Abstract
Y box-binding protein 1 (YB-1) is a multifunctional protein that can act as a regulator of transcription and of translation. In chicken embryo fibroblasts transformed by the oncoproteins P3k (phosphatidylinositol 3-kinase) or Akt, YB-1 is transcriptionally down-regulated. Expression of YB-1 from a retroviral vector induces a strong cellular resistance to transformation by P3k or Akt but does not affect sensitivity to transformation by other oncoproteins, such as Src, Jun, or Qin. The YB-1-expressing cells assume a tightly adherent, flat phenotype, with YB-1 localized in the cytoplasm, and show a greatly reduced saturation density. Both cap-dependent and cap-independent translation is inhibited in these cells, but the activity of Akt remains unaffected, suggesting that YB-1 functions downstream of Akt. A YB-1 protein with a loss-of-function mutation in the RNA-binding motif no longer binds to the mRNA cap structure, is localized in the cell nucleus, does not induce the flat cellular phenotype, and fails to interfere with P3k- or Akt-induced oncogenic transformation. This mutant also does not inhibit cap-dependent or cap-independent translation. These results suggest that YB-1 acts like a rapamycin mimic, inhibiting translational events that are required in phosphatidylinositol 3-kinase-driven oncogenic transformation.
Keywords: Akt, p3k, cell transformation, TOR
P3k and Akt were originally described as oncoproteins encoded by two highly tumorigenic retroviruses (1, 2). P3k is a homolog of the catalytic subunit p110 of phosphatidylinositol 3-kinase and controls the activity of the Ser/Thr protein kinase Akt, also referred to as protein kinase B. Both proteins are inherently oncogenic. The phosphatidylinositol 3-kinase–Akt-signaling pathway affects numerous and diverse cellular functions, many related to growth, survival, and differentiation (3–6). A downstream target of Akt is the Ser/Thr kinase TOR, which regulates translation by targeting two proteins: the translation initiation factor 4E-binding protein 1 (4E-BP1 or PHAS-1) and the p70 S6 kinase (S6K) (7–9). Hypophosphorylated 4E-BP1 binds to the initiation factor 4E and prevents the assembly of the translation initiation complex at the cap structure of mRNAs (10–12). After TOR-dependent phosphorylation, 4E-BP1 no longer binds to 4E, freeing it for assembly of the initiation complex and for cap-binding. Phosphorylation by TOR activates S6K, which then phosphorylates the ribosomal protein S6, controlling the translation of 5′TOP mRNAs (mRNAs that contain an oligopyrimidine tract at their 5′ termini) (13–16). This class of mRNAs encodes ribosomal proteins and translation elongation factors; the oligopyrimidine tract coordinates translation in a growth-dependent fashion. TOR plays a critical role in P3k- and Akt-induced oncogenic transformation. Inhibition of TOR by rapamycin induces cellular resistance to transformation by these two oncoproteins and reduces the growth of tumors that depend on a gain of function in the phosphatidylinositol 3-kinase pathway (17–19). These data suggest that the TOR-dependent stimulation of translation is an essential component of P3k/Akt-mediated oncogenesis.
In a survey of genes that are differentially expressed in Akt-transformed chicken embryo fibroblasts (CEF), we identified Y box-binding protein 1 (YB-1) as a potential Akt target. YB-1 is a multifunctional protein; it binds to DNA and RNA and controls both transcription and translation. As a transcription factor, YB-1 binds to the Y box promoter element CTGATTGGC/TC/T, containing an inverted CCAAT box, and either activates or represses gene expression. Examples of genes that are transcriptionally activated by YB-1 are multidrug resistance gene 1 (MDR-1), matrix metalloproteinase 2 (MMP-2), and protein tyrosine phosphatase 1B (PTP-1B) (20–22). Genes that are transcriptionally repressed by YB-1 are MHC class II genes and collagen α1(I) (23–25). YB-1 binds double-stranded DNA with low affinity; it has much higher affinities for single-stranded DNA and single-stranded RNA (21, 26, 27). The association with RNA is not sequence-specific, but sites with a high G content are preferred (28). YB-1 is the predominant protein component of messenger ribonucleoprotein particles and has a dose-dependent effect on translation (29–31). Low concentrations promote translation, high concentrations inhibit (31–33). YB-1 is composed of three domains, an amino-terminal alanine-proline-rich region, a centrally located cold shock domain (CSD), and a carboxyl-terminal region characterized by four alternating clusters of basic and acidic amino acids (34, 35). The function of the amino-terminal domain is not known. The CSD facilitates binding to nucleic acids. It forms an antiparallel β-barrel, enfolding the nucleic acid like a chaperone protein (36). CSDs are conserved in evolution from prokaryotes to higher vertebrates. The CSDs of YB-1 proteins are 100% identical in all vertebrates studied and are 45% identical to the Escherichia coli CspA protein. The carboxyl-terminal domain of YB-1 may contribute to DNA/RNA binding and may also be a docking site for other proteins that interact with YB-1 (34, 37). In vitro studies measuring translational activities attribute an inhibitory activity to this domain; the mechanism by which it interferes with translation remains unclear (33, 38).
Here, we report that YB-1 is down-regulated at the transcriptional level by the Akt and P3k oncoproteins. Overexpression of YB-1 results in a complete and specific cellular resistance to oncogenic transformation by P3k or Akt. Akt activity in the resistant cells remains unaffected, suggesting that YB-1 functions downstream of Akt. Vector-expressed YB-1 inhibits cap-dependent and cap-independent translation and induces a flat, adherent cellular phenotype. A YB-1 mutant that fails to bind to RNA does not interfere with P3k-and Akt-induced transformation and does not affect protein synthesis or the cellular phenotype. These results suggest that YB-1 acts as a rapamycin mimic, controlling translational events that are essential for P3kand Akt-induced oncogenic transformation.
