
Table 1. Body and WAT weights and serum measurements of PGC-1β mice.
| Wild-type control mice (high-fat diet; n = 6) | PGC-1β mice (high-fat diet; n = 6) | Wild-type control mice (chow diet; n = 5) | PGC-1β mice (chow diet; n = 4) | |
|---|---|---|---|---|
| Triglyceride, mg/dl | 51.2 ± 14.9 | 30 ± 2.7 | 48.2 ± 4.8 | 54.0 ± 5.1 |
| Nonesterized fatty acid, mEQ/l | 0.733 ± 0.069 | 0.61 ± 0.1 | 0.738 ± 0.1 | 0.64 ± 0.05 |
| Bilirubin, mg/dl | 0.53 ± 0.084 | 0.58 ± 0.18 | 0.7 ± 0.084 | 0.48 ± 0.1 |
| Blood glucose, mg/dl | 233 ± 26.5 | 195 ± 8.4 | 173 ± 8.6* | 183 ± 3.1 |
| Total cholesterol, mg/dl | 152 ± 9.3 | 138.7 ± 12 | 86.2 ± 2.6*** | 98.0 ± 5.4*** |
| Leptin, ng/ml | 25.0 ± 6.6 | 5.9 ± 3.5* | 11.4 ± 5.3 | 0.9 ± 0.64** |
| Insulin, ng/ml | 5.5 ± 1.4 | 1.3 ± 0.3** | 1.3 ± 0.5** | 0.33 ± 0.16*** |
| WAT weight, g | 2.0 ± 0.3 | 0.92 ± 0.28** | 1.68 ± 0.18 | 0.44 ± 0.18** |
| Body weight, g | 41.0 ± 2.0 | 32.1 ± 2.8** | 36.2 ± 1.1 | 28.6 ± 1.1** |
Sixteen-week-old male mice fed indicated diet underwent a 5-h fast before collection of serum. Data are expressed as mean ± SEM. P values between the group of wild-type controls on a high-fat diet and the group of PGC-1β mice on a high-fat diet, or between the group of wild-type controls on a high-fat diet and the group of wild-type control mice on a chow diet, or between the group of wild-type controls on a high-fat diet and the group of PGC-1β mice on a chow diet (*, P < 0.05; **, P < 0.01; ***, P < 0.001) were calculated by Fisher's PLSD test.
Blood was collected from PGC-1β and control mice, and serum components were analyzed (Table 1). The decrease in adipose mass appeared to lead to a decrease in the amount of leptin in the serum. As leptin is an antiappetite hormone secreted from WAT (21), the decreased level of leptin observed in PGC-1β mice is consistent with the observation that they are hyperphagic. Furthermore, decreased levels of insulin, which were equivalent to the normal levels, were observed in transgenic mice compared with control mice when fed a high-fat diet. Mice, when fed a high-fat diet, generally develop resistance to insulin and impaired glucose uptake, even with a high plasma insulin concentration (22). A similar state, which is called insulin resistance, is well observed in humans with obesity or who are suffering from type 2 diabetes (23). The decreased body weight and decreased insulin levels observed in PGC-1β mice indicate that the increased expression of PGC-1β can antagonize obesity and contribute to an antidiabetic state without suffering insulin resistance, even taking a high-calorie diet.
Increased Energy Expenditure in PGC-1β Mice. We next examined the energy expenditure of PGC-1β mice. Gas analyses of respiration chambers, each containing a single mouse, were carried out, and energy expenditure was calculated. Indeed, the energy expenditures of PGC-1β mice were significantly higher than those of control mice at 12 weeks of age (resting energy expenditure: PGC-1β mice, 126.3 ± 3.8 kcal/day/kg0.75; control mice, 101.5 ± 6.7 kcal/day/kg0.75; P < 0.05, n = 6, per group; and total energy expenditure: PGC-1β mice, 211.2 ± 10.7 kcal/day/kg0.75; control mice, 155.9 ± 7.8 kcal/day/kg0.75; P < 0.01, n = 6, per group), except for marginal difference in resting energy expenditure by another calculation without considering body weights (resting energy expenditure: PGC-1β mice, 7.8 ± 0.46 kcal/day; control mice, 7.2 ± 0.71 kcal/day; P = 0.62, n = 6, per group (not shown); and total energy expenditure: PGC-1β mice, 13.0 ± 0.19 kcal/day; control mice, 11.1 ± 0.80 kcal/day; P < 0.05, n = 6, per group) (Fig. 4 G–I). There were no significant differences in respiratory quotient (PGC-1β mice, 0.79 ± 0.01; control mice, 0.76 ± 0.02, P = 0.22 and n = 6 per group). Previous reports showed that respiratory quotient is generally low (0.7–0.8) in high-fat-fed rodents (24). This may be a reason why we could not observe significant differences in respiratory quotient between PGC-1β and control mice, because both mice were fed on a high-fat diet. Locomotor activity was not significantly different (PGC-1β mice, 7,338 ± 700 counts; control mice, 5,338 ± 390 counts, the values representing the sum of all transits over a 24-h period, P = 0.08). These results indicate that the differences in body weights between control and PGC-1β mice are most likely to be due to a difference in their energy expenditure.
PGC-1β Expression Can Antagonize Genetically Programmed Obesity. We next bred PGC-1β mice and KKAy mice to examine whether the obese phenotype of KKAy mice could be counteracted by increased expression of PGC-1β. KKAy mice have a mutation in Ay, which leads to ectopic expression of the agouti coat-color protein, causing a dominantly inherited syndrome of obesity and yellow fur (25). The agouti protein acts as an antagonist for the melanocortine receptor, which is considered to act as an appetite repressor in the central nervous system (25). KKAy males were bred with PGC-1β females, and their offspring, were selected by coat color [yellow, KKAy(+); black, KKAy(-)] and PGC-1β transgene expression. Comparison of body weights of each group revealed that the PGC-1β transgene significantly reduced the body weight increases of KKAy(+) mice (Fig. 5), demonstrating that increased PGC-1β expression can even antagonize obesity caused by a genetic abnormality.
Fig. 5.
Breeding of PGC-1β mice and genetically obese KKAy mice. Body weights of KKAy (+) PGC-1β (-)(□; n = 8), KKAy (-) PGC-1β (-)(○; n = 5), KKAy (+) PGC-1β (+) (▪; n = 5), and KKAy (-) PGC-1β (+) (•; n = 5) male mice were measured weekly. Values represent mean body weight ± SEM. For some data points, the error bars are smaller than the symbols. Body weight curves of each group were compared by one-way repeated-measures ANOVA. *, P < 0.05; **, P < 0.01.
Discussion
In this article, we present lines of evidence demonstrating a previously undescribed category of proteins that activate nuclear receptors via direct binding, in a way similar to the activation of nuclear receptors by classical lipophilic ligands. We therefore propose that these proteins are protein ligands of the nuclear receptors. These protein ligands include at least two related proteins, PGC-1β/ERRL1 and PGC-1α. PGC-1-related coactivator may be a third member, but this possibility remains to be tested.
PGC-1β appears to specifically function as an ERR ligand in our experiments. PGC-1β expression profile was quite similar to those of ERR1 and MCAD, the gene encoding for a pivotal enzyme of mitochondrial β-oxidation and a known target gene of ERR1. Consistent with their similar expression profiles, PGC-1β was found to be able to directly bind to ERRs without the help of any exogenous lipophilic hormones in vitro and is able to activate ERR-mediated transcription through the ERR-responsive element from the MCAD gene promoter in a transient Luc assay.