Materials and Methods
Cell Culture and Viral Infection. Primary cultures of CEF were prepared from White Leghorn embryos obtained from SPAFAS (Preston, CT). Stable transfections using 5 μg of plasmid DNA were carried out as described (39). Immunofluorescent staining was performed as described (40). The following viruses and oncogenes have been used in this study: PR-A (Prague strain of Rous sarcoma virus), v-src; and ASV17, v-jun (41, 42). The oncogenes c-qin, myr-p3kΔ72, and myr-aktΔ11–60 were expressed by using the avian retroviral vector RCAS (43–46).
Total Gene Analysis (TOGA) Assay and RT-PCR. Total RNA was isolated from CEF transformed by myr-p3kΔ72 and myr-aktΔ11–60 by using RNA STAT from Tel-Test (Friendswood, TX) according to the manufacturer's instructions. Cells from the same embryo transfected with only RCAS were used as a negative control. TOGA was performed at Digital Gene Technologies as described (47, 48). The relative expression levels of YB-1 in normal and transformed cells were determined by quantitative real-time RT-PCR with the ABI PRISM 7700 sequence detection system (PE Biosystems, Wellesley, MA). Each reaction mixture contained 50 pg of cDNA template, 5 μM forward and reverse primers, AmpliTaq Gold DNA polymerase, and the SYBR Green PCR master mix (Applied Biosystems). The primers were as follows: YB-1 5′-TGAACAAAAGAATTGGAACTGAAGA-3′ (forward) and YB-1 5′-CAACAGCAAAAAGCAAGCACTT-3′ (reverse). Each sample was amplified for 40 cycles; RT-PCRs were carried out three times. For each cDNA template, the cycle threshold necessary to detect the amplified product was normalized to the cycle-threshold values of a control gene, glyceraldehyde-3-phosphate dehydrogenase.
Plasmids. pHCV-IRES encoding the bicistronic reporter CAT/luc has been described (49) and was kindly provided by Y. Nakamura (University of Tokyo). pFBnFLAG was generated by replacing the SalI/NotI fragment containing the multiple cloning site of pFB (Stratagene) with a multicloning site (MCS) encoding the FLAG epitope flanked by SfiI(A) and SfiI(B) sites. cDNA encoding the chicken full-length YB-1 protein was isolated from CEF by PCR using the oligonucleotides 5′-GCGGCCGCCACCATGAGCAGCGAGGCCGAGACCCAGCCGCCC-3′ and 5′-CTCGAGTTACTCAGCCCCGCCCTGCTCGGCCTCGGG-3′. The PCR fragment was cloned into TOPO (Invitrogen), excised with EcoRI, and cloned into the EcoRI sites of the adaptor vectors pBSFI (44) and pFBnFLAG, resulting in pBSFI-YB-1 and pFBnFLAG-YB-1, respectively. RCAS(B)-YB-1 and RCAS(B)-FLAG/YB-1 were generated by insertion of the SfiI fragment of pBSFI-YB-1, and pFBnFLAGYB-1 into the SfiI site of RCAS(B). A YB-1 mutant bearing two point mutations within the RNP-1 motif (amino acids 68–72, GYGFI → GAGAI) was constructed by using the pFBnFLAGYB-1 as a template for mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit, Stratagene). The YB-1 sequence containing an amino-terminal FLAG tag and the mutated RNP-1 motif was excised with SfiI and cloned into RCAS(B), resulting in RCAS(B)-FLAG/YB-1*.
Protein Analyses. Western blotting was done as described (50). The following primary antibodies were used: α-phospho-GSK-3β (Ser-9, Cell Signaling Technology, Beverly, MA), α-phospho-S6K (Thr-389, Cell Signaling Technology), α-phospho-4E-BP1 (Ser-65, NEB, Beverly, MA), α-hemagglutinin (monoclonal, Babco/Covance, Berkeley, CA), α-FLAG M5 (Sigma), α-4E (Cell Signaling Technology). 7-Methylguanosine triphosphate (m7GTP)-pull-down assays using 500 μg of protein together with 50 μl of m7GTP beads (Amersham Biosciences) were performed as published (51). For chloramphenicol acetyltransferase (CAT) and luciferase assays, 1.5 × 105 cells stably transfected with RCAS vectors were seeded into MP12 wells, grown overnight, and transfected (Lipofectamine Plus, GIBCO/BRL), each with 500 ng of pHCV-IRES, containing the bicistronic reporter CAT/luc and 5 ng of the Renilla luciferaseencoding vector pRLCMV (Promega). After overnight incubation in medium with 7.5% serum, cells were serum-starved for 24 h (0.25% serum) and incubated in medium with 15% serum for another 24 h. Cell lysates were harvested by using the Passive Lysis Buffer of the dual-luciferase reporter assay system from Promega. CAT assays (Quan-T-CAT kit from Amersham Biosciences) and luciferase assays were performed with 10 μg of protein.
Results
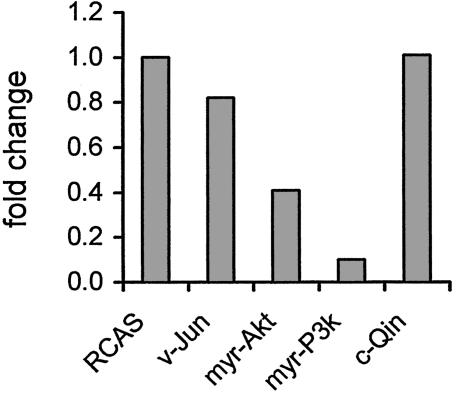
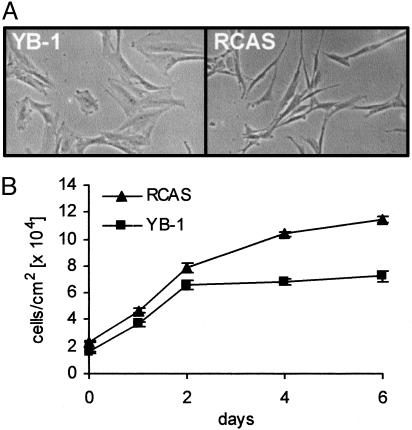
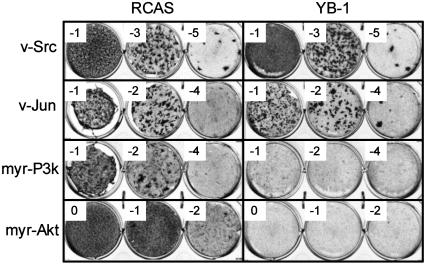
YB-1 Affects Cellular Morphology, Growth Potential, and Susceptibility to Oncogenic Transformation. We have determined the expression profiles of CEF transformed by the P3k or Akt oncoproteins by using the TOGA technology and identified YB-1 among the transcriptionally down-regulated genes. The TOGA result was confirmed by quantitative real-time PCR with the RNA used in the TOGA screen and RNA from a different chicken embryo (Fig. 1). The functional significance of reduced YB-1 expression in P3k- or Akt-induced oncogenic transformation was explored by expressing YB-1 with an amino-terminal FLAG tag from the RCAS retroviral vector. Control experiments had shown that the tagged construct and untagged YB-1 constructs behaved identically. Expression of the YB-1 protein was confirmed by Western blotting and immunofluorescent staining (data not shown). CEF expressing YB-1 changed shape and behavior. They became flat, showed fewer filopodia and increased spreading, and were more tightly attached to the supporting substrate (Fig. 2A). Although their initial growth rates did not differ significantly from that of control CEF, their final saturation density was only about half that of the controls (Fig. 2B). This result suggests that YB-1 enhances the cellular contact inhibition of growth. YB-1-expressing CEF were also used in an assay for oncogenic transformation. They proved solidly resistant to the P3k and Akt oncoproteins, while retaining undiminished sensitivity to the oncogenicity of Src, Jun, or Qin (Fig. 3 and Table 1). This resistance to transformation correlates with the expression profile of YB-1 in transformed cells, down-regulated by Akt and P3k but unaffected or enhanced by Src, Jun, or Qin, suggesting a specific inhibitory role for YB-1 in the P3k/Akt-signaling pathway.
Fig. 1.
Transcriptional expression of YB-1 in CEF transformed by various oncoproteins. Real-time PCR measuring YB-1 mRNA levels was performed as described in Materials and Methods. YB-1 mRNA expression in RCAS-transfected cells was designated 1.0.
Fig. 2.
Morphology and growth of YB-1-expressing cells. (A) A micrograph of YB-1-expressing CEF is shown (phase-contrast optics, ×6.3 objective lens). (B) Proliferation of YB-1-expressing cells. CEF were stably transfected with RCAS-YB-1 (▪) or with RCAS (▴) and cultured for 10 days. Then, equal numbers of cells were seeded onto 35-mm wells. The next day (day 0), and on days as indicated, cells were counted. The results represent the mean of three independent experiments; error bars mark the standard deviation.
Fig. 3.
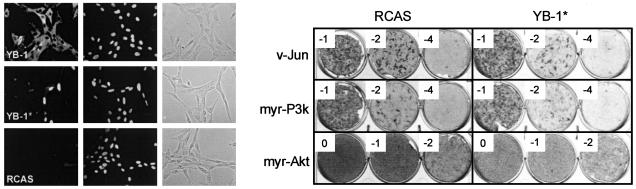
YB-1 mediated interference with oncogenic transformation. CEF were transfected with RCAS(B)-YB-1 and RCAS(B) and cultured for 10 days. Then, equal numbers of cells were seeded onto 35-mm wells and infected with oncogenic viruses carrying subgroup A envelope proteins as indicated. The log10 of the virus dilution is shown in the corner of each well. Cells were fed with agar medium every 2 days and stained with 2% crystal violet after 13 (v-Src) and 17 (v-Jun, myr-P3k, and myr-Akt) days, respectively.
Table 1. Effects of YB-1 on transforming activities of various oncoproteins in CEF.
| RCAS
|
YB-1
|
|||
|---|---|---|---|---|
| Oncoprotein | FFU/ml | EOT | FFU/ml | EOT |
| v-Jun | 2.3 × 105 | 1.00 | 2.3 × 105 | 1.00 |
| myr-P3k | 1.7 × 105 | 1.00 | 1.0 × 103 | 0.01 |
| myr-Akt | 6.5 × 105 | 1.00 | 4.0 × 104 | 0.06 |
| c-Qin | 1.8 × 104 | 1.00 | 2.4 × 104 | 1.33 |
| v-Src | 5.7 × 105 | 1.00 | 7.5 × 105 | 1.32 |
FFU, focus-forming units; EOT, efficiency of transformation (FFU/ml on YB-1-expressing cells divided by FFU/ml on RCAS controls).