These results easily lead to the idea that PGC-1β is a regulator of ERR-mediated transcription in the control of energy expenditure. This idea is further supported by our in vivo studies using transgenic mice overexpressing PGC-1β. PGC-1β mice exhibited significantly higher energy expenditure and weighed significantly less than the control mice, although they are hyperphagic. Consistently, PGC-1β mice had less WAT than control mice. Furthermore, the endogenous MCAD gene was up-regulated in skeletal muscle in PGC-1β mice where the PGC-1β expression was elevated.
In contrast to PGC-1β, PGC-1α can potentially activate many nuclear receptors, from the classical estrogen receptors, RARs, RXRs, PPARs, and pregnane X receptor to the orphan HNF-4α, SF1, and ERRs, and is thus predicted to function as a wide-range nuclear receptor agonist. One of the most drastic changes usually encountered by mammals is changes in environmental temperature, which fluctuate seasonally, daily, and even hourly. It is easily imaginable that the endocrine system contributes toward adaptation to seasonal temperature changes. It is reasonable to speculate that PGC-1α plays a pivotal role in adapting to a rapid temperature change via activating these receptors simultaneously by simply augmenting its expression level. Consistently, PGC-1α transgenic mice are also lean (Y.K. and A.K., unpublished work).
These observations clearly show that a protein ligand for nuclear receptors is defined as a protein whose expression overlaps with its target receptor(s) and can directly bind to its target and activate its target receptor(s)-mediated transcription. This protein ligand system could be a yet-unknown common mechanism underlying the control of many orphan receptors. One of the prominently advantageous aspects of such protein ligands is the ability to control their expression levels more easily compared with classical lipophilic ligands (see the introduction), allowing animals to adapt to rapid changes in internal and external environments. Given that both PGC-1α and PGC-1β also show constitutive expression in certain tissues, protein modifications such as phosphorylation and acetylation, are another potential mechanism to quickly activate nuclear receptors via protein ligands. Such a mutually compensatory nature of two ligand systems (using lipophilic and protein ligands) allows a single receptor to respond to both quick and long-lasting changes in the environment. The other advantageous aspect of protein ligands is the ability to control their expression in restricted tissues.
Finally, this study may provide a concept of a novel category of drugs that contribute to antiobese and antidiabetic functions. When the expression of PGC-1β in skeletal muscle is slightly augmented, as is typically observed in PGC-1β mice, total energy expenditures are increased up to 1.3 times, and, as a result, less fat is accumulated and stored. Then, the pharmacological activation of PGC-1β is expected to allow individuals to lose weight while maintaining a normal or even advanced caloric intake, which hence is an ideal target for the development of new antiobesity therapeutics. How then should such antiobesity drugs work? One potential mode of action would be to augment PGC-1β expression. Another potential mode of action would be the direct activation of ERRs, in the same way that classical lipophilic ligands act. A third potential role for such drugs would be to augment the interaction between PGC-1β and ERRs, because PGC-1β and ERRs are already expressed at significant levels in skeletal muscle, for example.
Acknowledgments
We thank T. Nakane, K. Okazaki, A. Fujimoto, Y. Kai, S. Ikeda, N. Tsuboyama-Kasaoka, S. Miura, Y. Yamamoto, N. Sarukura, and K. Yasuda for technical assistance. We thank H. A. Popiel for proofreading this article. We dedicate this article to K. Umesono.
Abbreviations: ERR, estrogen receptor-related receptor; ERRL1, ERR ligand 1; PPAR, peroxisome proliferator-activated receptor; PGC-1, PPARγ coactivator 1; MCAD, medium-chain acyl CoA dehydrogenase; BAT, brown adipose tissue; WAT, white adipose tissue; RAR, retinoic acid receptor.