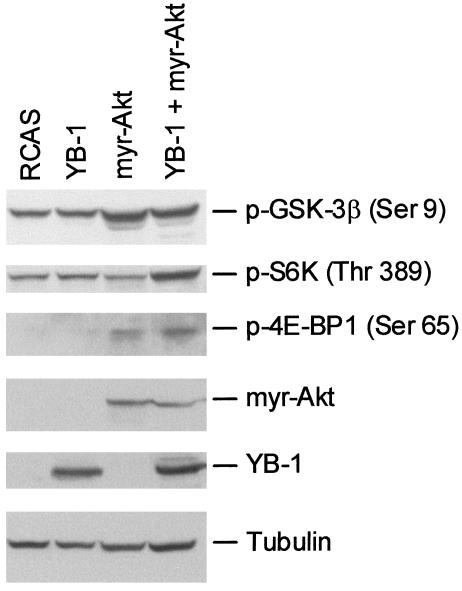
Expression of YB-1 Does Not Interfere with the Activity of Akt. The P3k and Akt oncoproteins lie in the same signaling pathway, with Akt positioned downstream of P3k. P3k-induced oncogenic transformation depends on Akt; a dominant negative Akt interferes with that transformation (44). The specific cellular resistance against P3k and Akt seen in YB-1-overexpressing CEF could therefore result from an inhibition of Akt. We evaluated Akt function by checking the extent of phosphorylation of Akt targets. These tests compared cells infected with vector alone, cells expressing YB-1, cells expressing myr-Akt, and cells expressing both YB-1 and myr-Akt. Fig. 4 shows that the levels of phosphorylation in the Akt targets GSK-3β, S6K, and 4E-BP1 were not detectably reduced by the expression of YB-1. These data suggest that Akt remains active in YB-1-expressing cells. We conclude that YB-1 interferes with the oncogenic signal downstream of Akt and downstream of principal Akt targets.
Fig. 4.
Phosphorylation of Akt targets in YB-1-expressing cells. Protein extracts were taken from actively dividing CEF transfected with RCAS expressing YB-1 or myr-Akt inserts, and from CEF transfected with RCAS-YB-1 plus RCAS-myr-Akt. Lysates were probed with phosphospecific antibodies directed against proteins as shown. Overexpression of myr-Akt and YB-1 proteins was confirmed by using hemagglutinin- and FLAG-specific antibodies, respectively. Tubulin served as loading control.
The RNA-Binding Motif of YB-1 Is Required for Interference with P3k and Akt. Oncogenic transformation by P3k or Akt depends on the activity of the TOR kinase, which controls rates of translation through the activities of 4E-BP1 and S6K (10, 12, 15). Inhibition of TOR by rapamycin induces a specific cellular resistance to transformation by P3k and Akt (17). This resistance probably reflects defective signaling from TOR to the translational machinery. YB-1 also has a controlling function in protein synthesis (31–33). The similarities between rapamycin- and YB-1-induced resistance to transformation prompted us to check whether YB-1 might affect cellular susceptibility to P3k and Akt through its control of translation. We therefore generated a YB-1 mutant that fails to bind to mRNA. Following published studies, we introduced two amino acid substitutions in the RNA-binding motif of YB-1, replacing the sequence GYGFI with GAGAI (52, 53). This mutant will be referred to as YB-1*. We expressed YB-1 and YB-1* in CEF, confirmed the expression by Western analysis (data not shown), and performed immunofluorescent staining to determine subcellular localization. As expected of a protein that is abundant in messenger ribonucleoprotein particles, YB-1 was found almost exclusively in the cytoplasm, which agrees with earlier reports (Fig. 5 Left) (54–56). In contrast, YB-1* was exclusively nuclear. This observation suggests that the ability to bind to mRNA is necessary for the cytoplasmic localization of YB-1. Without a functional RNA-binding motif, YB-1 translocates to the nucleus. The carboxyl-terminal domain of YB-1 contains arginine-rich sequences that could function as nuclear translocation signals. The mutant YB-1* also failed to induce the flat, adherent cellular phenotype, did not affect cellular saturation density, and no longer induced resistance to oncogenic transformation by P3k and Akt (Fig. 5 Right). These observations suggest that binding to mRNA and, by implication, control of translation, is indispensable for the observed effects of YB-1 on the cellular phenotype: increased adherence, reduced growth potential, and complete resistance to P3k- and Akt-induced transformation.
Fig. 5.
YB-1* with a mutated RNA-binding motif. (Left) CEF expressing either FLAG-tagged YB-1 or YB-1* proteins were fixed in 3.7% formaldehyde and treated with anti-FLAG and FITC-conjugated secondary antibody as shown on the left. 4′,6-Diamidino-2-phenylindole (DAPI)-stained nuclei and phase-contrast images are shown in the center and right, respectively. RCAS-transfected cells were used as a negative control. (Right) YB-1* in a transformation interference assay. Equal numbers of CEF transfected with RCAS-YB-1* and vector only were seeded onto 35-mm wells and challenged with transforming viruses encoding v-Jun, myr-Akt, and myr-P3k. Numbers in the corner of each well indicate the log10 of viral dilutions.
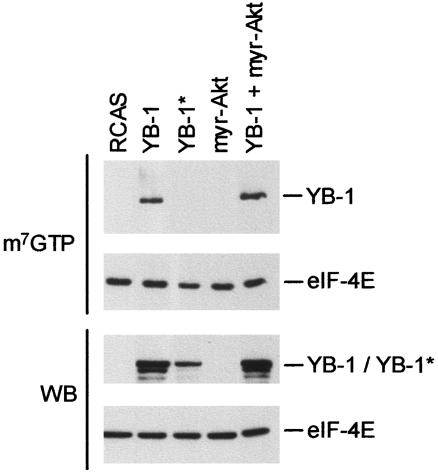
YB-1 Binds to the Cap Structure of mRNA. A recent study found that YB-1 can bind to the cap structure of mRNA in vitro, competing with the cap-binding translational initiation factor 4E (38). We tested for competition between 4E and YB-1 in cap-binding by using a m7GTP pull-down assay. m7GTP-Sepharose beads mimic the cap region of mRNA, are recognized by cap-binding proteins, and can be used for the isolation of such proteins (51, 57). We performed this procedure with cells expressing YB-1 and YB-1* and identified the m7GTP-bound proteins in Western blots with anti-FLAG and anti-4E antibodies. As can be seen in Fig. 6, YB-1 binds to m7GTP. In contrast, YB-1* fails to bind. Even long exposures of the blot did not reveal YB-1* pulled down by the m7GTP beads, yet both YB-1 and YB-1* were abundantly present in the total cell lysates used in the pull-down experiments. Binding of 4E to the beads was also found in extracts of YB-1- and YB-1*-expressing cells. Compared with control cells transfected with empty vector, the lysates of YB-1- and YB-1*-expressing cells showed no detectable reduction in the 4E–m7GTP interaction. This apparent failure of YB-1 to compete with 4E could be due to the excess of m7GTP beads used in the test or to the different affinities of YB-1 and 4E for the cap structure. However, the fact that YB-1 binds to the cap makes it a potential competitor of 4E.
Fig. 6.
Cap-binding activity of YB-1. Lysates of CEF expressing the indicated proteins and CEF transfected with RCAS only were analyzed in a m7GTP pull-down assay. Bound FLAG-YB-1 and 4E proteins were detected by Western blotting. A Western blot (WB) analysis using 40 μg of total protein was carried out to confirm the expression of nonbound proteins. eIF, eukaryotic initiation factor.
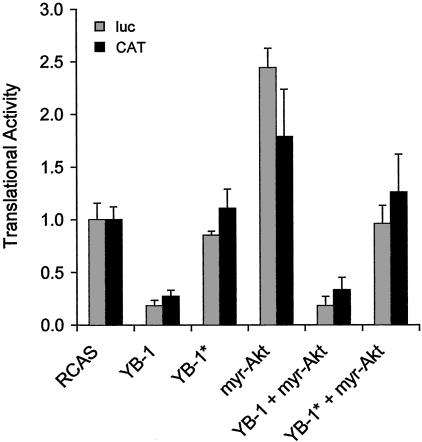
YB-1 Inhibits Cap-Dependent and Cap-Independent Translation. YB-1 can inhibit protein synthesis in vivo and in vitro, but available information does not discriminate between effects on cap-dependent and cap-independent translation (31–33). We measured the effect of YB-1 on cap-dependent and cap-independent translation with the bicistronic reporter construct pHCV-IRES (49). In this reporter, the CAT gene is expressed from the cytomegalovirus promoter by cap-dependent translation; the luciferase gene is translated cap-independently by using an internal ribosome entry site (IRES). We transiently transfected pHCV-IRES into CEF expressing various combinations of YB-1, YB-1*, and Akt and determined efficiencies of translation for the cap-dependent CAT and the cap-independent luciferase (Fig. 7). YB-1 reduced both cap-dependent and cap-independent translation ≈4-fold. Even in the presence of myristylated Akt, translation remained diminished, indicating that the YB-1-induced phenotype prevailed and confirming the conclusion that YB-1 functions downstream of Akt and of principal Akt targets. Because CAT/luc mRNA levels remained unchanged, a possible inhibitory effect on the transcriptional expression of the reporter could be excluded (data not shown). The highly oncogenic myristylated form of Akt enhanced translation, activating cap-dependent and cap-independent mechanisms. The increase in cap-dependent translation agrees with the observation that Akt induces a TOR-mediated inactivation of 4E-BP1, thereby promoting initiation of translation. In contrast to YB-1, YB-1* did not reduce translation, suggesting that mRNA binding is required for the inhibitory effect of YB-1. In the presence of YB-1*, however, Akt did not enhance translation. This finding could be due to a remaining inhibitory activity of YB-1*, because the carboxyl terminus of YB-1 itself is able to reduce translation in vitro (33).
Fig. 7.
YB-1 inhibits cap-dependent and cap-independent translation. CEF, expressing proteins encoded by RCAS vectors as indicated, were transiently transfected with pHCV-IRES containing the bicistronic reporter CAT/luc. Luciferase activity reflects internal ribosome entry site-dependent translation ( ); CAT activity represents cap-dependent translation (▪). Experiments were carried out three times; standard deviations are shown as error bars. The translational activity of cells transfected with the empty vector was designated 1.0.
); CAT activity represents cap-dependent translation (▪). Experiments were carried out three times; standard deviations are shown as error bars. The translational activity of cells transfected with the empty vector was designated 1.0.