References
- 1.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P. & Evans, R. M. (1995) Cell 83, 835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giguere, V., Yang, N., Segui, P. & Evans, R. M. (1988) Nature 331, 91-94. [DOI] [PubMed] [Google Scholar]
- 3.Shigeta, H., Zuo, W., Yang, N., DiAugustine, R. & Teng, C. T. (1997) J. Mol. Endocrinol. 19, 299-309. [DOI] [PubMed] [Google Scholar]
- 4.Eudy, J. D., Yao, S., Weston, M. D., Ma-Edmonds, M., Talmadge, C. B., Cheng, J. J., Kimberling, W. J. & Sumegi, J. (1998) Genomics 50, 382-384. [DOI] [PubMed] [Google Scholar]
- 5.Hong, H., Yang, L. & Stallcup, M. R. (1999) J. Biol. Chem. 274, 22618-22626. [DOI] [PubMed] [Google Scholar]
- 6.Sladek, R., Bader, J. A. & Giguere, V. (1997) Mol. Cell. Biol. 17, 5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vega, R. B. & Kelly, D. P. (1997) J. Biol. Chem. 272, 31693-31699. [DOI] [PubMed] [Google Scholar]
- 8.Kamei, Y., Xu, L., Heinzel, T., Torchia, J., Kurokawa, R., Gloss, B., Lin, S. C., Heyman, R. A., Rose, D. W., Glass, C. K. & Rosenfeld, M. G. (1996) Cell 85, 403-414. [DOI] [PubMed] [Google Scholar]
- 9.Glass, C. K. & Rosenfeld, M. G. (2000) Genes Dev. 14, 121-141. [PubMed] [Google Scholar]
- 10.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829-839. [DOI] [PubMed] [Google Scholar]
- 11.Yoon, J. C., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C. R., Granner, D. K., et al. (2001) Nature 413, 131-138. [DOI] [PubMed] [Google Scholar]
- 12.Lehman, J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M. & Kelly, D. P. (2000) J. Clin. Invest. 106, 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tontonoz, P., Hu, E. & Spiegelman, B. M. (1994) Cell 79, 1147-1156. [DOI] [PubMed] [Google Scholar]
- 14.Takada, I., Yu, R. T., Xu, H. E., Lambert, M. H., Montana, V. G., Kliewer, S. A., Evans, R. M. & Umesono, K. (2000) Mol. Endocrinol. 14, 733-740. [DOI] [PubMed] [Google Scholar]
- 15.Komenami, N., Yamamoto, Y. & Miyoshi, M. (1995) J. Nutr. Sci. Vitaminol. (Tokyo) 41, 395-407. [DOI] [PubMed] [Google Scholar]
- 16.Odle, J., Benevenga, N. J. & Crenshaw, T. D. (1991) J. Nutr. 121, 605-614. [DOI] [PubMed] [Google Scholar]
- 17.Lin, J., Puigserver, P., Donovan, J., Tarr, P. & Spiegelman, B. M. (2002) J. Biol. Chem. 277, 1645-1648. [DOI] [PubMed] [Google Scholar]
- 18.Andersson, U. & Scarpulla, R. C. (2001) Mol. Cell. Biol. 21, 3738-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentschke, M., Susens, U. & Borgmeyer, U. (2002) Biochem. Biophys. Res. Commun. 299, 872-879. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193-200. [DOI] [PubMed] [Google Scholar]
- 21.Friedman, J. M. (2000) Nature 404, 632-634. [DOI] [PubMed] [Google Scholar]
- 22.Li, B., Nolte, L. A., Ju, J. S., Han, D. H., Coleman, T., Holloszy, J. O. & Semenkovich, C. F. (2000) Nat. Med. 6, 1115-1120. [DOI] [PubMed] [Google Scholar]
- 23.Lovejoy, J. C. (1999) Curr. Atheroscler. Rep. 1, 215-220. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell, G. M., Nobbs, S. & Bates, D. J. (1987) Am. J. Physiol. 253, E264-E270. [DOI] [PubMed] [Google Scholar]
- 25.Siracusa, L. D. (1994) Trends Genet. 10, 423-428. [DOI] [PubMed] [Google Scholar]