Discussion
YB-1 can function as a regulator of transcription and of translation. It is also involved in the cellular responses to stress and DNA damage; a possible role in DNA repair and apoptosis has been suggested (54, 58). Our data suggest that an inhibitory effect of YB-1 on translation and protein synthesis mediates interference with oncogenic transformation. The resistance to transformation is correlated with the ability of YB-1 to bind to the cap structure of mRNA and with inhibition of translation; it also requires an intact RNA-binding motif in YB-1. The mechanism by which YB-1 affects protein synthesis is not completely understood. In rabbit reticulocyte lysates, low concentrations of YB-1 promote translation; high concentrations block it (31, 33). YB-1 may prevent the assembly of the translational initiation complex 4F at the mRNA by competing with the 4E initiation factor for binding to the cap structure (33, 38). This activity would explain interference with cap-dependent but not with cap-independent translation. Both types of translation were inhibited in our reporter assays. Besides competing with 4E, YB-1 must therefore affect other essential components of the translational machinery. Pull-down experiments with m7GTP beads showed cap binding by YB-1 but did not reveal competition between YB-1 and 4E for the cap structure. Further work is required to explain this unexpected result.
Transcriptional regulatory activities require nuclear localization, yet, in the YB-1-expressing CEF that are resistant to P3k and Akt-induced oncogenic transformation, YB-1 is exclusively cytoplasmic. Cytoplasmic localization of YB-1 may be a direct consequence of binding to mRNA. When RNA binding is impaired as in YB-1*, the protein translocates to the nucleus. The carboxyl-terminal region of YB-1 contains several potential nuclear localization signals. A recent study describes a truncated YB-1 protein of Chironomus tentans, termed p40, that lacks ≈50 amino acids at the carboxyl terminus (59). This protein becomes associated with pre-mRNA concomitantly with transcription and remains loaded onto the transcript during nucleocytoplasmic transport and during translation. YB-1 may behave in a similar fashion. This finding suggests that mRNA acts as the carrier molecule that transports YB-1 into the cytoplasm. Evidence exists that nuclear localization of YB-1 can result from association with p53, modification by kinases, or cleavage induced by thrombin (54–56). The decision between the principal activities of YB-1 appears to be governed by cellular localization, nuclear for transcriptional effects and cytoplasmic for translation.
YB-1 shows aberrant activity in human cancers, for instance in carcinomas of the breast and of the lung (60–62). In both types of cancers, YB-1 levels are increased. Yet these cancers also commonly show a gain of function in the phosphatidylinositol 3-kinase signaling pathway. The apparent contradiction to the results reported here is resolved by the predominantly nuclear localization of YB-1 in these human tumors. YB-1 may therefore function as transcriptional regulator in these situations. Among the transcriptional targets of YB-1 are the multidrug resistance gene 1 and matrix metalloproteinase 2, two genes important in cancer progression. On the other hand, adenocarcinomas of the lung in mice show transcriptional down-regulation of YB-1 (63). Differential expression of YB-1 in cancers may therefore be positive or negative, depending on the prevailing function of YB-1, transcriptional vs. translational, in a particular cell system.
YB-1 mimics some effects of rapamycin, presumably without affecting TOR directly. Both TOR and YB-1 are regulators of translation. TOR acts by phosphorylating 4E-BP1 and S6K, enhancing cap-dependent translation and translation of 5′TOP mRNAs. YB-1 regulates protein synthesis in a dose-dependent fashion by an unknown mechanism. TOR and YB-1 are both functionally dysregulated in P3k- or Akt-induced oncogenic transformation. TOR undergoes gain of function; YB-1 suffers loss of function. We speculate that the down-regulation of YB-1 facilitates protein synthetic activities that are required for P3kor Akt-induced oncogenic transformation.
Acknowledgments
We thank M. Aoki, B. Hilbush, and G. Denning for helpful comments, D. Lo and A. Warren for discussion, and H. Filoteo, A. Middlecamp, M. Callahan, I. Cisnero, and the TOGA Technology group at Digital Gene Technologies for excellent technical support. Y. Nakamura kindly provided the bicistronic reporter construct pHCV-IRES. This work was supported by National Institutes of Health Research Grants CA78230 and CA79616. A.G.B. is the recipient of Erwin Schrödinger Fellowships J2101 and J2278-B04 provided by the Austrian Science Foundation. This is manuscript no. 15887-MEM of The Scripps Research Institute.
Abbreviations: CEF, chicken embryo fibroblasts; CSD, cold shock domain; YB-1, Y box-binding protein 1; TOGA, total gene analysis; m7GTP, 7-methylguanosine triphosphate; CAT, chloramphenicol acetyltransferase; myr, myristyl.
References
- 1.Bellacosa, A., Testa, J. R., Staal, S. P. & Tsichlis, P. N. (1991) Science 254, 274-277. [DOI] [PubMed] [Google Scholar]
- 2.Chang, H. W., Aoki, M., Fruman, D., Auger, K. R., Bellacosa, A., Tsichlis, P. N., Cantley, L. C., Roberts, T. M. & Vogt, P. K. (1997) Science 276, 1848-1850. [DOI] [PubMed] [Google Scholar]
- 3.Alessi, D. R. & Downes, C. P. (1998) Biochim. Biophys. Acta 1436, 151-164. [DOI] [PubMed] [Google Scholar]
- 4.Wymann, M. P. & Pirola, L. (1998) Biochim. Biophys. Acta 1436, 127-150. [DOI] [PubMed] [Google Scholar]
- 5.Scheid, M. P. & Woodgett, J. R. (2001) Nat. Rev. Mol. Cell Biol. 2, 760-768. [DOI] [PubMed] [Google Scholar]
- 6.Brazil, D. P., Park, J. & Hemmings, B. A. (2002) Cell 111, 293-303. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S. & Schreiber, S. L. (1994) Nature 369, 756-758. [DOI] [PubMed] [Google Scholar]
- 8.Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P. & Snyder, S. H. (1994) Cell 78, 35-43. [DOI] [PubMed] [Google Scholar]
- 9.Sabers, C. J., Martin, M. M., Brunn, G. J., Williams, J. M., Dumont, F. J., Wiederrecht, G. & Abraham, R. T. (1995) J. Biol. Chem. 270, 815-822. [DOI] [PubMed] [Google Scholar]
- 10.Sonenberg, N. & Gingras, A. C. (1998) Curr. Opin. Cell Biol. 10, 268-275. [DOI] [PubMed] [Google Scholar]
- 11.Raught, B. & Gingras, A. C. (1999) Int. J. Biochem. Cell Biol. 31, 43-57. [DOI] [PubMed] [Google Scholar]
- 12.Gray, N. K. & Wickens, M. (1998) Annu. Rev. Cell Dev. Biol. 14, 399-458. [DOI] [PubMed] [Google Scholar]
- 13.Chung, J., Kuo, C. J., Crabtree, G. R. & Blenis, J. (1992) Cell 69, 1227-1236. [DOI] [PubMed] [Google Scholar]
- 14.Price, D. J., Grove, J. R., Calvo, V., Avruch, J. & Bierer, B. E. (1992) Science 257, 973-977. [DOI] [PubMed] [Google Scholar]
- 15.Dufner, A. & Thomas, G. (1999) Exp. Cell Res. 253, 100-109. [DOI] [PubMed] [Google Scholar]
- 16.Jefferies, H. B., Fumagalli, S., Dennis, P. B., Reinhard, C., Pearson, R. B. & Thomas, G. (1997) EMBO J. 16, 3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki, M., Blazek, E. & Vogt, P. K. (2001) Proc. Natl. Acad. Sci. USA 98, 136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohga, T., Uchiumi, T., Makino, Y., Koike, K., Wada, M., Kuwano, M. & Kohno, K. (1998) J. Biol. Chem. 273, 5997-6000. [DOI] [PubMed] [Google Scholar]
- 21.Mertens, P. R., Harendza, S., Pollock, A. S. & Lovett, D. H. (1997) J. Biol. Chem. 272, 22905-22912. [DOI] [PubMed] [Google Scholar]
- 22.Fukada, T. & Tonks, N. K. (2003) EMBO J. 22, 479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didier, D. K., Schiffenbauer, J., Woulfe, S. L., Zacheis, M. & Schwartz, B. D. (1988) Proc. Natl. Acad. Sci. USA 85, 7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ting, J. P., Painter, A., Zeleznik-Le, N. J., MacDonald, G., Moore, T. M., Brown, A. & Schwartz, B. D. (1994) J. Exp. Med. 179, 1605-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norman, J. T., Lindahl, G. E., Shakib, K., En-Nia, A., Yilmaz, E. & Mertens, P. R. (2001) J. Biol. Chem. 276, 29880-29890. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald, G. H., Itoh-Lindstrom, Y. & Ting, J. P. (1995) J. Biol. Chem. 270, 3527-3533. [DOI] [PubMed] [Google Scholar]
- 27.Izumi, H., Imamura, T., Nagatani, G., Ise, T., Murakami, T., Uramoto, H., Torigoe, T., Ishiguchi, H., Yoshida, Y., Nomoto, M., et al. (2001) Nucleic Acids Res. 29, 1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zasedateleva, O. A., Krylov, A. S., Prokopenko, D. V., Skabkin, M. A., Ovchinnikov, L. P., Kolchinsky, A. & Mirzabekov, A. D. (2002) J. Mol. Biol. 324, 73-87. [DOI] [PubMed] [Google Scholar]
- 29.Blobel, G. (1972) Biochem. Biophys. Res. Commun. 47, 88-95. [DOI] [PubMed] [Google Scholar]
- 30.Barrieux, A., Ingraham, H. A., Nystul, S. & Rosenfeld, M. G. (1976) Biochemistry 15, 3523-3528. [DOI] [PubMed] [Google Scholar]
- 31.Minich, W. B. & Ovchinnikov, L. P. (1992) Biochimie 74, 477-483. [DOI] [PubMed] [Google Scholar]
- 32.Davydova, E. K., Evdokimova, V. M., Ovchinnikov, L. P. & Hershey, J. W. (1997) Nucleic Acids Res. 25, 2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nekrasov, M. P., Ivshina, M. P., Chernov, K. G., Kovrigina, E. A., Evdokimova, V. M., Thomas, A. A., Hershey, J. W. & Ovchinnikov, L. P. (2003) J. Biol. Chem. 278, 13936-13943. [DOI] [PubMed] [Google Scholar]
- 34.Wolffe, A. P. (1994) BioEssays 16, 245-251. [DOI] [PubMed] [Google Scholar]
- 35.Evdokimova, V. M. & Ovchinnikov, L. P. (1999) Int. J. Biochem. Cell Biol. 31, 139-149. [DOI] [PubMed] [Google Scholar]
- 36.Kloks, C. P., Spronk, C. A., Lasonder, E., Hoffmann, A., Vuister, G. W., Grzesiek, S. & Hilbers, C. W. (2002) J. Mol. Biol. 316, 317-326. [DOI] [PubMed] [Google Scholar]
- 37.Sommerville, J. & Ladomery, M. (1996) FASEB J. 10, 435-443. [DOI] [PubMed] [Google Scholar]
- 38.Evdokimova, V., Ruzanov, P., Imataka, H., Raught, B., Svitkin, Y., Ovchinnikov, L. P. & Sonenberg, N. (2001) EMBO J. 20, 5491-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bos, T. J., Monteclaro, F. S., Mitsunobu, F., Ball, A. R., Jr., Chang, C. H., Nishimura, T. & Vogt, P. K. (1990) Genes Dev. 4, 1677-1687. [DOI] [PubMed] [Google Scholar]
- 40.Bos, T. J., Bohmann, D., Tsuchie, H., Tjian, R. & Vogt, P. K. (1988) Cell 52, 705-712. [DOI] [PubMed] [Google Scholar]
- 41.Maki, Y., Bos, T. J., Davis, C., Starbuck, M. & Vogt, P. K. (1987) Proc. Natl. Acad. Sci. USA 84, 2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duff, R. G. & Vogt, P. K. (1969) Virology 39, 18-30. [DOI] [PubMed] [Google Scholar]
- 43.Li, J., Chang, H. W., Lai, E., Parker, E. J. & Vogt, P. K. (1995) Cancer Res. 55, 5540-5544. [PubMed] [Google Scholar]
- 44.Aoki, M., Batista, O., Bellacosa, A., Tsichlis, P. & Vogt, P. K. (1998) Proc. Natl. Acad. Sci. USA 95, 14950-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoki, M., Schetter, C., Himly, M., Batista, O., Chang, H. W. & Vogt, P. K. (2000) J. Biol. Chem. 275, 6267-6275. [DOI] [PubMed] [Google Scholar]
- 46.Hughes, S. H., Greenhouse, J. J., Petropoulos, C. J. & Sutrave, P. (1987) J. Virol. 61, 3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo, D., Hilbush, B. & Sutcliffe, J. G. (2001) Eur. J. Pharm. Sci. 14, 191-196. [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe, J. G., Foye, P. E., Erlander, M. G., Hilbush, B. S., Bodzin, L. J., Durham, J. T. & Hasel, K. W. (2000) Proc. Natl. Acad. Sci. USA 97, 1976-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svitkin, Y. V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H. D. & Sonenberg, N. (2001) RNA 7, 1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 50.Bader, A. G., Schneider, M. L., Bister, K. & Hartl, M. (2001) Oncogene 20, 7524-7535. [DOI] [PubMed] [Google Scholar]
- 51.Barash, I. (1999) Mol. Cell. Endocrinol. 155, 37-49. [DOI] [PubMed] [Google Scholar]
- 52.Bouvet, P., Matsumoto, K. & Wolffe, A. P. (1995) J. Biol. Chem. 270, 28297-28303. [DOI] [PubMed] [Google Scholar]
- 53.Schroder, K., Graumann, P., Schnuchel, A., Holak, T. A. & Marahiel, M. A. (1995) Mol. Microbiol. 16, 699-708. [DOI] [PubMed] [Google Scholar]
- 54.Koike, K., Uchiumi, T., Ohga, T., Toh, S., Wada, M., Kohno, K. & Kuwano, M. (1997) FEBS Lett. 417, 390-394. [DOI] [PubMed] [Google Scholar]
- 55.Stenina, O. I., Shaneyfelt, K. M. & DiCorleto, P. E. (2001) Proc. Natl. Acad. Sci. USA 98, 7277-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y. F., Homer, C., Edwards, S. J., Hananeia, L., Lasham, A., Royds, J., Sheard, P. & Braithwaite, A. W. (2003) Oncogene 22, 2782-2794. [DOI] [PubMed] [Google Scholar]
- 57.Lin, T. A., Kong, X., Haystead, T. A., Pause, A., Belsham, G., Sonenberg, N. & Lawrence, J. C., Jr. (1994) Science 266, 653-656. [DOI] [PubMed] [Google Scholar]
- 58.Uramoto, H., Izumi, H., Ise, T., Tada, M., Uchiumi, T., Kuwano, M., Yasumoto, K., Funa, K. & Kohno, K. (2002) J. Biol. Chem. 277, 31694-31702. [DOI] [PubMed] [Google Scholar]
- 59.Soop, T., Nashchekin, D., Zhao, J., Sun, X., Alzhanova-Ericsson, A. T., Bjorkroth, B., Ovchinnikov, L. & Daneholt, B. (2003) J. Cell Sci. 116, 1493-1503. [DOI] [PubMed] [Google Scholar]
- 60.Janz, M., Harbeck, N., Dettmar, P., Berger, U., Schmidt, A., Jurchott, K., Schmitt, M. & Royer, H. D. (2002) Int. J. Cancer 97, 278-282. [DOI] [PubMed] [Google Scholar]
- 61.Yahata, H., Kobayashi, H., Kamura, T., Amada, S., Hirakawa, T., Kohno, K., Kuwano, M. & Nakano, H. (2002) J. Cancer Res. Clin. Oncol. 128, 621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibahara, K., Sugio, K., Osaki, T., Uchiumi, T., Maehara, Y., Kohno, K., Yasumoto, K., Sugimachi, K. & Kuwano, M. (2001) Clin. Cancer Res. 7, 3151-3155. [PubMed] [Google Scholar]
- 63.Yao, R., Wang, Y., Lubet, R. A. & You, M. (2002) Oncogene 21, 5814-5821. [DOI] [PubMed] [Google Scholar]